Abstract
The pigmentation patterns of shells in the genus Conus can be generated by a neural-network model of the mantle. We fit model parameters to the shell pigmentation patterns of 19 living Conus species for which a well resolved phylogeny is available. We infer the evolutionary history of these parameters and use these results to infer the pigmentation patterns of ancestral species. The methods we use allow us to characterize the evolutionary history of a neural network, an organ that cannot be preserved in the fossil record. These results are also notable because the inferred patterns of ancestral species sometimes lie outside the range of patterns of their living descendants, and illustrate how development imposes constraints on the evolution of complex phenotypes.
Keywords: pattern formation, developmental evolution, phylogenetics, ancestral inference
Pigmentation patterns on mollusk shells are typical complex phenotypes. They differ substantially among closely related species, but the complexity of the patterns makes it difficult to characterize their similarities and differences. Consequently, it has proven difficult to describe the evolution of pigmentation patterns or to draw inferences about how natural selection might affect them. In this report, we present an attempt to resolve this problem by combining phylogenetic methods with a realistic developmental model that can generate pigmentation patterns of shelled mollusks in the diverse cone snail genus Conus. The model is based on the interactions between pigment-secreting cells and a neuronal network whose parameters are measurable physiological quantities. The neural model used here is a generalization of models proposed earlier by Ermentrout et al. (1) and Boettiger et al. (2). Furthermore, the species have a well supported phylogeny that allows us to infer rates and patterns of parameter evolution.
We chose 19 species in the genus Conus for which Nam et al. have presented a resolved phylogeny (3). For each species, we found a model parameter set that matched the observed pigmentation pattern. Then we applied likelihood-based phylogenetic methods to measure phylogenetic signal in the model parameters, compare possible evolutionary models, estimate the model parameters of ancestral species, and then use these to infer the pigmentation patterns of ancestral species.
Neural Model
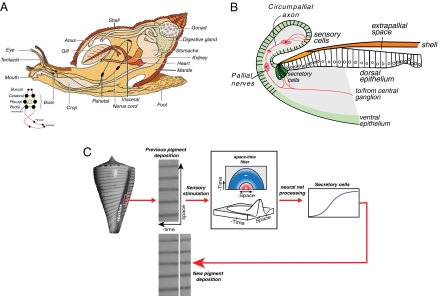
Fig. 1 shows a schematic of the mantle geometry and illustrates the basic principle of the neural model. The mathematical details are described in SI Appendix, Supplement A. The model is built on two general properties of neural networks: spatial lateral inhibition (also called center-surround), and “delayed temporal inhibition.” The latter can be viewed as “lateral inhibition in time” (4–6), as illustrated in Fig. 1C, Center.
Fig. 1.
The neural-network model of the mantle. (A) Rough anatomy of a generic shelled mollusk. Note the “brain,” where the neural patterns are processed consists of a ring of ganglia. (B) Cross-section of the mantle showing how the sensory cells “taste” the previously laid pigment patterns that are processed by the central ganglion and sent to the mantle network that controls the pigment-secreting cells. (C) Simple pattern on a Conus shell and how the model extrapolates the previous pattern to produce the current day's pigment secretion. The pigmentation pattern is read by the sensory cells in the mantle. This activity is then passed through the space–time filter of neural activation and inhibition. Here, time represents the pigmentation pattern that was laid down in previous bouts, whereas space is the dimension along the growing edge of the cell. The resulting filtered activity is passed through nonlinearities for excitation and inhibition, and this net activity drives the secretory cells that lay down the new pigmented shell material. The spatial filter, shown in top and perspective views, has the form of a Mexican hat, in which excitatory activity stimulates a surrounding inhibitory field. The temporal filter that implements delayed inhibition is half a Mexican hat. It generates a refractory period following a period of activity. The pigment secreting cells have a sigmoidal stimulus response curve. Feedback occurs as the current pigment deposition becomes part of the input to the sensory cells for the next secretion bout.
