Abstract
Recently we reported that the BH3-only proteins Bim and Noxa bind tightly but transiently to the BH3-binding groove of Bak to initiate Bak homo-oligomerization. However, it is unclear how such tight binding can induce Bak homo-oligomerization. Here we report the ligand-induced Bak conformational changes observed in 3D models of Noxa·Bak and Bim·Bak refined by molecular dynamics simulations. In particular, upon binding to the BH3-binding groove, Bim and Noxa induce a large conformational change of the loop between helices 1 and 2 and in turn partially expose a remote groove between helices 1 and 6 in Bak. These observations, coupled with the reported experimental data, suggest formation of a pore-forming Bak octamer, in which the BH3-binding groove is at the interface on one side of each monomer and the groove between helices 1 and 6 is at the interface on the opposite side, initiated by ligand binding to the BH3-binding groove.
Paralogs of the B-Cell lymphoma 2 (BCL-2) gene express proteins that regulate the cellular life-or-death switch by controlling mitochondrial outer membrane permeabilization (MOMP)1,2,3,4,5. Sharing up to four conserved BCL-2–homology (BH) domains (viz., BH1, BH2, BH3, and BH4)1,6, these proteins can be divided into anti- and pro-apoptotic subgroups. The anti-apoptotic members such as Bcl-2 contain four BH domains and inhibit MOMP. The pro-apoptotic members, all of which promote MOMP, can be further divided based on structural and functional differences into effectors and BH3-only proteins. Effectors, including Bax (Bcl-2-associated x protein) and Bak (Bcl-2 antagonist killer), contain BH1, BH2 and BH3 domains and are capable of directly causing MOMP. In contrast, the BH3-only proteins exhibit homology to Bcl-2 only in their BH3 domains, produce MOMP indirectly, and can be subcategorized into activators and sensitizers7. The activators such as Bim (Bcl-2-interacting mediator of cell death) appear to bind Bax or Bak to trigger homo-oligomerization8,9. The sensitizers such as Bad (Bcl-2 antagonist of cell death) are thought to indirectly promote MOMP by binding and neutralizing the anti-apoptotic members1.
Protein-protein interactions involving various Bcl-2 paralogs play a critical role in their function. The binding of BH3-only proteins to the anti-apoptotic proteins involves a surface groove called the BH3-binding groove as revealed by crystal or nuclear magnetic resonance (NMR) structures of anti-apoptotic protein complexes. The effectors also have a groove akin to the BH3-binding groove of the anti-apoptotic proteins. Whether this groove in Bak or Bax binds BH3 domains of direct activators and contributes to MOMP has previously been unclear.
Although the BH3-only protein Noxa (Latin for damage) was widely considered to be a sensitizer7,10,11,12, it has been recently reported that Noxa can function as a direct activator of Bak and Bax13,14. We have further reported that, unlike the sensitizer Bad, Noxa and Bim bind tightly (KD values of 20–30 nM) but transiently to the BH3-binding groove of Bak, thereby triggering the formation of a Bak homo-oligomer of ∼200 kDa, MOMP, and killing of intact cells14.
Despite the fact that Bak and Bax play pivotal roles in apoptotic responses and new information about their regulation by BH3-only proteins has become available13,14,15, there has been no structural information on interactions of BH3-only proteins with the BH3-binding groove of Bak or Bax except for a brief report of a computational three-dimensional (3D) model of the Noxa·Bak complex14. It has been unclear how a high-affinity activator can bind Bak transiently and trigger Bak homo-oligomerization, as one would expect hetero-oligomerization to be induced by binding of Noxa to Bak with a KD value of 24 nM.
In this report, we describe the ligand-induced Bak conformational changes observed in computational 3D models of Noxa·Bak and Bim·Bak that were refined by near-microsecond-scale molecular dynamics simulations. In the context of these complex models and the Bak conformational changes, we discuss the nature of interactions of BH3-only proteins with the Bak BH3-binding groove and a possible mechanism for Bak homo-oligomerization induced by tight but transient binding of activator BH3-only proteins.
Results
The Noxa·Bak Complex Model
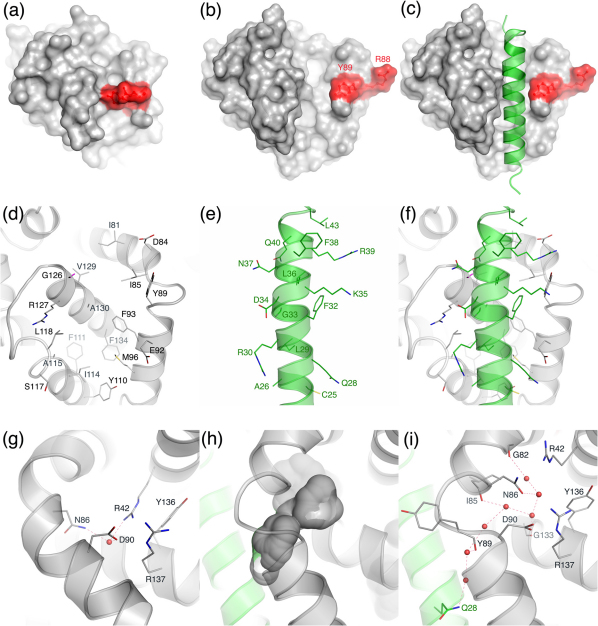
Unless specified otherwise, human Bcl-2 paralogs were used in our studies and the Bak helical nomenclature of reference16 is used in this report. Our homology modeling using the SWISS-MODEL program17 predicted that residues 19–45 of Noxa18 adopt a helical conformation, a finding that has subsequently been confirmed in an unpublished crystal structure of the helical Noxa binding at the BH3-binding groove of the anti-apoptotic paralog Bcl-2–related protein A1 (BFL-1)19,20 released after this study (Protein Data Bank ID: 3MQP). Based on our initial site-directed mutagenesis data involving residues at the Bak BH3-binding groove14 and the structural similarity between the apo Bak crystal structure and the NMR structure of mouse anti-apoptotic protein Mcl-1 (myeloid cell leukemia-1) whose BH3-binding groove is occupied by a mouse Noxa BH3 domain (mNoxa)21, we conjectured that the helical Noxa would dock at the BH3-binding groove of Bak, even though the groove is occluded by R88Bak and Y89Bak in all the reported apo Bak crystal structures (Figure 1a)22,23. We then generated the Noxa·Bak complex by manually docking the helical Noxa atop the BH3-binding groove of a Bak conformer (residues 21–183) that was taken from a crystal structure of an apo Bak homo-dimer with the C-terminal transmembrane domain excised22. The manual docking was performed in such a way that L29Noxa was placed as close to I114Bak and L118Bak as possible and that L36Noxa was as close to V129Bak and I85Bak as possible. This model was refined first by energy minimization, which moved Noxa closer to the BH3-binding groove and pushed R88Bak out of the groove. The resulting complex was further refined by multiple molecular dynamics simulations using a published protocol24. Seventy-two independent 10-ns simulations were carried out, thus offering collectively 0.72-μs sampling for the Noxa·Bak conformations. Although Y89Bak was still within the BH3-binding groove and underneath Noxa in the energy-minimized Noxa·Bak model, as described below, the model refined by the simulations has Y89Bak pushed out of the groove and the BH3-binding groove expanded to accommodate Noxa fully (Figures 1b and 1c). To provide the information on the protein dynamics and the solvent effect on complex stability as described below, we release coordinates of average and energy-minimized average conformations of Noxa·Bak (datasets S1 and S2, respectively) as well as coordinates of a water-containing instantaneous conformation of Noxa·Bak that has the smallest root mean square deviation (RMSD) to the average conformation (dataset S3).
