Abstract
Plant genomes contain large numbers of cell surface leucine-rich repeat (LRR) and intracellular nucleotide binding (NB)-LRR immune receptors encoded by resistance (R) genes that recognize specific pathogen effectors and trigger resistance responses. The unregulated expression of NB-LRR genes can trigger autoimmunity in the absence of pathogen infection and inhibit plant growth. Despite the potential serious consequence on agricultural production, the mechanisms regulating R-gene expression are not well understood. We identified microRNA (miRNA) progenitor genes precursor transcripts, and two miRNAs [nta-miR6019 (22-nt) and nta-miR6020 (21-nt)] that guide cleavage of transcripts of the Toll and Interleukin-1 receptor-NB-LRR immune receptor N from tobacco that confers resistance to tobacco mosaic virus (TMV). We further showed that cleavage by nta-miR6019 triggers RNA-dependent RNA polymerase 6- and ribonuclease Dicer-like 4-dependent biogenesis of 21-nt secondary siRNAs “in phase” with the 22-nt miR6019 cleavage site. Furthermore, we found that processing of the 22-nt nta-miR6019 depended on an asymmetric bulge caused by mismatch in the nta-miR6019 precursor. Interestingly, coexpression of N with nta-miR6019 and nta-miR6020 resulted in attenuation of N-mediated resistance to TMV, indicating that these miRNAs have functional roles in NB-LRR regulation. Using a bioinformatics approach, we identified six additional 22-nt miRNA and two 21-nt miRNA families from three Solanaceae species—tobacco, tomato, and potato. We show that members of these miRNA families cleave transcripts of predicted functional R genes and trigger production of phased secondary 21-nt siRNAs. Our results demonstrate a conserved role for miRNAs and secondary siRNAs in NB-LRR/LRR immune receptor gene regulation and pathogen resistance in Solanaceae.
Two major classes of plant innate immune receptors include pattern recognition receptors (PRRs) and resistance (R) proteins (1, 2). PRRs recognize conserved pathogen-associated molecular patterns (PAMPs) and activate PAMP-triggered immunity (PTI), whereas R proteins recognize divergent pathogen effectors and trigger the hypersensitive cell death resistance response.
The majority of R genes encode intracellular innate immune proteins with nucleotide binding (NB) and leucine-rich repeat (LRR) domains. Some NB-LRR genes encode proteins with an N-terminal domain similar to the Toll and Interleukin-1 receptors that mediate innate immunity in animals (TIR-NB-LRR), whereas others encode proteins with a coiled-coil domain at the N terminus (CC-NB-LRR) (3, 4). Another class of R genes encodes cell surface innate immune receptors with a transmembrane domain and an extracellular LRR domain (termed receptor-like proteins, RLPs) (5). Most active R genes are found within tandemly repeated arrays that arose through duplication and positive selection over the course of plant–pathogen interactions (6–8).
In plants and other organisms, small RNA (sRNA) systems mediate gene silencing and affect genome integrity, gene regulation, and antiviral defense. Different classes of sRNAs have been characterized (9). The DICER-LIKE 1 (DCL1) enzyme cleaves long RNA precursors that fold into hairpins to generate 21- and 22-nt mature microRNAs (miRNAs). Functionally unique 22-nt miRNAs are required to generate a specialized class of secondary small interfering RNAs called transacting siRNAs (tasiRNAs) from TAS transcripts. Secondary siRNA production also requires RNA-dependent RNA polymerase 6 (RDR6) to produce double-stranded RNA (dsRNA) from miRNA-cleaved transcripts. The subsequent processing of dsRNA by Dicer-like 4 (DCL4) yields 21-nt secondary siRNAs in register, or in phase, with the miRNA cleavage site. tasiRNAs have been demonstrated to act noncell autonomously and are hypothesized to reinforce silencing of multicopy loci (10–12).
sRNA-mediated transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS) have been implicated to regulate host defense against pathogens (13). However, we lack mechanistic understanding of the impact of TGS and PTGS in plant innate immunity. Here, we describe identification of two miRNAs, nta-miR6019 (22-nt) and nta-miR6020 (21-nt), that guide sequence-specific cleavage of transcripts of the TIR-NB-LRR immune receptor N that confers resistance to tobacco mosaic virus (TMV). We found that N gene-specific 21-nt sRNAs are in phase with the nta-miR6019 cleavage site in the N gene mRNA. Biogenesis of these sRNAs depends on both RDR6 and DCL4. Moreover, using transient coexpression assays, we demonstrated that synthesis of these sRNAs depends on the presence of 22-nt nta-miR6019, indicating that these sRNAs are secondary (potentially transacting) siRNAs. We show that transient expression of N-targeted miRNAs in Nicotiana benthamiana attenuates N-mediated resistance to TMV, demonstrating that miRNAs play an important role in regulating disease resistance in Solanaceae. We also discovered eight unique families of miRNAs that target members of immune receptors from nine R-gene families in two crop species of Solanaceae, potato and tomato. Many of these miRNAs trigger the biogenesis of secondary 21-nt siRNAs from their R-gene targets. Thus, we propose that there is a conserved role for miRNAs and secondary siRNAs in regulating NB-LRR and LRR innate immune receptor gene expression and pathogen resistance in plants.
Results
Identification and Characterization of miRNAs Targeting N Transcripts for Silencing.