The neural field equations describe the local pattern of neuron spiking. Local activity of excitatory neurons induces the activity of inhibitory interneurons in the surrounding tissue. The net spatial activity has “Mexican hat” shape, as shown in Fig. 1C (5–7). As shell material and pigment are laid down in periodic bouts of secretion, the surface pigment pattern is a space–time record of the animal's secretory activity, in which distance from the shell aperture is proportional to the number of bouts of secretion. Excitation of a cell during a bout inhibits its excitation for some future number of bouts, so that an active neuron will eventually be inhibited and remain inactive for a “refractory” period. Thus, “delayed inhibition” is equivalent to “half a Mexican hat backward in time.” Finally, the secretory activity of pigment granule secretory cells depends sigmoidally on the difference between the activities of the excitatory and inhibitory cells, as shown in Fig. 1C. The logic of the model is that the sensory cells read the history of pigmentation and send this to the neural net that uses this history to “predict” the next increment of pigmentation and instruct the secretory cells to deposit accordingly. This feedback from output to input distinguishes the neural model from models whose future state depends only on their current state (e.g., diffusible morphogens and cellular automata).
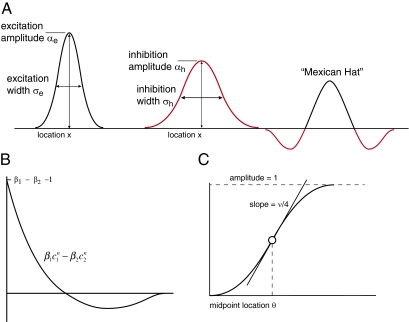
The neural field model is characterized by 17 free parameters, each of which has a concrete physiological interpretation, as described in Fig. 2. In effect, there are four cell types: sensory cells, excitatory neurons, inhibitory neurons, and secretory cells; their effective connectivity relationship is shown in SI Appendix, Supplement A. The behavior of each cell type is given by its input/output relationships, as shown in Fig. 2. Each excitatory and inhibitory neuron is described by a Gaussian spatial synaptic weight kernel described by two parameters (amplitude and width), and a temporal kernel described by four parameters. As several of the parameters appear in products with other parameters, we can normalize their magnitudes and thereby reduce them to three free parameters each describing the spatial and temporal ranges of excitation and inhibition. The precise parameter reduction procedure is described in SI Appendix, Supplement A.
Fig. 2.
Definition of cell specific model parameters. (A) Gaussian excitation and inhibition kernels whose difference creates the Mexican-hat spatial field. (B) Temporal filter implementing delayed inhibition. β1 (β2) is the strength of the temporal excitation (inhibition) and c1 (c2) is the decay in “time” of the excitation (inhibition), wherein time is measured discretely in secretory bouts, denoted by n (0 < c1 < c2 < 1, so that the inhibition decays more slowly in time; thus, the most recent activity is excitatory and more distant activity is inhibitory). (C) Sigmoid response function of the secretory cells; ν is the sharpness of the nonlinearity and θ is the midpoint (there is one nonlinearity for excitation and one for inhibition).
Imbued with these properties, the neural network drives secretory cells to lay down both the shell material and pigment. Thus, the model can reproduce both the shell shape and the surface pattern for many shelled mollusk species, as described previously (2). The present model differs in several essential ways from that proposed previously (2); this is also discussed in SI Appendix, Supplement A.
The basic neural model consists of a simple feedback circuit that spatially and temporally filters previous activity and feeds the result through a nonlinear function to produce the next bout of pigment. One needs 17 parameters to specify the shape of the functions in Fig. 2. By varying the 17 parameters in the model, we were able to produce a wide variety of cone shell patterns. Some of these patterns are very sensitive to the initial conditions (i.e., “chaotic dynamics”), and thus small changes in the initial pattern or small amounts of noise give rise to diversity among individuals while still maintaining the same qualitative pattern. Fig. 3A provides an example showing multiple instances of a simulation of Conus crocatus such that there are small differences in initial data or the addition of a small amount of noise. The overall look of the pattern is the same, but there are clear individual differences.
Fig. 3.
(A) Both noise and chaos generate within-species pattern diversity. a, Three real C. crocatus shells. b, Three shells generated with 1% noise only. c, Three shells generated with slightly different initial conditions, but no noise. d, Three shells with both 1% noise and slightly different initial conditions. (B) Two examples of “unknown” patterns having too-wide inhibition fields.
Somewhat surprisingly, the regions of parameter space that correspond to cone shell patterns are fairly restricted and almost always require that the effective spatial interaction be lateral inhibition. When we chose parameters outside this range, we produced shell patterns that do not correspond to any known species (Fig. 3B).