Figure 1. Bak conformational changes induced by binding of Noxa to the BH3-binding groove.
(a) The BH3-binding groove of the unbound Bak showing the blockage of the groove by R88bak and Y89bak. (b) The BH3-binding groove of the bound Bak showing the vacated groove by R88bak and Y89bak revealing two hydrophobic holes. (c) The helical Noxa anchored at the Bak BH3-binding groove showing complementarity between Noxa and the groove. (d) The Bak residues that interact with Noxa. (e) The Noxa residues that interact with Bak or solvent. (f) Intermolecular interactions between Noxa and the Bak BH3-binding groove and between F38Noxa and solvent molecules (not shown). (g) The unbound Bak with a water molecule hydrogen-bonding to Bak residues underneath the turn between two helices. (h) The bound Bak with a water-filled cavity underneath the turn between two helices. (i) The bound Bak with a 7-water hydrogen-bond network involving residues of Bak and Noxa in the cavity underneath the turn between two helices. The unbound Bak is taken from the crystal structure of Protein Data Bank code of 2IMT, and the bound Bak is taken from the simulation-refined Noxa·Bak model. For clarity hydrogen atoms are not displayed except for the nonpolar hydrogen atoms of Gly. Atoms of oxygen, nitrogen, sulfur, hydrogen, carbon of Bak, and carbon of Noxa are colored in red, blue, yellow, magenta, gray, and green, respectively. Dashed lines denote hydrogen bonds.
Computational Support for the Noxa·Bak Model
As a control study, we performed 11 10-ns simulations of the first model of the NMR structure of mouse Mcl-1 in complex with mNoxa (mNoxa·mMcl-1)21 using the same simulation protocol as the one for Noxa·Bak. We also carried out 11 10-ns simulations of the first model of the NMR structure of mouse Mcl-1 in complex with mouse Puma (p53 up-regulated modulator of apoptosis; mPuma·mMcl-1)21. These simulations rendered two average conformers with alpha carbon root mean square deviations (ACRMSDs) of 1.5 Å for mNoxa·mMcl-1 and 1.8 Å for mPuma·mMcl-1 relative to the respective average NMR structures (Table 1). Each of the two computational average conformers was obtained by averaging all 11,000 trajectories saved at 1.0-ps intervals during the last 1-ns period of the 11 10-ns simulations. Reassuringly, these ACRMSDs are less than the corresponding ones of the 20 NMR models of the mNoxa·mMcl-1 and the 20 NMR models of mPuma·mMcl-1 (2.5 Å for mNoxa·mMcl-1 and 2.0 Å for mPuma·mMcl-1). These results suggest that the 0.72-μs molecular dynamics simulation protocol is applicable to Noxa·Bak, because the sizes of mNoxa·mMcl-1 (189 residues) and mPuma·mMcl-1 (189 residues) are comparable to that of Noxa·Bak (190 residues).
Table 1. Interaction Energies and Root Mean Square Deviations of Bak or mMcl-1 Complexes.
| Complex | E (kcal/mol) | E / BH3 residue (kcal/mol) | CRMSD (Å) | ACRMSDMMDS (No. of conformers) (Å) | ACRMSDNMR (No. of conformers) (Å) |
|---|---|---|---|---|---|
| Noxa·Bak | −132 | −4.9 | 1.2b | 2.7 (5,100)e | NA |
| A1–26K27·Bak | −110a | −4.1a | 1.8c | 2.4 (2,000)f | NA |
| mNoxa·mMcl-1 | −176a | −6.5a | 1.5d | 2.8 (11,000)g | 2.5 (20)h |
| mPuma·mMcl-1 | −152a | −5.8a | 1.8d | 2.6 (11,000)g | 2.0 (20)h |
aE: the intermolecular interaction energy and the intermolecular interaction energy per residue of BH3-only protein were obtained using the same method as the ones for Noxa·Bak except that 11 10-ns–long simulations were performed. The initial structure of mNoxa·mMcl-1 or mPuma·mMcl-1 used for the simulations was taken from the first NMR model of Protein Data Bank code of 2ROD or 2ROC, respectively. The initial structure of A1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26K27·Bak was obtained from mutation from the simulation-refined Noxa·Bak model.
bCRMSD: alpha carbon root mean square deviation between the average structure of 5,100 similar conformers identified by cluster analysis from the second-round simulation of Noxa·Bak and the energy-minimized average structure of 271 similar conformers identified by cluster analysis from the first-round simulation of Noxa·Bak, where the average structure of the 5,100 conformers was not subjected to any energy minimization.
cCRMSD: alpha carbon root mean square deviation between the average structure of 2,000 similar conformers identified by cluster analysis from the simulations of A1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26K27·Bak and the energy-minimized structure of A1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26K27·Bak derived from mutations of the energy-minimized average structure of the 5,100 Noxa·Bak conformers.
dCRMSD: alpha carbon root mean square deviation between the average structure of 11,000 similar conformers identified by cluster analysis from the simulation of mNoxaA·Mcl-1 (or Puma·Mcl-1) and the average structure of the 20 NMR models of mNoxaA·Mcl-1 (or Puma·Mcl-1), where the two average structures were not subjected to any energy minimization and the cluster analysis identified only one cluster of conformers.
eACRMSDMMDS: average alpha carbon root mean square deviation among the 5,100 similar conformers of Noxa·Bak.
fACRMSDMMDS: average alpha carbon root mean square deviation among the 2,000 similar conformers of A1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26K27·Bak.
gACRMSDMMDS: average alpha carbon root mean square deviation among the 11,000 similar conformers of mNoxa·Mcl-1 (or Puma·Mcl-1).
hACRMSDNMR: average alpha carbon root mean square deviation among the 20 NMR conformers of mNoxa·Mcl-1 (or Puma·Mcl-1).
Because the Bak BH3-binding groove is mainly hydrophobic, helices such as polyalanine can bind to the groove with favorable intermolecular interaction energies. To be biologically relevant, the intermolecular interaction energy of Noxa with the Bak groove should intuitively be lower than that of polyalanine. To confirm this, we calculated intermolecular interaction energies of the Noxa·Bak complex and a reference complex of A1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26K27·Bak that was generated by mutating residues 19–45 of Noxa in the Noxa·Bak complex model to A19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44K45. The mutation to A19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44K45 rather than A19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45 was to keep the net charge of the mutant peptide identical to that of Noxa. To obtain lower intermolecular interaction energy for the mutant complex, we refined the complex by running 11 10-ns simulations using the same simulation protocol for Noxa·Bak. Reassuringly again, the intermolecular interaction energy and the interaction energy per residue of the reference complex are −110.0 kcal/mol and −4.1 kcal/mol, respectively (Table 1), while the corresponding energies for Noxa·Bak are −132.0 and −4.9 kcal/mol, respectively. These energies suggest that the Noxa·Bak model is worthy of further experimental testing.