In a previous study, we identified a 21-nt sRNA complementary to the N gene from a N. benthamiana sRNA library (14). Searches of tobacco sRNA libraries using the 21-nt N-related sRNA identified a matching 22-nt sRNA in wild-type tobacco libraries (ref. 15 and http://somart.ist.berkeley.edu). This sRNA was complementary to positions 125–146 bp in exon 1 of N (GenBank U15605) (Fig. 1A and Table 1), which is a conserved sequence among N-homologs in tobacco. Based on its length, abundance, and complementarity to sequences conserved among N homologs, we hypothesized that this 22-nt sRNA potentially targets the N gene for silencing.
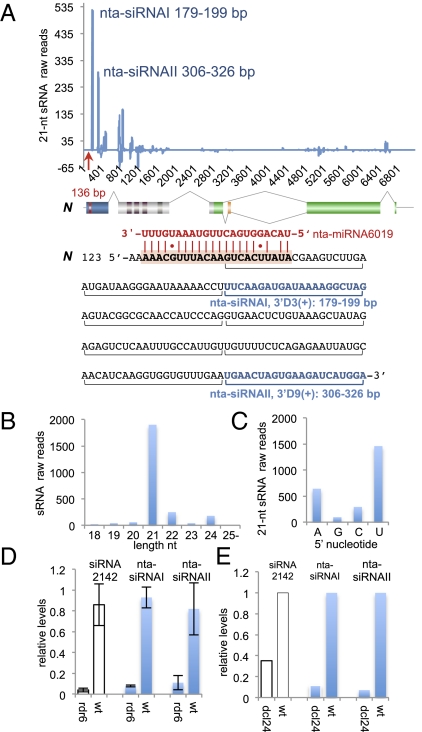
Fig. 1.
nta-miR6019 and nta-miR6020-directed N cleavage. (A) nta-miR6019a (bold red font), nta-miR6020a (dark red font), and N target sites (bold, black and shaded font) with N cleavage sites (Clv) indicated in base pairs (bp). N and N-homolog cleavage product sequences (black font), shown aligned with N. N (U15605) map, below with encoded protein domains are indicated as filled shaded rectangles: TIR, blue; NB, gray; LRR, green. The MiS1 transposon (alternative exon of N), filled orange triangle (14); the N miRNA target, red vertical line. (B) Predicted foldback structure of nta-premiR6019,6020a. (C) Ethidium bromide stained agarose gel of 5′ RACE products of RNA from N-CFPT2T1 and N-CFPt2t1 tobacco lines generated using N-CFP primers (Upper) and N-primers (Lower). (D) Maps of N-CFPT2T1 and N-CFPt2t1 transgenes with the N TIR domain, CFP gene, and miRNA target regions indicated as filled dark blue, cyan, and red rectangles, respectively. miRNA target sequences in N-CFPT2T1 and N-CFPt2t1 are shown in black (wild-type) and gray (mutated) bold font shown below maps. nta-miR6019 (red font) and nta-miR6020 (dark red font) shown above and below N sequences respectively. Number of nta-miR6020 cleavage products is indicated in dark red font and arrow.
Table 1.
miRNAs and R-gene targets
| miRNA | Sequence | L | R* | C† | Clv‡§ |
| nta-miR6019 | UACAGGUGACUUGUAAAUGUUU | 22 | N | T | 136 |
| nta-miR6020 | AAAUGUUCUUCGAGUAUCUUC | 21 | N | T | 122 |
| nta-miR6021 | UUGGAAGAGGCTGCUAUUGGA | 21 | Hcr9 | R | 347 |
| stu-miR6022 | UGGAAGGGAGAAUAUCCAGGA | 21 | Hcr9 | R | 1121 |
| sly-miR6022 | UGGAAGGGAGAAUAUCCAGGA | 21 | Hcr9 | R | 1157 |
| stu-miR6023 | UUCCAUGAAAGUGUUUUUGGAU | 22 | Hcr9 | R | 106 |
| sly-miR6023 | UUCCAUGAAAGAGUUUUUGGAU | 22 | Hcr9 | R | 142 |
| stu-miR6024 | UUUUAGCAAGAGUUGUUUUCCC | 22 | Rx1 | C | 535 |
| Rpivnt1 | C | 678 | |||
| sly-miR6024 | UUUUAGCAAGAGUUGUUUUACC | 22 | Tm2 | C | 765 |
| stu-miR482b | UUACCGAUUCCCCCCAUUCCAA | 22 | NL25 | T | 792 |
| Ry | T | 1796 | |||
| stu-miR482c | UUUCCUAUUCCACCCAUGCCAA | 22 | RB | C | 1048 |
| stu-miR482d | UCUUGCCUACACCGCCCAUGCC | 22 | R2 | C | 576 |
| R3a | C | 597¶ | |||
| stu-miR482e | UCUUGCCAAUACCGCCCAUUCC | 22 | R3a | C | 597¶ |
| nta-miR6025 | UACCAACAAUUGAGAUAACAUC | 22 | R1 | C | 654 |
| stu-miR6026 | UUCUUGGCUAGAGUUGUAUUGC | 22 | Rpivnt1 | C | 679 |
| sly-miR6026 | UUCUUGGCUAGAGUUGUAUUGC | 22 | Tm2 | C | 767 |
| sly-miR6027 | UGAAUCCUUCGGCUAUCCAUAA | 22 | Sw5 | C | 2427 |
*R-gene names, sequences, accessions, and references in Dataset S1.