Although our basic model is capable of producing many of the observed patterns, there are some species (e.g., Conus textile) in which we had to assume that some of the parameters were modulated in space and “time” to specify prepatterns. The prepatterns generally are periodic or consist of a localized region where the parameter is greater or smaller than that of the surrounding region. Such prepatterns could be hard-wired into the network or could themselves be produced by another neural network in the central ganglia (further details are provided in SI Appendix, Supplement A).
Finally, we should point out some important differences between the morphogen models for shell patterns developed by Meinhardt and coworkers (8, 9) and the neural network model used here (1, 2). Structural studies provide strong evidence that shell patterns are a neurosecretory phenomenon rather than a diffusing morphogen phenomenon (2). However, from a theoretical viewpoint, morphogen models can be viewed as an approximation to the neural net model when the range of communication between neurons is short (9, 10). Therefore, in principle, morphogen models could have been used instead of the neural model (11). From a practical viewpoint, however, this would be considerably more difficult because a separate morphogen model is required for each shell pattern, whereas the neural model has a single set of parameters that are varied to match each pattern. Also, as the neural models are more general, they can generate a wider variety of patterns than can diffusible morphogen models. One other difference is fundamental. Morphogen models described by diffusion-reaction dynamics unfold with no “memory” of the system state other than the current state. The neural model, however, is a sensory feedback system in which the current secretion depends on sensing the history of the pattern before the current state.
Phylogenetic Analyses
Inferred Parameter Values for Each Species.
We chose 19 species from the phylogeny published by Nam et al. (3) based on mitochondrial cytochrome C oxidase subunit I and rDNA sequences and on internal transcribed spacer 2 sequences from nuclear ribosomal DNA. There were sufficient data that the order of branching events in the phylogeny could be completely determined with a high degree of statistical confidence.
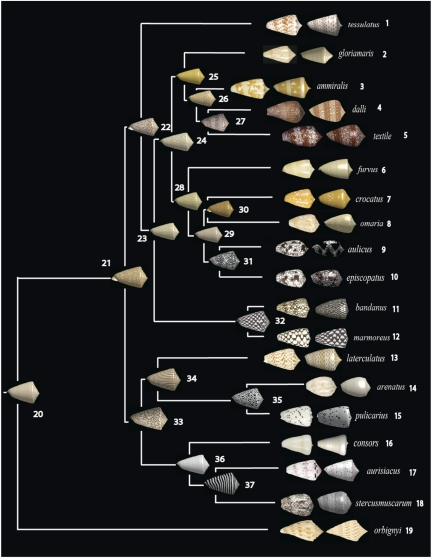
The neural network model was fit to each living species in the phylogenetic tree. Nine species can be reproduced using the basic model (i.e., a single neural network). Six species (Conus tessulatus, Conus aurisiacus, Conus ammiralis, Conus orbignyi, Conus stercusmuscarum, and Conus laterculatus) require a spatial prepattern (generated by a “hidden” network), and four species (Conus dalli, C. textile, Conus aulicus, and Conus episcopatus) require spatiotemporal prepatterns (generated by one or two hidden networks). In phylogenetic analyses of these shell parameters, we focus on the primary network, which can be compared across all species. The fitted parameters for each species are shown in SI Appendix, Supplement C. Images of real shells and their corresponding simulated ones are shown in Fig. 4.
Fig. 4.
Maximum-likelihood estimates of ancestral shell patterns. Shells of living species are displayed at the tips; to the right of these are shells “grown” in the computer by using the neural-network model and the fitted parameters. By using a Brownian motion model for the evolution of continuous traits, the maximum-likelihood value was estimated for each neural network parameter at each node. The neural network model was used to produce the shells using the estimated parameters for each node. Color is not part of the neural network model, so it was added independently to the models of living shells, and then mapped onto the phylogeny (using maximum likelihood) as a binary trait (black/white or brown/white). The text includes further details.
Test for Phylogenetic Signal in Estimated Parameter Values.
Phenotypic traits like body size and shape typically exhibit a substantial degree of “phylogenetic signal,” meaning that they are inherited, and the phenotypes of closely related species are strongly correlated (12). One purpose of the present study is to determine whether parameters of the neural-network model exhibit a phylogenetic signal. They will if the construction of the model accurately approximates the real developmental process of shell patterning. Therefore, we tested for a phylogenetic signal when the model parameters are fitted to the observed pigmentation patterns. A basic test for phylogenetic signal in traits is to compare the observed data to a null model in which all phylogenetic signal are obliterated by randomly shuffling the species names or trait values at the tips of the phylogenic tree (13). To test for a phylogenetic signal in the neural network parameters, we constructed a neighbor-joining phylogeny of the 19 species based on the parameter values alone and compared it with the DNA phylogeny of Nam et al. (3). The parameter-based phylogeny was obtained as described in SI Appendix, Supplement B.