Experimental Support for the Noxa·Bak Model
Our sequence analysis identified L29 and D34 as two highly conserved residues in the Noxa BH3 domain and L29, F32, and L36 as the hydrophobic residues in this domain14. The double (L29A and D34A) and triple (L29E, F32E, and L36E) mutants of Noxa showed diminished binding to Bak14, suggesting that the identified residues are crucial for Bak binding. Our sequence analysis also identified R127 as a highly conserved residue in the BH3-binding grooves of Bak, Bax, Bcl-2, Bcl-xL (Bcl-2 X protein), and Mcl-1. The Bak R127A mutant exhibited markedly reduced binding to Noxa14, indicating that BH3-binding groove is involved in the binding of Bak with Noxa. Acquired prior to the Noxa·Bak complex prediction, these mutation data are consistent with the Noxa·Bak model in view of the pair-wise intermolecular interactions between Noxa and Bak as detailed in Table 2 and Figures 1d–1f. In addition, the model suggested that F93, I114, and G126 of Bak directly interact with Noxa (Table 2 and Figures 1d–1f), and subsequent single mutations of these residues (F93E; I114E; G126S) markedly reduced the binding of Bak with Noxa14. Although NMR or X-ray crystal structure of Noxa·Bak is required to further validate the model, the mutagenesis data suggest that the computational model can be used with care for obtaining insight into Noxa-induced Bak conformational changes that are crucial to the Bak homo-oligomerization.
Table 2. Residues Involving Favorable Intermolecular Interactions between Noxa and Bak.
| IDNoxa-– IDBak | Interaction | IDNoxa-– IDBak | Interaction |
|---|---|---|---|
| C25—M96 | van der Waals | F32—I85 | van der Waals |
| C25—Y110 | van der Waals | F32—Y89 | pi-pi |
| C25—I114 | van der Waals | G33—L118 | van der Waals |
| A26—I114 | van der Waals | G33—R127 | van der Waals |
| A26—S117 | van der Waals | G33—A130 | van der Waals |
| Q28—E92 | van der Waals | D34—R127 | ionic |
| Q28—M96 | van der Waals | K35—Y89 | cation-pi |
| L29—M96 | van der Waals | L36—I85 | van der Waals |
| L29—F111 | van der Waals | L36—G126 | van der Waals |
| L29—I114 | van der Waals | L36—V129 | van der Waals |
| L29—A115 | van der Waals | L36—A130 | van der Waals |
| L29—L118 | van der Waals | N37—G126 | van der Waals |
| L29—A130 | van der Waals | N37—G126 | hydrogen bond |
| L29—L131 | van der Waals | R39—D84 | ionic |
| L29—F134 | van der Waals | Q40—W125 | hydrogen bond |
| R30—L118 | van der Waals | L43—I81 | van der Waals |
| F32—F93 | pi-pi | L43—D84 | van der Waals |
Bak Conformational Changes Induced by Noxa
Expanding the Bak BH3-Binding Groove
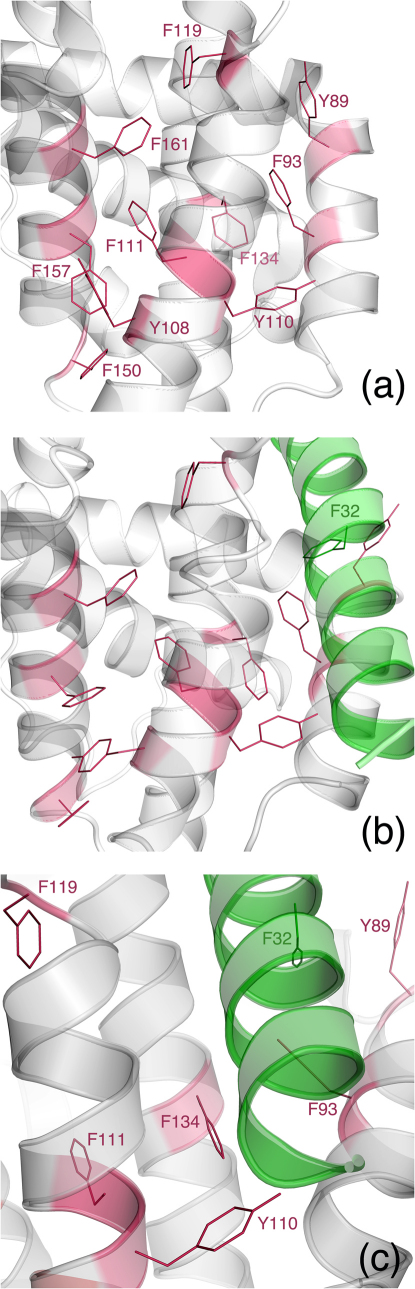
Compared to the two reported crystal structures of apo Bak22,23, one noticeable change of the Bak in complex with Noxa is main-chain and side-chain conformational changes of R88Bak and Y89Bak (Figure 1). In the apo Bak crystal structures, Y89 is in a network of 10 aromatic residues that interact with one another via pi-pi interactions (Figure 2a). This aromatic network, which includes Y89, Y108, Y110, F93, F111, F119, F134, F150, F157, and F161, presumably provides structural stability to Bak in water. Hence one would expect that, upon Noxa binding, R88Bak might move away from the BH3-binding groove and Y89Bak might remain in the groove to maintain the integrity of the aromatic core. Surprisingly, Noxa pushed Y89Bak out of the groove through main-chain and side-chain conformational changes of Bak. These changes are accompanied by expansion of the groove and formation of two hydrophobic holes in the groove (Figure 1b), one comprised of I81Bak, I85Bak, and V129Bak, and the other made of F93Bak, I114Bak, L118Bak, and A130Bak. Consequently, Noxa snugs down to the groove with its L29Noxa and L36Noxa plugged into the two holes. This groove expansion also creates a cavity (Figure 1h) allowing seven water molecules to form a hydrogen-bond network underneath the turn that connects helices α2 and α3 of Bak (Figure 1i; dataset S3), whereas only one water molecule is present underneath the turn (Figure 1g) as revealed by the high-resolution crystal structure of the apo Bak22. The 7-water hydrogen-bond network, which involves residues from both Noxa and Bak (Q28Noxa, G82Bak, I85Bak, N86Bak, Y89Bak, and G133Bak; Figure 1i), contributes to the affinity of Noxa for Bak.
Figure 2. Expansion and side-chain dynamics of the aromatic core inside the Noxa·Bak complex.