†Classes of R-genes: T, TIR-NB-LRR; C, CC-NB-LRR; R, RLP.
‡Predicted miRNA cleavage sites. Cleavage confirmed by 5' RACE or dRNA analysis indicated by bold and underlined font, respectively.
§R-gene 21-nt secondary siRNAs detected for all miRNAs except nta-miR6020, nta-miR6021, and nta-miR6025.
¶Cleavage by siRNA 3'D2[−] at target site 625 bp.
To investigate the origin of this sRNA, we searched tobacco genome survey sequences and identified two potential precursors with sequence identity to the 22-nt sRNA (GenBank FH007932 and FH689777, 90% sequence identity to each other). We investigated each candidate precursor by analysis of predicted transcript secondary structures, the distribution of matching tobacco sRNAs, and expression of each precursor. Using mfold (16) for RNA secondary structure predictions and sRNA mapping, we found that transcripts of each candidate MIR sequence formed two stem-loop structures with candidate 22-nt miRNA and miRNA* (termed nta-miR6019 and nta-miR6019*) sequences mapping to the second stem of each (Fig. 1B and Fig. S1). Two other sRNAs mapped to the first stem of each MIR precursor. Sequence analysis suggested that one of these sRNAs was a distinct miRNA that potentially targeted N transcripts at position 112–132 bp in N, 13 base pairs upstream and overlapping the nta-miR6019 target site by 9 base pairs (Fig. 1A). We termed the leftward stem sRNAs as nta-miR6020 and nta-miR6020* (Fig. 1B and Fig. S1). Tobacco N gene target sequences of nta-miR6019 and nta-miR6020 encode conserved TIR domain amino acids 20–30 that were originally identified in comparisons of the N and the Toll and IL-IR (TIR) protein domains (3, 4, 17) for which the TIR domain name is derived.
MIR progenitors were further validated by sequence analysis of rapid amplification of cDNA 5′- and 3′-ends (5′ and 3′ RACE) and showed that 5′ RACE products matched candidate nta-MIRR6019,6020 precursors and that 3′ RACE products extended both MIR loci (Fig. S2 A and B and Dataset S1). Our results also suggested that the pri-miRNA transcripts of both precursors are alternatively spliced (Fig. S2 A and B). We termed these precursors nta-MIR6019,6020a and nta-MIR6019,6020b. Thus, we concluded that the tobacco genome encodes at least two copies of progenitor nta-MIR6019,6020 and that each progenitor encodes two miRNAs, nta-miR6019 and nta-miR6020, predicted to guide sequence-specific cleavage of N and N homologs.
We validated nta-miR6019–, and nta-miR6020–guided specific cleavage of N and N homologs by analyses of 5′ RACE product sequences. 5′ RACE products were generated by using mRNA isolated from a well-characterized transgenic tobacco line, TG34, which expresses the N gene in an SR1:nn background (3) and confers complete resistance to TMV. Primers used for amplification were designed to amplify products downstream of the predicted nta-miR6019 and nta-miR6020 cleavage sites. Sequences of 18 products showed cleavage at the predicted nta-miR6019 or nta-miR6020 target sites, with 1 derived from nta-miR6019 cleavage of N and the other 17 corresponding to 5 unique sequences derived from the cleavage of N-homologs by nta-miR6020 (Fig. 1A). These results suggest that nta-miR6019 and nta-miR6020 cleave N- and N-homologous transcripts in vivo.
To further validate nta-miR6019 and nta-miR6020 target specificity, we generated transgenic plants expressing N-CFPT2T1 and N-CFPt2t1, chimeric N-Cyan Fluorescent Protein (N-CFP) miRNA sensors carrying wild-type (T2T1) or inactivated (t2t1) N-miRNA target sites (Fig. 1D). We performed 5′ RACE assays on RNA isolated from these lines by using primers designed to detect nta-miR6019– and nta-miR6020–directed cleavage of N-CFP and, as controls, primers to detect cleavage of endogenous N transcripts. Agarose gel fractionation of cleavage products showed that the N-CFP–specific primers amplified a fragment of the expected size from the N-CFPT2T1 line, whereas no cleavage products were detected in samples prepared from the mutant N-CFPt2t1 line (Fig. 1C). In contrast, the N-specific primers amplified cleavage products from endogenous N-homologs in both N-CFPT2T1 and N-CFPt2t1 tobacco lines (Fig. 1C). Sequenced cleavage products prepared from N-CFPT2T1 showed nta-miR6020 directed cleavage at the predicted target site in 17 of the 20 clones sequenced (Fig. 1D and Fig. S1). Although nta-miR6019–directed cleavage was not detected among the sequenced products, nta-miR6019–directed cleavage of N-CFPT2T1 was confirmed in transient coexpression assays of N-CFPT2T1 and 35S:nta-MIR6019,6020, as described below. These results validated the sequence-specificity of nta-miR6019 and nta-miR6020 targeted cleavage of N.
Identification of N Gene 21-nt siRNAs in Phase with the nta-miR6019 Cleavage Site.