(a) The 10-residue aromatic core of the apo Bak crystal structure (Protein Data Bank code: 2IMT). (b) The 11-residue aromatic core of the Noxa·Bak model obtained with energy minimization after molecular dynamics simulations. (c) Close up view of the highly mobile aromatic residues of the Noxa·Bak model obtained without energy minimization after molecular dynamics simulations.
Expansion of the Aromatic Network of Noxa·Bak
As shown by the energy-minimized average conformation of the Noxa·Bak model (Figure 2b and dataset S2), F32Noxa occupies the groove region where Y89Bak resides in the apo Bak conformation. Consequently, the 10-residue aromatic network is expanded to an 11-residue network after the groove is occupied by Noxa. Furthermore, as revealed by the average conformation of the Noxa·Bak model (dataset S1), 8 of the 11 aromatic residues (F32Noxa, Y89Bak, Y108Bak, F93Bak, F111Bak, F134Bak, F150Bak, and F157Bak) showed ring contraction due to ring spin with or without side-chain motion. In particular, the side chains of F32Noxa, Y89Bak, F93Bak, and F134Bak are markedly contracted relative to the side chain of Y110Bak (Figure 2c). These observations suggest that the high affinity of the Noxa·Bak complex stems from both enthalpy and configurational entropy25.
Exposing the Groove between Helices α1 and α6
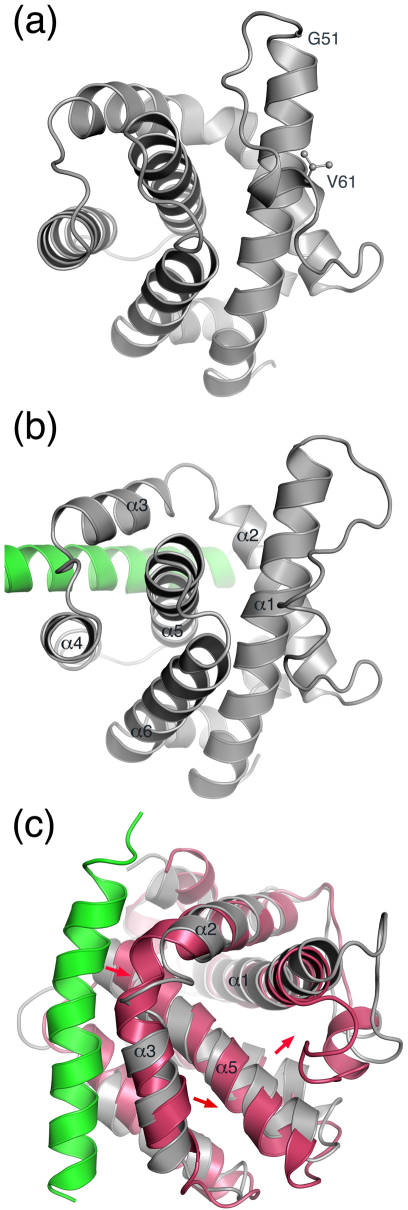
The most noticeable Bak conformational change induced by Noxa occurs in the loop between helices α1 and α2 (residues 51–61; Figures 3a and 3b), which in turn partially exposes the groove between helices α1 and α6 that is located on the opposite side of Bak from the BH3-binding groove (Figure 3b). This is caused by the Bak BH3-binding groove expansion, which pushes helix α5 slightly away from helix α3 (Figure 3c). In the apo Bak crystal structure, because the BH3-binding groove is contracted, there is room between helices α1 and α5 to accommodate residues 51–61 (Figure 3c). In the bound Bak computational model (Figure 3b), because the BH3-binding groove is expanded, there is no room between helices α1 and α5 for residues 51–61, which leads to the large conformational change of residues 51–61 and in turn partially exposes the groove between α1 and α6 (Figure 3c). As discussed below, partial exposure of this groove might be one of the key steps in Bak homo-oligomerization.
Figure 3. Main-chain conformational changes of Bak induced by Noxa binding.
(a) Residues 51–61 blocking the groove between helices 1 and 6 in the unbound Bak crystal structure (Protein Data Bank code: 2IMT; Bak in gray). (b) Residues 51–61 partially vacating the groove between helices 1 and 6 in the Noxa·Bak model obtained with energy minimization after molecular dynamics simulations (Bak and Noxa are in gray and green, respectively). (c) Overlay of the unbound Bak (magenta) and the Bak (gray) in complex with Noxa green) with arrows showing that Noxa slightly pushes helices α3 and α5 away, resulting in moving residues 51–61 away from helix 5.
The Bim·Bak Model and Bim-Induced Bak Conformational Changes
The computational 3D model of the Bim·Bak complex was subsequently generated by mutating Noxa (residues 19–45) of the Noxa·Bak model to Bim (residues 143–166) followed by refinement with energy minimization and then 100 10-ns molecular dynamics simulations using the same simulation protocol as the Noxa·Bak model. Coordinates of average and energy-minimized average conformations of the Bim·Bak model are provided in datasets S4 and S5, respectively; coordinates of a water-containing instantaneous conformation of Bim·Bak that has the smallest RMSD to the average conformation are provided in dataset S6. As apparent from datasets S4–S6, induced by binding of Bim to the Bak BH3-binding groove, the Bim·Bak model has almost the same Bak conformational changes as those of the Noxa·Bak model, except that the Bim-induced conformational change of the loop between helices α1 and α2 exposes more of the groove between α1 and α6 in two Boltzmann-weighted minor conformations than the Noxa-induced loop conformational change (datasets S7 and S8).
Discussion
Computational Models of Noxa·Bak and Bim·Bak. To date there has been no experimental structure of Bak or Bax with its BH3-binding groove occupied by a BH3-only protein. An NMR structure has been reported for Bax with its C-terminal transmembrane domain bound at the BH3-binding groove and a conformationally constrained peptide of the Bim BH3 domain docked at a groove between two helices that correspond to helices α1 and α6 of Bak15. Derived from manual docking and extensive refinement with near-microsecond-scale molecular dynamics simulations, the computational model of Noxa bound in the BH3-binding groove of Bak is supported by the computational self-consistency tests in terms of conformational sampling and intermolecular interaction energy and by site-directed mutagenesis studies. This model is also supported by an unpublished crystal structure of Noxa in complex with BFL-1 (Protein Data Bank ID: 3MQP; released after our model prediction); the alpha-carbon and heavy-atom RMSDs of Noxa (residues 19–40) between the crystal structure and the energy-minimized average conformation of the model (dataset S2) are 0.9 and 2.3 Å, respectively. Energetically, one may have a concern with the Noxa·Bak model in which F38Noxa is exposed to solvent rather than being buried in the BH3-binding groove (Figures 1e and 1f). In the 3MQP crystal structure at a resolution of 2.24 Å, F38Noxa is also exposed to solvent, despite a penalty of the solvation energy of the solvent-exposed phenyl group. While verification by X-ray crystallographic or NMR studies is necessary, the computational Noxa·Bak model and its derivative model of Bim·Bak offer insights as discussed below.