In addition to miRNAs targeting the N gene, we identified other sRNAs that perfectly matched both strands of the N-gene sequence and mapped primarily to exons (Fig. 2A). Most sRNAs were 21-nt-long and terminated in a 5′-U residue (Fig. 2 B and C) (12). We observed that the two 5′-proximal and most abundant sRNAs, termed nta-siRNAI and nta-siRNAII, were 42 bp (N coordinates, 179–199 bp) and 169 bp (N coordinates, 306–326 bp) downstream of the 22-nt nta-miR6019 cleavage site at position 136 bp in the N gene (Fig. 2A). The positions of nta-siRNAI and nta-siRNAII correspond to three and nine 21-nt registers [3′D3(+) and 3′D9(+)] from the terminus of the N transcript 3′ cleavage fragment, suggesting that they were in phase with the 22-nt miRNA nta-miR6019 cleavage site. The structure of these N-specific sRNAs and their phasing with respect to the nta-miR6019 cleavage site suggested that they were secondary siRNAs, similar to tasiRNAs and, therefore, likely generated by a process initiated by 22-nt miRNA-guided cleavage and dependent on RDR6 and DCL4 for biogenesis (10–12).
Fig. 2.
N 21-nt secondary siRNAs are phased and require RDR6 and DCL4 for biogenesis. (A) Plot of secondary siRNAs (blue line) as the number of tobacco 21-nt sRNA raw reads (y axis) along the N gene (U15605 coordinates, x axis). A map of N (Fig. 1A) and N and nta-miR6019 sequences in black and red, respectively, are shown below. Horizontal brackets below N sequence indicate 21-nt siRNA phasing. nta-siRNAI and nta-siRNAII registers and coordinates are indicated in blue. (B) N siRNA length [nucleotide (nt), x axis] and number (raw reads, y axis) in wild-type tobacco sRNA library. (C) 5′-terminal nucleotide (x axis) of N 21-nt siRNAs (in raw reads, y axis). (D) Relative levels of nta-TAS3 siRNA2142, nta-siRNAI, and nta-siRNAII in TG34 (wt) and TG34::nta-amiR:RDR6 tobacco lines (rdr6), measured by quantitative miR-ID analysis. Data normalized to miRNA390 levels. Quantitative analysis was repeated with two biological replicas and three technical replicas for each. (E) Relative level of nta-TAS3 siRNA2142, nta-siRNAI, and nta-siRNAII in TG34 tobacco (wt) compared with levels in TG34::nta-RNAi:DCL2,DCL4 (dcl24). Data was normalized to miRNA390 levels in each library.
To test this possibility, we determined the expression levels of nta-siRNAI, nta-siRNAII, and a tobacco homolog of the TAS3 secondary siRNA2142 (nta-siRNA2142; Table S1) in TG34 tobacco and TG34-expressing nta-amiR:RDR6 (TG34::nta-amiR:RDR6). We confirmed that RDR6 expression in TG34::nta-amiR:RDR6 lines was reduced (Fig. S3B). We found that the levels of nta-siRNAI, nta-siRNAII, and nta-siRNA2142 were reduced in the TG34::nta-amiR:RDR6 line by approximately 10-, 7-, and 20-fold, respectively, compared with their levels in TG34 (Fig. 2D). We also compared the levels of these siRNAs in a TG34 sRNA library and a library from an nta-RNAi:DCL2,DCL4 line in the TG34 background (TG34::nta-RNAi:DCL2,DCL4) (14). We found that the relative levels of nta-siRNAI, nta-siRNAII, and nta-siRNA2142 were reduced by 9-, 14-, and 3-fold in the TG34::nta-RNAi:DCL2,DCL4 line compared with TG34 (Fig. 2E). These results indicate that cleavage by 22-nt nta-miR6019 might trigger production of phased secondary 21-nt siRNAs from cleaved N transcripts.
Cleavage by 22-nt nta-miR6019 Is Required to Trigger Secondary siRNA Synthesis.
Based on the RDR6- and DCL4-dependent production of N-secondary siRNAs in phase with the nta-miR6019 cleavage site, we hypothesized that 22-nt nta-miR6019 cleavage triggered secondary siRNA biogenesis. We also postulated that 21-nt siRNAs could be triggered from a N-CFP miRNA sensor with a wild-type nta-miR6019 target sequence, but not from a N-CFP sensor with mutations that prevented binding of nta-miR6019. We further reasoned that repair of the mismatch in the nta-miR6019/N duplex (Fig. 3A, row 1) would enhance nta-miR6019/N pairing, increase nta-miR6019 cleavage, and possibly increase the accumulation of miRNA-dependent secondary siRNA.
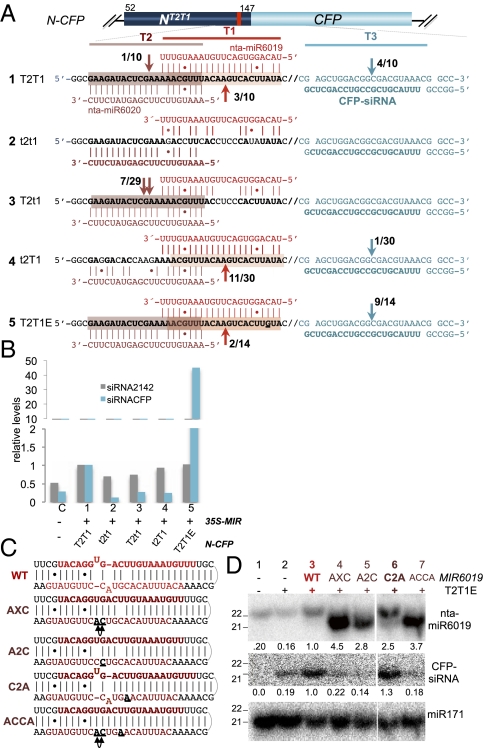
Fig. 3.
nta-miR6019 (22-nt) triggers N secondary siRNA biogenesis. (A) Map of N-CFP miRNA sensor (top) and sensor sequences (1–5) with N (black bold) shown aligned with nta-miR6019 (red) and nta-miR6020 (dark red). CFP is shown in cyan, and CFP-siRNA is indicated in bold cyan. Arrows indicate cleavage positions at targets T1, T2, and T3, and numbers indicate cloning frequency. (B) The relative levels of CFP-siRNA and tasiRNA2142 measured by quantitative miR-ID analysis in N. benthamiana samples coinfiltrated with indicated N-CFP miRNA sensors and 35S:nta-MIR6019,6020, normalized to levels of miRNA390. (C) Sequences and predicted secondary structures of wild-type (WT) nta-premiR6019 (red) and mutant nta-premiR6019 (AXC, A2C, C2A, and ACCA, dark red) constructions. (D) Northern blot hybridization of sRNAs isolated from N. benthamiana leaves coinfiltrated with indicated 35S:nta-MIR6019,6020 vectors and N-CFPT2TE1 miRNA sensor. Hybridization probes, miR6019, CFP-siRNA, and miR171 (Table S1) are indicated.
To address these predictions, a N-CFPT2T1E miRNA sensor with a predicted nta-miR6019-enhanced binding site was constructed (Fig. 3A, row 5). We tested whether cleavage guided by 22-nt nta-miR6019, 21-nt nta-miR6020, or both triggered N cleavage and 21-nt secondary siRNA biogenesis by using transient coexpression of wild-type or mutated N-CFP miRNA sensors with 35S-nta-MIR6019,6020 in N. benthamiana (validated for overexpression of nta-miR6019; Fig. S3A). We analyzed the cleavage products in coinfiltrated samples by gel electrophoresis and sequencing. Staining of agarose gels detected larger fragments in the size range of those predicted for cleavage by nta-miR6019 and nta-miR6020 in samples with nonmutated target sites. We also detected a strongly staining shorter fragment in the N-CFPT2T1E sample and more faintly staining diffuse bands in the other samples. Sequence analysis of gel fractions enriched for longer cleavage products confirmed cleavage by nta-miR6019 at wild-type N-CFP T1 targets (Fig. 3A, rows 1, 4, and 5) and cleavage by miRNA6020 at wild type N-CFP T2 targets (Fig. 3A, rows 1 and 3), whereas no cleavage products were detected in samples expressing N-CFP with mutated t1 (Fig. 3A, rows 2 and 3) and/or mutated t2 targets (Fig. 3A, rows 2 and 4). We sequenced cleavage products of samples enriched for shorter fragments and detected cleavage at an additional target (T3), 72 bp downstream of the miR6019 cleavage site directed by a predicted complementary in-phase CFP-siRNA in samples expressing N-CFP with a wild-type T1 or an enhanced T1E target (Fig. 3A, rows 1, 4, and 5).
Using Northern blot analysis and quantitative sRNA analysis (18) we showed an ≈50-fold increase in the level of 21-nt CFP-siRNA in samples coinfiltrated with N-CFPT2T1E compared with levels of CFP-siRNA measured in wild-type N-CFPT2T1 coinfiltrated samples, whereas the level of nta-TAS3 siRNA2142 was the same in both samples (Fig. 3B and Fig. S3A). These results indicated that nta-miR6019 cleavage contributes to increased secondary siRNA accumulation from the enhanced N-CFPT2T1E sensor and suggested that cleavage of N transcripts by 22-nt nta-miR6019 triggered 21-nt secondary siRNA production.
We next tested whether 21-nt nta-miR6019 was capable of cleaving the N gene and/or triggering secondary siRNA biogenesis. Using site-directed mutagenesis, we engineered nta-MIR6019 mutants, MIR6019AXC, MIR6019A2C, and MIR6019ACCA with altered nta-miR6019* sequences to remove mismatches and, thus, potentially alter the size of the nta-miRNA6019 while allowing the miRNA sequence to bind and cleave the N gene. A fourth mutant, MIR6019C2A, was engineered to repair the symmetric mismatch found in the wild-type nta-premiR6019 structure while retaining the predicted crucial asymmetric mismatch, and was expected to produce 22-nt miRNAs in a similar fashion to wild-type nta-premiR6019 (Fig. 3C).
We determined whether the mutated nta-miRNAs were capable of cleaving the N gene and/or triggering secondary siRNA biogenesis by Northern blot hybridization analysis. Hybridization using the nta-miR6019 probe confirmed that MIR6019AXC, MIR6019A2C, and MIR6019ACCA generated 21-nt nta-miR6019 (Fig. 3D, lanes 4, 5, and 7) and that the levels of secondary CFP-siRNAs detected in these samples were not above the background level detected in the control sample infiltrated with N-CFPT2T1E alone (Fig. 3D, lane 2). In contrast, Northern blot analysis showed that MIR6019C2A produced higher levels of 22-nt nta-miR6019 than wild-type MIR6019 (Fig. 3D, lane 6; compare lanes 6 and 3) and higher levels of CFP-siRNA compared with the wild-type control (Fig. 3D, Middle, lane 6; compare lanes 6 and 3) or the 21-nt producing nta-miR6019 samples (Fig. 3D, Middle, lanes 4, 5, and 7; compare lane 6 with lanes 4, 5, and 7). We postulate that the increase in 22-nt nta-miR6019 from MIR6019C2A is responsible for the observed higher levels of secondary CFP-siRNA detected in this sample. These results reinforce the model that DCL1-mediated biogenesis of 22-nt miRNAs depends on asymmetric bulges in the duplex structure of the miRNA precursor and that the length of miRNAs plays an important role in the secondary siRNA biogenesis.