Binding of BH3-Only Proteins to the BH3-Binding Groove. Binding of BH3-only proteins to the BH3-binding groove of anti-apoptotic proteins has been extensively studied. According to current literature the interactions with the anti-apoptotic BH3-binding groove include hydrophobic and ionic interactions5,21,26,27,28,29. In the Noxa·Bak model, Noxa interacts with hydrophobic residues of I81Bak, I85Bak, F93Bak, V129Bak, I114Bak, L118Bak, and A130Bak and ionic residues of D84Bak, E92Bak, and R127Bak at the BH3-binding groove (Figures 1d–1f). These hydrophobic and ionic interactions are consistent with the interactions involving the anti-apoptotic BH3-binding groove.
Two other types of interactions also appear to contribute to the tight binding between Noxa and Bak. First, Noxa also interacts with Bak via pi-pi interactions involving the 11-residue aromatic network of the complex. Second, cation-pi interactions also contribute. In the Noxa·Bak model, Y89Bak, R88Bak, K35Noxa, and R39Noxa are exposed to solvent. It is, therefore, difficult to discern from an instantaneous conformation or an energy-minimized average conformation whether Y89Bak forms cation-pi interactions with R88Bak, K35Noxa, and/or R39Noxa. In the average conformation of the Noxa·Bak model without energy minimization (dataset S1), the shortest distances of Y89Bak to R88Bak, K35Noxa, and R39Noxa are 6.4, 3.7, and 7.6 Å, respectively. These distances indicate cation-pi interactions of Y89Bak with R88Bak, K35Noxa, and R39Noxa, according to the survey of cation-pi interactions in protein crystal structures30. Therefore, the binding of Noxa to the Bak BH3-binding groove involves pi-pi interactions of F32Noxa and cation-pi interactions of K35Noxa and R39Noxa with the respective Bak residues at the groove in addition to the anticipated hydrophobic and ionic interactions.
In the BH3 domain of Bim, residues that correspond to F32Noxa, K35Noxa, and R39Noxa are I155Bim, E158Bim, and Y162Bim respectively, raising the possibility that the pi-pi and cation-pi interactions are unique to the Noxa·Bak complex. Interestingly, the Bim·Bak model refined by microsecond-scale molecular dynamics simulations shows that F159Bim, Y162Bim, and Y163Bim form pi-pi interactions with the Bak BH3-binding groove and extend the 10-residue aromatic network of apo Bak to a 13-residue aromatic network in the Bim·Bak complex (datasets S4–S6). These residues also have cation-pi interactions with R88Bak bridged by Y89Bak (datasets S4–S6).
In examining structures of anti-apoptotic proteins in complex with BH3-only proteins, we also found pi-pi and cation-pi interactions of residues at the BH3-binding groove of the following proteins: mPuma·mMcl-1 with a 7-residue aromatic network (Protein Data Bank ID: 2ROC21), mNoxa·mMcl-1 with an 8-residue aromatic network (Protein Data Bank ID: 2ROD21), mouse Bcl-XL in complex with mouse Bim possessing an 18-residue aromatic network (Protein Data Bank ID: 1PQ131), and Norway rat Mcl-1 bound with Bim stabilized by a 9-residue aromatic network (Protein Data Bank ID: 2NL928). However, the pi-pi and cation-pi interactions are missing in Bfl-1 in complex with Noxa (Protein Data Bank ID: 3MQP) and mouse Mcl-1 bound to an alternative mouse Noxa BH3 domain (Protein Data Bank ID: 2JM628). These observations suggest that interactions of BH3-only proteins with the Bak BH3-binding groove as well as some of the anti-apoptotic Bcl-2 paralogs involve not only hydrophobic and ionic interactions but also pi-pi and cation-pi interactions.
Bak Homo-Oligomerization Induced by Bim or Noxa. Recently we reported the experimental observation that a high-affinity activator such as Noxa or Bim can bind Bak transiently to trigger Bak homo-oligomerization14. Knowing that the KD values of Noxa and Bim for binding to Bak in the presence of CHAPS are 24 nM and 29 nM, respectively14, one would expect hetero-oligomerization induced by these high-affinity binders. A homo-oligomerization mechanism at the structural level is, therefore, needed to resolve the paradox of tight but transient binding.
Bcl-2 paralogs have been proposed to be part of a putative pore structure for cytochrome c release32,33,34. Three hypothetical homo-oligomerization mechanisms (asymmetric–single-conformer, asymmetric–two-conformer, and symmetric–single-conformer) have also been proposed for Bak16,35,36. To resolve the paradox described above, we investigated these mechanisms by building respective pore models of Bak homo-oligomers. According to the symmetric–single-conformer mechanism, Bak everts its helix α2 and subsequently forms a symmetric and reciprocal homo-dimer as a subunit of a Bak homo-oligomer16. However, we could not build a pore model for this mechanism for two reasons. First, residues 51–71 of the first Bak clash with helices α1, α4, α5, and α6 of the second Bak, unless a large portion of helix α1 and/or helix α2 of the first Bak is uncoiled. Second, the direction toward which the C-terminal transmembrane helix of one Bak points is the opposite of the other, which prevents half of the Bak transmembrane helices of the oligomer from being anchored in the membrane. We could not build a pore model for the asymmetric–two-conformer mechanism either. Alternatively, given the Bak conformational changes induced by Bim or Noxa, we built a Bak octamer pore model for the asymmetric–single-conformer mechanism that can resolve the paradox.
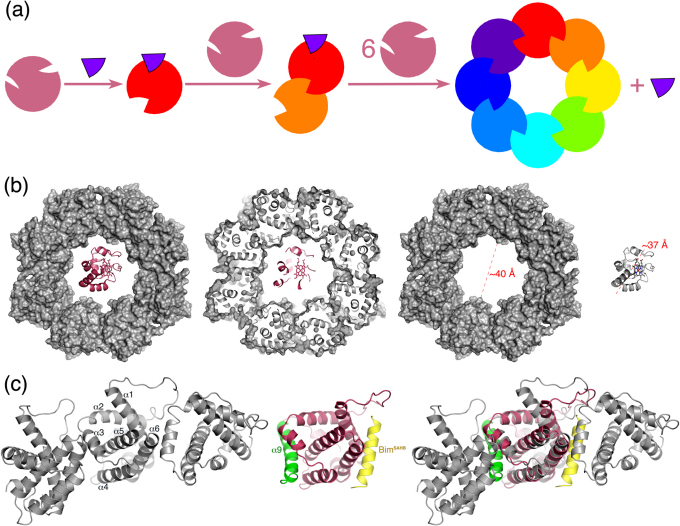
To simplify the description of the Bak octamer model, we refer the two grooves between helices α3 and α4 and between helices α1 and α6 to as BH3front-binding and BH3back-binding grooves, respectively, because the groove between α1 and α6 is on the opposite side of Bak from the canonical BH3-binding groove. Formation of the Bak octamer presumably requires three steps (Figure 4a). Step 1, Noxa (indigo) binds an apo Bak (magenta) to partly expose the BH3Back-binding groove of BakI (red). Step 2, the resulting Noxa·Bak1 complex binds another apo Bak to have the turn that joins helices α2 and α3 of BakII (orange) to be docked at the BH3back-binding groove of BakI. This complexation involving the BH3back-binding groove of BakI partly exposes the BH3back-binding groove of BakII. Step 3, repeating Step 2 six times results in a hetero-nonamer (Noxa·BakI·BakII·BakIII·BakIV·BakV·BakVI·BakVII·BakVIII) to partly expose the BH3back-binding groove of BakVIII, which ejects Noxa and accommodates the turn that joins helices α2 and α3 of BakI to form a donut-shaped homo-octamer that is akin to the pore model mentioned in references32,33,34,35.