Overexpression of nta-MIR6019,6020 Attenuates N-Mediated Resistance to TMV.
To understand the potential impact of nta-miR6019 and nta-miR6020 on N-mediated resistance to TMV, we used Agrobacterium tumefaciens infiltration to coexpress nta-MIR6019,6020, the N gene, and a TMV-GFP amplicon in N. benthamiana leaves. N-mediated TMV resistance was assessed by TMV-GFP localization, the inhibition of TMV spread, and induction of the hypersensitive cell death response in Agrobacterium-inoculated leaves.
The nta-MIR6019,6020 precursor vectors used in these assays overexpressed wild-type 22-nt nta-miR6019 (35S:nta-MIR6019,6020) or 21-nt nta-miR6019 (35S:nta-MIR6019AXC,6020) (Fig. 3D). A control vector, 35S:nta-fb6025.0, expressed a tobacco foldback sequence containing nta-miR6025a but does not produce sRNAs. Binary vectors expressed the N gene or the Rx1 (resistance to potato virus X) from native tobacco or potato promoter sequences, respectively (19, 20).
At 6 d after infiltration, TMV-GFP fluorescence was distributed to small punctate areas (Fig. 4C, lane 2) in leaves coinfiltrated with N and 35S:nta-fb6025.0, indicating restricted spread of the virus. In contrast, we observed that the number and size of TMV-GFP fluorescent regions significantly increased in leaves coexpressing N and either 35S:nta-MIR6019AXC,6020 or wild-type 35S:nta-MIR6019,6020 (Fig. 4C, lanes 3 and 4) compared with N coexpressed with the 35S:nta-fb6025.0 control (compare Fig. 4C, lanes 3 and 4 with lane 2). Comparable patterns of widespread GFP fluorescence were observed in leaves expressing TMV-GFP without any resistance gene (Fig. 4C, lane 8) and in leaves coexpressing Rx1 with 35S:nta-fb6025.0, 35S:nta-MIR6019AXC,6020 or 35S:nta-MIR6019,6020 indicating that TMV-GFP spread was not restricted in any of these control samples (Fig. 4C, lanes 5–7, respectively).
Fig. 4.
Overexpression of nta-MIR6019,6020 attenuates N-mediated resistance to TMV. (A) A. tumefaciens expression vectors used in N. benthamiana coexpression assays, 35S:nta-fb6025, 35S:nta-MIR6019AXC,6020, and 35S:nta-MIR6019,6020 are indicated as nta-fb6025.0, miR6019 AXC, and miR6019 WT, respectively. (B) Coinfiltrated leaves and control untreated leaves photographed under bright light at 6 d after infiltration. (C) Same leaves as in B, photographed under UV light. (D) Coinfiltrated leaves and control untreated leaves photographed under bright light at 7 d after infiltration. The number of plants of six plants tested that displayed representative phenotypes are indicated below each lane.
At 7 d after infiltration, we observed the formation of the hypersensitive response in leaves of two of the six plants coinfiltrated with TMV-GFP, N, and 35S:nta-fb6025.0 (Fig. 4D, lane 2), whereas the hypersensitive response was not observed in leaves of plants coinfiltrated with 35S:nta-MIR6019AXC,6020 or 35S:nta-MIR6019,6020. The hypersensitive response was not detected in control leaves coexpressing Rx and MIR vectors or in leaves expressing TMV-GFP alone (Fig. 4D, lanes 5–8, respectively). These results suggest that N-mediated restriction of TMV-GFP spread and induction of the hypersensitive response was attenuated in leaves overexpressing wild-type 22-nt nta-miR6019 or nta-miR6019AXC with nta-miR6020.
Families of 22-nt miRNAs Target Solanaceae R genes and Trigger siRNA Production.
To assess the potential broader impact of R-gene regulation by miRNAs in tobacco, potato, and tomato, we used a bioinformatic pipeline developed to predict R-gene miRNAs, MIR progenitors, and miRNA cleavage products and identified 10 Solanaceae miRNA families (including nta-miR6019 and nta-miR6020 described above) that target 10 Solanaceae R-gene families (Table 1 and Table S2; bioinformatic pipeline available at http://somart.ist.berkeley.edu/). The sequences of characterized miRNAs and MIR precursors and the sequences of predicted R-gene targets are described in Table S2 and Dataset S1, respectively. The miRNA/R–gene-target duplex sequences predicted by the bioinformatic pipeline are in Dataset S2.