Figure 4. Proposed pore-forming Bak homo-oligomerization mechanism and its support.
(a) A hypothetic pore-forming Bak homo-oligomerization scheme (the apo Bak is represented by a magenta sphere with the BH3-binding groove on the right and the groove between helices 1 and 6 on the left; the bound Bak is shown by a sphere in other colors; Noxa is depicted by a purple wedge). (b) An energy-minimized atomic model of Bak homo-octamer showing a cavity diameter suitable for the passage of the crystal structure of bovine cytochrome C (Protein Data Bank ID: 2B4Z; the heme ring is in stick). (c) Close-up view of three Bak units of the homo-octamer showing two interfaces of each unit of the Bak octamer that resemble the two interfaces of the Bax NMR structure (Protein Data Bank ID: 2K7W) with its C-terminal transmembrane domain (α9 in green) bound at the BH3-binding groove and a conformationally constrained peptide of the Bim BH3 domain (BimSAHB in yellow) docked at a groove between two helices that correspond to helices α1 and α6 of Bak.
Derived from the Noxa·Bak model with energy minimization and 12 10-ns molecular dynamics simulations in explicit water, the Bak octamer pore model is supported by several experimental observations. First, we have reported that the apparent molecular weight of the Bak homo-oligomer induced by Noxa is ∼200 kDa, ∼8 times of the monomeric Bak molecular weight of 25 kDa14. Second, the Bak octamer is consistent with the report that detergent exposure induces formation of a Bax oligomer composed of 6–8 Bax monomers33. Although early indirect structural data34,37,38,39 suggested that a Bax oligomer with ∼4 or 7 Bax monomers might be responsible for the efflux of cytochrome C, a recent high-resolution (1.5-Å) crystal structure of bovine cytochrome C40 and studies using hydrodynamic analysis and atomic force microscopy34 unequivocally show that the Stokes diameter of cytochrome C is much greater than the earlier reported diameter of 17 ± 3 Å. Given the 90% sequence identity between human and bovine cytochrome C (National Center Biotechnology Information session numbers: NP_061820 and NP_001039526, respectively), the diameter of human cytochrome C is at least 36.84 Å according to the oval-shaped bovine cytochrome C crystal structure. A Bak heptamer has a calculated cavity diameter of ∼26 Å that is too narrow for the passage of cytochrome C. The cavity diameter of the Bak octamer pore model is calculated to be ∼40 Å (Figure 4b), hence indicating that the donut-shaped octamer has a cavity suitable for the passage of cytochrome C with a diameter in the range of 37–40 Å and lending further credence to the Bak octamer model.
Given the debate on whether the Bax/Bak-induced MOMP is selective41,42,43,44,45,46 or nonselective47,48 for effluxes of cytochrome C and other intermembrane-space proteins, and, more importantly, given the fact that other larger proteins than cytochrome C are also released from mitochondria during apoptosis, it is important to note the possibility that a series of ∼40-Å holes punched by the Bak octamers aligned next to each other or aligned in a circle pattern can make a larger hole to release larger proteins such as Smac (second mitochondria-derived activator of caspases).
Unlike the symmetric–single-conformer model, the octamer pore model permits all of the C-terminal transmembrane helices to point in the same direction and be orthogonal to the ring structure, thus enabling the ring structure to be attached to a membrane. Each Bak in the proposed octamer has two interfaces to make the pore structure (Figure 4c), the BH3front-binding groove at the interface on one side of each Bak and the BH3back-binding groove at the interface on the opposite side. Consistent with this model, both front and back grooves (helices α1, α3, α4, and α6) are reportedly involved in Bak or Bax oligomerization5,14,15. While our experimental studies showed that mutations of the BH3front-binding groove can prevent Bak oligomerization14, others reported that the Bak activation also involves α15 and α649 and that the corresponding BH3back-binding groove (α1 and α6) of a Bax NMR structure binds BimSAHB, a stapled peptide that is thought to model the Bim BH3 domain, while the corresponding BH3front-binding groove (α3 and α4) of Bax is occupied by its C-terminal transmembrane domain (α9)15. The two interfaces of the Bax NMR structure resemble the two interfaces of each Bak in the octamer (Figure 4c). This resemblance further strengthens the likelihood that Bak homo-oligomerization involves a pore-forming Bak homo-octamer.
While computational refinement and further experimental verification of this Bak octamer pore model are required, the proposed homo-oligomerization mechanism explains how high-affinity BH3-only activators can induce Bak homo-oligomerization. Because of the geometric restraint on both ends of the hetero-nonamer and the partial opening of the BH3back-binding groove of BakVIII, the proximity and orientation effect50 (viz., the entropic effect in a modern term) and the allosteric modulation greatly increase the affinity of the BH3back-binding groove of BakVIII for the turn that joins helices α2 and α3 of BakI thus enabling the replacement of Noxa by the back groove of BakVIII even though Noxa has a higher affinity for the BH3front-binding groove of BakI than apo BakVIII. In other words, when BakI and BakVIII are restrained at both ends of the nonamer, the affinity of the BH3back-binding groove of BakVIII for the turn of Bak1becomes higher than that of Noxa due to the partial opening of the back groove of BakVIII and the avoidance of the entropy loss for the complexation of Bak1 with BakVIII. This explains how a high-affinity activator such as Noxa or Bim can bind Bak transiently and trigger Bak homo-oligomerization rather than hetero-oligomerization.
In conclusion, Simulation-refined 3D models of Noxa·Bak and Bim·Bak show pi-pi and cation-pi interactions in addition to the anticipated hydrophobic and ionic interactions between BH3-only protein and Bak. The models also show that binding of Noxa or Bim to the BH3-binding groove exposes the groove between helices α1 and α6 that is on the opposite side of the Bak molecule from the BH3-binding groove. These observations, coupled with the reported experimental data, suggest that a BH3-only protein induces formation of a pore-forming Bak homo-octamer through tight but transient binding to the canonical BH3-binding groove.