Seven of the 10 miRNA families identified were 22 nt in length (Table 1 and Table S2). Structural analyses of the 22-nt miRNA precursors showed that predicted foldback sequences contained an asymmetric bulge in the 22-nt miRNA strand and, in most cases, an asymmetric bulge in the 22-nt miRNA* strand (nta-premR6019a, Fig. 1B; nta-premiR6019b, Fig. S1; sly-premiR6023, -miR6024, -miR6026, and stu-premiR6027, Fig. S3C; stu-premiR6024, Fig. S4; stu-premiR482d, Fig. S4). sRNA mapping confirmed the locations of miRNA and miRNA* sequences in the stem regions of the predicted foldback secondary structures of precursor miRNAs (premiRNAs) (Figs. S1, S4, and S5).
We identified two 21-nt miRNA families, sly-, stu-miR6022 and nta-miR6021, predicted to target members of the tomato Hcr9 (Homologs of Cladosporium fulvum resistance 9) family (21) (Table 1, Table S2, and Fig. S5). The sly-pri-miR6022 precursor was identified in the 3′UTR of a cloned tomato full-length cDNA (AK327901) predicted to encode a hydrolase-like protein (Fig. S5), and sRNA mapping confirmed the locations of miRNA and miRNA* sequences in the stem of the predicted precursor (Fig. S5). The tomato Hcr9 gene family is targeted by another miRNA, sly-miR6023 (22-nt) at position 142 bp, located >1,000 base pairs upstream of the sly-miR6022 target site at 1,157 bp (Table 1 and Fig. S5).
We validated the activity of predicted miRNAs by sequencing products of cleavage assays by using RNA isolated from Solanaceae species corresponding to predicted miRNAs and R-gene targets shown in Table 1 (5′ RACE primers; Table S1). Sequence analysis confirmed cleavage of tomato Hcr9 by sly-miR6022 (Fig. S5), potato Rx1 by stu-miR6024 (Fig. S4), potato R2 (22) by stu-miR482d (Fig. S4), and a tobacco R1-gene homolog (R1-GH) by nta-miR6025a (Table 1, cleavage product sequences in Dataset S2). Although predicted cleavage products of R3a (23) by stu-miR482d and stu-miR482d were not identified, products were detected at a site corresponding to cleavage by an in-phase minus stand R3a siRNA (3′D2[-]) at position 625 bp (Table 1).
To validate the activity of other predicted R-gene miRNAs, we searched dRNA libraries (24) for cleavage products with 90% or more sequence identity to predicted 3′ cleaved transcripts of R genes (http://somart.ist.berkeley.edu/). We identified candidate cleavage products of tomato Hcr9-0 by sly-miR6023 (Fig. S5) and potato Hcr-9 homologs by stu-miR6023 at predicted miRNA target sites (Table 1). We also identified dRNA candidate cleavage products of potato R-genes NL25 (25) and RB (26) that corresponded to predicted stu-miR482b and stu-miR482c cleavage sites, respectively. Predicted dRNA cleavage products of tomato Sw5 by sly-miR6027 were also identified (Table 1; http://somart.ist.berkeley.edu/).
Using small RNA mapping we identified potato, tomato, and tobacco candidate secondary 21-nt siRNAs with 100% sequence identify to R genes cleaved by 22-nt miRNAs with the exception of R1-GH from tobacco (Table 1).
Members of the 22-nt miRNA482 families target conserved sequences encoding the P-loop of the NB protein domain in all three Solanaceae species studied and are related to miR472, which was previously identified in Arabidopsis and other eudicots. In Arabidopsis, the 22-nt miR472 targets functionally uncharacterized R-gene sequences of the CC-NB-LRR class and triggers the production of secondary siRNA (10, 11). Progenitors of tomato and potato miR482b, miR482c, miR482d, and miR482e are clustered on chromosome 6 (Table S2). This large family of miRNAs targets several R-gene families including the TIR-NB-LRR genes Ry (Resistance to potato virus Y) (27), NL25, and N, and the CC-NB-LRR genes RB, R2, and R3a, conferring resistance to the potato late blight oomycete pathogen Phytophthora infestans (Table 1 and Dataset S2). A summary of the identified Solanaceae miRNAs and innate immune receptor gene targets is presented in Fig. S6.
Discussion
In this study, we identified 10 predominantly 22-nt miRNA families targeting R genes. Structural analyses showed that the 22-nt miRNA precursors bear an asymmetric bulge in the miRNA and miRNA* foldback precursor. The stability of the asymmetric bulge conformation may be affected by the paired nucleotide. We found that removing this asymmetric bulge conformation by site-directed mutagenesis was sufficient to cause 21-nt nta-miR6019 synthesis instead of the usual 22-nt nta-miR6019, confirming the importance of this conformation. By removing other mismatches from the nta-premiR6019 structure, however, we were able to engineer a mutant, termed nta-miR6019C2A, which generates both 21-nt and 22-nt miRNAs, demonstrating that, with reduced mismatches in the miRNA/miRNA* duplex, the premiRNA can adopt both double mismatches and double asymmetric bulge conformations, leading to the accumulation of both 21- and 22-nt miRNA species.
Sequence comparisons between the nta-MIR6019,6020a precursor and the N gene identified discrete stretches of homology between nta-MIR6019,6020a intron and exon sequences, and coding and noncoding sequences of N. (Fig. S2). Sequence comparisons between the tomato sly-premiR6022 inverted repeat sequences and the tomato Hcr9-0 gene targeted by sly-miR6022 identified homology between sly-premiR6022 and Hcr9-0 (Fig. S2). These results suggest that R-gene miRNAs could arise by the insertion of fragments derived from R-gene sequences into new sites, including genes. This model is consistent with conclusions drawn from comparison of Arabidopsis thaliana and A. lyrata genomes that showed gene mobility is promoted by tandemly repeated gene sequences, and that classes of genes that tend to form tandem duplications, for example the F-box protein genes and R-genes, are more likely to transpose than other gene classes (28). The potential to create new chimeric genes by shuffling existing sequences is also mediated by repetitive elements in the genome including retrotransposons and PACK MULE elements (29, 30). Recombination events facilitated by repeated sequences might have mediated R-gene fragment transposition to unlinked genome sites, which subsequently evolved into novel miRNA precursors.
One important question that arises from this study is: what potential role(s) do these R-gene–targeting miRNAs play? Plants have evolved hundreds, even thousands of innate immune receptor genes and, although they provide protection from diverse, evolving pathogen effectors, their large numbers and inherent activity in triggering cell death are potential threats to plant fitness (31). It is reasonable to speculate that miRNA-mediated attenuation of R-gene expression may have coevolved with multicopy R-gene loci and might be one of several mechanisms that contribute to limiting potential fitness costs associated with their evolutionary trajectories. This process would facilitate the continuing amplification and diversification of R genes. Both bacteria and viruses have evolved effectors that can suppress miRNA and siRNA pathways (32, 33). In addition to contributing to pathogen spread, the temporal shutdown of miRNA and siRNA function by pathogen effectors might also enhance R-gene function by blocking the formation or activity of the R-gene targeting miRNAs and, thus, provide some balance to resistance and susceptibility during host–microbe interactions. Thus, we propose that R-gene miRNAs could also function in fine-tuning R-gene function during host–microbe coevolution.
The potential wide distribution of R-gene miRNAs and siRNAs among Solanaceae species suggests that this two-part silencing system might be a conserved mechanism for regulating expression of multiple members of large R-gene families. This conserved mechanism could have broader implications for the regulation of R-gene families in other species. It could allow fine-tuned regulation of R-gene expression through altered biogenesis, function, or movement of small RNAs, which play crucial roles not only in disease resistance, but also in hybridization, autoimmunity, and plant fitness (34).
Materials and Methods
Vector constructions, primers and probes.
The sequences of expression vector inserts are in Dataset S1. Oligonucleotide primers and hybridization probes are in Table S1.
Further experimental details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Hantz and J. Calfas for care of plants. National Science Foundation Plant Genome Research Program Grant DBI-0218166 and US Department of Agriculture Current Research Information System Grant 5335-22000-007-00D supported this work.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences of all mature miRNAs and foldback precursors reported in this paper have been deposited in miRBase, www.mirbase.org.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118282109/-/DCSupplemental.
References
- 1.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Whitham S, et al. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 4.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP. Signaling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 5.Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- 6.Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: Evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 7.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 8.Friedman AR, Baker BJ. The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007;17:493–499. doi: 10.1016/j.gde.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Baulcombe D. RNA silencing. Trends Biochem Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Cuperus JT, et al. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol. 2010;17:997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HM, et al. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuang H, et al. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: New functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahalingam G, Meyers BC. Computational methods for comparative analysis of plant small RNAs. Methods Mol Biol. 2010;592:163–181. doi: 10.1007/978-1-60327-005-2_12. [DOI] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinesh-Kumar SP, et al. Transposon tagging of tobacco mosaic virus resistance gene N: Its possible role in the TMV-N-mediated signal transduction pathway. Proc Natl Acad Sci USA. 1995;92:4175–4180. doi: 10.1073/pnas.92.10.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P, Johnston BH, Kazakov SA. miR-ID: A novel, circularization-based platform for detection of microRNAs. RNA. 2011;17:365–380. doi: 10.1261/rna.2490111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendahmane A, Kanyuka K, Baulcombe DC. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell. 1999;11:781–792. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parniske M, Jones JD. Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc Natl Acad Sci USA. 1999;96:5850–5855. doi: 10.1073/pnas.96.10.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park TH, et al. Characterization and high-resolution mapping of a late blight resistance locus similar to R2 in potato. Theor Appl Genet. 2005;111:591–597. doi: 10.1007/s00122-005-2050-4. [DOI] [PubMed] [Google Scholar]
- 23.Huang S, et al. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 2005;42:251–261. doi: 10.1111/j.1365-313X.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- 24.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hehl R, et al. TMV resistance gene N homologues are linked to Synchytrium endobioticum resistance in potato. TAG Theoretical and Applied Genetics. 1999;98:379–386. [Google Scholar]
- 26.Song J, et al. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci USA. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidal S, Cabrera H, Andersson RA, Fredriksson A, Valkonen JP. Potato gene Y-1 is an N gene homolog that confers cell death upon infection with potato virus Y. Mol Plant Microbe Interact. 2002;15:717–727. doi: 10.1094/MPMI.2002.15.7.717. [DOI] [PubMed] [Google Scholar]
- 28.Woodhouse MR, Pedersen B, Freeling M. Transposed genes in Arabidopsis are often associated with flanking repeats. PLoS Genet. 2010;6:e1000949. doi: 10.1371/journal.pgen.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elrouby N, Bureau TE. Bs1, a new chimeric gene formed by retrotransposon-mediated exon shuffling in maize. Plant Physiol. 2010;153:1413–1424. doi: 10.1104/pp.110.157420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 31.Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- 32.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Ding SW. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bomblies K, Weigel D. Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet. 2007;8:382–393. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.