Methods
Model Preparation
All His, Glu, Asp, and Cys residues were treated as HIP, GLU, ASP, and CYS, respectively, for Bak in complex with Noxa or Bim and for the Bak octamer using the AMBER force field (ff99SB)51,52. The Noxa·Bak and Bim·Bak complexes were generated by manual docking as described above and by mutating Noxa to Bim, respectively. The Bak octamer was generated by manual alignment. The crystallographically determined water molecules were removed from the Bak crystal structure before the manual docking or alignment. The topology and coordinate files of the Noxa·Bak and Bim·Bak complexes were generated by the PREP, LINK, EDIT, and PARM modules of the AMBER 5 program (University of California, San Francisco). The topology and coordinate files of the Bak octamer were generated by the xLeap module of the AmberTools 1.5 program (University of California, San Francisco). The energy minimization was performed by using the SANDER modules of the AMBER programs Version 5 (for Noxa·Bak and Bim·Bak) and Version 11 (for the Bak octamer) with a dielectric constant of 1.0 and 500 cycles of steepest-descent minimization followed by 10,000 cycles of conjugate-gradient minimization.
For Noxa·Bak, the docking-generated complex and the average complex structure of cluster 6 from the first-round simulations (Table S1) were used for the first- and second-round simulations, respectively; the energy-minimized complex was solvated by using EDIT with 5,897 and 6,744 TIP3P water molecules53 for the first- and second-round simulations, leading to a system of 20,703 and 23,244 atoms, respectively. The mutation-derived Bim·Bak complex was solvated by using EDIT with 6,599 TIP3P water molecules leading to a system of 22,773 atoms. The energy-minimized Bak octamer was solvated by using the xLeap module with 59,580 TIP3P water molecules leading to a system of 198,828 atoms. The water molecules were obtained from solvating the complex using a pre-equilibrated box of TIP3P molecules, whose hydrogen atom charge was set to 0.4170, where any water molecule was removed if it had an oxygen atom closer than 2.2 Å to any solute atom or a hydrogen atom closer than 2.0 Å to any solute atom, or if it was located further than 10.0 Å along the x-, y-, or z-axis from any solute atom.
Multiple Molecular Dynamics Simulations
The solvated system was energy-minimized for 100 cycles of steepest-descent minimization followed by 100 cycles of conjugate-gradient minimization to remove close van der Waals contacts in the system, then heated from 0 to 300 K at a rate of 10 K/ps under constant temperature and volume, and finally simulated independently with a unique seed number for initial velocities at 300 K under constant temperature and pressure using the PMEMD module of the AMBER program (University of California, San Francisco). For the AMBER program, Version 8 was used for Noxa·Bak and Bim·Bak, and Version 11 was for the Bak octamer. All simulations used (1) a dielectric constant of 1.0, (2) the Berendsen coupling algorithm54, (3) a periodic boundary condition at a constant temperature of 300 K and a constant pressure of 1 atm with isotropic molecule-based scaling, (4) the Particle Mesh Ewald method to calculate long-range electrostatic interactions55, (5) a time step of 1.0 fs, (6) the SHAKE-bond-length constraints applied to all the bonds involving the H atom, (7) saving the image closest to the middle of the “primary box” to the restart and trajectory files, (8) formatted restart file, and (9) default values of all other inputs of the PMEMD module. All simulations were performed on a cluster of Apple Mac Pros with 92 Intel Xeon cores (3.0 GHz), a cluster of Apple Xserves with 590 G5 processors (2.2/2.4 GHz), and the Intel Xeon clusters at the University of Minnesota Supercomputing Institute.
Data Analysis
Average structures were obtained by using the CARNAL module of AMBER 5. Cluster analyses were performed by using the PTRAJ module of AmberTools 1.5 (University of California, San Francisco). RMSDs and ACRMSDs were calculated by using the McLachlan algorithm56 as implemented in ProFit V2.6 (http://www.bioinf.org.uk/software/profit/) and the CARNAL module, respectively.
For each of the 20 (round 1) or 52 (round 2) simulations of Noxa·Bak, 200 (round 1) or 100 (round 2) instantaneous conformations were saved at 5-ps (round 1) or 10-ps (round 2) intervals during the last 1-ns period. A total of 4,000 (round 1) or 5,200 (round 2) instantaneous conformations of Noxa·Bak from the 20 (round 1) or 52 (round 2) simulations were subjected to a cluster analysis using the averagelinkage algorithm [epsilon = 3.0 Å; RMS on Y89Bak, F93Bak, F111Bak, F134Bak, and F32Noxa for Noxa·Bak or Y89Bak, F93Bak, F111Bak, F134Bak, and F159Bim for Bim·Bak]57 implemented in the PTRAJ module of the AmberTools package (Version 1.4 for Noxa·Bak and Version 1.5 for Bim·Bak; University of California, San Francisco). Seven (round 1) or 2 (round 2) clusters of the Noxa·Bak conformations were identified (Table S1). The average conformation of each cluster was energy-minimized using the SANDER module of the AMBER 5.0 program with a dielectric constant of 40.0 and 500 cycles of steepest-descent minimization followed by 10,000 cycles of conjugate-gradient minimization. The interaction energy between BH3-only protein and Bak of the energy-minimized complex was then calculated using an in-house progm with a dielectric constant of 40.0 and a nonbonded cutoff of 50,000 Å (Table S1).
Graphics
All figures were generated by using the MacPyMOL V1.5.0 (Schrödinger LLC, Portland, OR), AutoCAD 2012 for Mac Version F.51.M.57 (Autodesk Inc, San Rafael, CA), and Adobe Photoshop CS5 Extended Version 12.1 x64 (Adobe Systems Incorporated, San Jose, CA) programs.
Author Contributions
Conceived and designed the experiments: YPP SHK HD. Performed the experiments: YPP HD AS XWM PAS. Analyzed the data: YPP HD AS XWM PAS SHK. Wrote the paper: YPP. Contributed with revisions: YPP HD AS XWM PAS SHK.
Supplementary Material
Supplementary Information
Supplementary Information
Acknowledgments
This work was supported by the Mayo Foundation for Medical Education and Research and in part by grants R01 CA69008 and T32 GM072474. The computational studies were supported in part by the University of Minnesota Supercomputing Institute.
References
- Cory S. & Adams J. M. The BCL-2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2, 647–656 (2002). [DOI] [PubMed] [Google Scholar]
- Jiang X. & Wang X. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 (2004). [DOI] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L. & Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163 (2007). [DOI] [PubMed] [Google Scholar]
- Youle R. J. & Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 (2008). [DOI] [PubMed] [Google Scholar]
- Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J. & Green D. R. The BCL-2 family reunion. Mol. Cell 37, 299–310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanave C., Santamaria M. & Saccone C. Comparative genomics: the evolutionary history of the BCL-2 family. Gene 333, 71–79 (2004). [DOI] [PubMed] [Google Scholar]
- Letai A. et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2, 183–192 (2002). [DOI] [PubMed] [Google Scholar]
- Walensky L. D. et al. A stapled BID BH3 helix directly binds and activates BAX. Mol. Cell 24, 199–210 (2006). [DOI] [PubMed] [Google Scholar]
- Kim H. et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell 36, 487–499 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T. et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell 17, 525–535 (2005). [DOI] [PubMed] [Google Scholar]
- Kim H. et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8, 1348–1358 (2006). [DOI] [PubMed] [Google Scholar]
- Martin S. J. Opening the Cellular Poison Cabinet. Science 330, 1330 (2010). [DOI] [PubMed] [Google Scholar]
- Du H. et al. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J. Biol. Chem. 286, 491–501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H. et al. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J. Cell Biol. 194, 38–47 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E. et al. BAX activation is initiated at a novel interaction site. Nature 455, 1076–1081 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewson G. et al. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell 30, 369–380 (2008). [DOI] [PubMed] [Google Scholar]
- Kiefer F., Arnold K., Künzli M., Bordoli L. & Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Sato T., Kagami Y. & Shimotohno K. Molecular cloning and characterization of a cDNA for a novel phorbol-12-myristate-13-acetate-responsive gene that is highly expressed in an adult T-cell leukemia cell line. J. Virol. 64, 4632–4639 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. S. et al. A novel Bcl-2 related gene, Bfl-1, is overexpressed in stomach cancer and preferentially expressed in bone marrow. Oncogene 11, 1693–1698 (1995). [PubMed] [Google Scholar]
- Brien G., Trescol-Biemont M.-C. & Bonnefoy-Bérard N. Downregulation of Bfl-1 protein expression sensitizes malignant B cells to apoptosis. Oncogene 26, 5828–5832 (2007). [DOI] [PubMed] [Google Scholar]
- Day C. L. et al. Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J. Mol. Biol. 380, 958–971 (2008). [DOI] [PubMed] [Google Scholar]
- Moldoveanu T. et al. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol. Cell 24, 677–688 (2006). [DOI] [PubMed] [Google Scholar]
- Wang H. et al. Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator. J. Struct. Biol. 166, 32–37 (2009). [DOI] [PubMed] [Google Scholar]
- Pang Y.-P. Three-dimensional model of a substrate-bound SARS chymotrypsin-like cysteine proteinase predicted by multiple molecular dynamics simulations: catalytic efficiency regulated by substrate binding. Proteins. 57, 747–757 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian B. J. et al. Configurational entropy in protein-peptide binding: computational study of Tsg101 ubiquitin E2 variant domain with an HIV-derived PTAP nonapeptide. J. Mol. Biol. 389, 315–335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M. et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 (1997). [DOI] [PubMed] [Google Scholar]
- Petros A. M., Olejniczak E. T. & Fesik S. W. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta 1644, 83–94 (2004). [DOI] [PubMed] [Google Scholar]
- Czabotar P. E. et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits C., Czabotar P. E., Hinds M. G. & Day C. L. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure 16, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- Verdonk M. L., Boks G. J., Kooijman H., Kanters J. A. & Kroon J. Stereochemistry of charged nitrogen-aromatic interactions and its involvement in ligand-receptor binding. J. Comput.-Aided Mol. Design 7, 173–182 (1993). [DOI] [PubMed] [Google Scholar]
- Liu X., Dai S., Zhu Y., Marrack P. & Kappler J. W. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 19, 341–352 (2003). [DOI] [PubMed] [Google Scholar]
- Muchmore S. W. et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 381, 335–341 (1996). [DOI] [PubMed] [Google Scholar]
- Antonsson B., Montessuit S., Lauper S., Eskes R. & Martinou J. C. Bax oligomerization is required for channel-forming activity in liposomes and to trigger cytochrome c release from mitochondria. Biochem. J. 345 Pt 2, 271–278 (2000). [PMC free article] [PubMed] [Google Scholar]
- Saito M., Korsmeyer S. J. & Schlesinger P. H. BAX-dependent transport of cytochrome c reconstituted in pure liposomes. Nat. Cell Biol. 2, 553–555 (2000). [DOI] [PubMed] [Google Scholar]
- Reed J. C. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 13, 1378–1386 (2006). [DOI] [PubMed] [Google Scholar]
- O'Neill J. W., Manion M. K., Maguire B. & Hockenbery D. M. BCL-XL dimerization by three-dimensional domain swapping. J. Mol. Biol. 356, 367–381 (2006). [DOI] [PubMed] [Google Scholar]
- Wilmsen H. U., Leonard K. R., Tichelaar W., Buckley J. T. & Pattus F. The aerolysin membrane channel isformed by heptamerization of the monomer. EMBO J. 11, 2457–2463 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L. et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 (1996). [DOI] [PubMed] [Google Scholar]
- Lewis S., Bethell S. S., Patel S., Martinou J. C. & Antonsson B. Purification and biochemical properties of soluble recombinant human Bax. Protein Expr. Purif. 13, 120–126 (1998). [DOI] [PubMed] [Google Scholar]
- Mirkin N., Jaconcic J., Stojanoff V. & Moreno A. High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor. Proteins. 70, 83–92 (2008). [DOI] [PubMed] [Google Scholar]
- Adrain C., Creagh E. M. & Martin S. J. Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J. 20, 6627–6636 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springs S. L., Diavolitsis V. M., Goodhouse J. & McLendon G. L. The kinetics of translocation of Smac/DIABLO from the mitochondria to the cytosol in HeLa cells. J. Biol. Chem. 277, 45715–45718 (2002). [DOI] [PubMed] [Google Scholar]
- Pardo O. E. et al. Fibroblast growth factor 2-mediated translational control of IAPs blocks mitochondrial release of Smac/DIABLO and apoptosis in small cell lung cancer cells. Mol. Cell. Biol. 23, 7600–7610 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Ren X., Yang L., Lin Y. & Wu X. A JNK-dependent pathway is required for TNFalpha-induced apoptosis. Cell 115, 61–70 (2003). [DOI] [PubMed] [Google Scholar]
- Parone P. A. et al. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol. Cell. Biol. 26, 7397–7408 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhola P. D., Mattheyses A. L. & Simon S. M. Spatial and Temporal Dynamics of Mitochondrial Membrane Permeability Waves during Apoptosis. Biophys. J. 97, 2222–2231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T. et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111, 331–342 (2002). [DOI] [PubMed] [Google Scholar]
- Muñoz-Pinedo C. et al. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc. Natl. Acad. Sci. U.S.A. 103, 11573–11578 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewson G. et al. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol. Cell 36, 696–703 (2009). [DOI] [PubMed] [Google Scholar]
- Koshland D. E. The comparison of non-enzymic and enzymic reaction velocities. J. Theor. Biol. 2, 75–86 (1962). [Google Scholar]
- Hornak V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 65, 712–725 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrom L., Okur A. & Simmerling C. Evaluating the performance of the ff99SB force field based on NMR scalar coupling data. Biophys. J. 97, 853–856 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandreskhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1982). [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Di Nola A. & Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984). [Google Scholar]
- Darden T. A., York D. M. & Pedersen L. G. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- McLachlan A. D. Rapid comparison of protein structures. Acta Cryst. A38, 871–873 (1982). [Google Scholar]
- Shao J., Tanner S. W., Thompson N. & Cheatham III T. E. Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 3, 2312–2334 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information