For each method of measuring distances between trees, we constructed a null distribution on tree-to-tree distances by taking the parameter-based tree and randomly reshuffling the species names. The distances between the randomized null-parameter tree and the DNA tree were then calculated. This procedure was repeated 10,000 times to produce the null distribution.
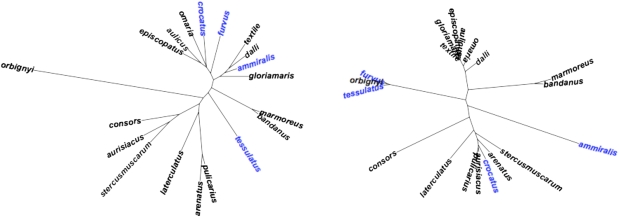
The trees are compared in Fig. 5. Despite several dissimilarities between the DNA- and parameter-based trees, the observed distance between the trees is much less than expected under the null hypothesis of only random similarity between the trees (SI Appendix, Fig. S7). The differences are statistically significant—P = 0.0146 for the topology-based distance measure and P = 0.0001 the branch-length-based distance measure—indicating that the observed distance was smaller than all the 10,000 null distances generated. We conclude that there is a phylogenetic signal in the parameter values, despite the fact that they do not perfectly reflect the phylogenetic relationships of the group.
Fig. 5.
Comparison of the DNA-based phylogeny of cone snails (Left, after Nam et al. (3), unrooted for display) and the parameter-based tree (Right, present study). Species labeled in blue exhibit major changes in topological position in the parameter-based tree. The observed tree-to-tree distances are significantly shorter than expected under a null hypothesis of random similarity (SI Appendix, Fig. S7).
Similarity of DNA- and Parameter-Based Trees.
Looking more closely at the parameter and DNA trees, we can see there is broad similarity but with notable exceptions. In both trees, there are two large clades, called arbitrarily clade 1 (C. stercusmuscarum, C. aurisiacus, Conus pulicarius, Conus arenatus, and C. laterculatus) and clade 2 (C. gloriamaris, C. dalli, C. textile, Conus omaria, C. episcopatus, and C. aulicus), that are nearly the same in both trees, although the detailed branching order differs slightly. In addition, Conus bandanus and Conus marmoreus are sister groups in both trees. There are some conspicuous differences, however. Most notably, Conus furvus, C. tessulatus, and C. orbignyi form a tight clade in the parameter tree yet are widely separated in the DNA tree. In fact, in the DNA tree, C. orbignyi is a well supported out-group to the other 18 species. C. ammiralis is part of clade 2 on the DNA tree but is quite separate on the parameter tree. C. crocatus is in clade 2 on the DNA tree and in clade 1 on the parameter tree (Fig. 5).
The overall similarity of the DNA-based and parameter-based trees is consistent with the hypothesis that the parameters of the developmental model evolved sufficiently slowly that sets of parameters in closely related species are similar. However, there are some exceptional lineages on which more rapid evolution of parameters seems to have occurred. The three species C. furvus, C. tessulatus, and C. orbignyi appear to have converged not only in pattern but in the developmental process that produces that pattern. C. crocatus appears to have shifted its pattern to become similar to species in clade 2, and both C. ammiralis and C. consors have undergone relatively rapid evolution that resulted in quite distinct patterns. The apparently higher rate of parameter evolution on these lineages is consistent with the action of natural selection either directly on pigmentation pattern or indirectly as a correlated response to selection on physiological processes that affect parameter values. In the absence of knowledge of the physiological basis of parameter values, we have no way to directly test for natural selection.
Parametric and nonparametric tests of the Brownian motion model.
The estimation of parameter values for ancestral species in the phylogeny is most easily done if the Brownian motion model of continuous trait evolution can be used. Therefore, when we had established that detectable phylogenetic signal existed in the neural network parameters, we conducted a series of tests to assess the utility of Brownian motion versus other models for modeling the evolution of neural network parameters, as recommended by Blomberg et al. (14). We concluded that Brownian motion was an overall reasonable first approximation for the evolution of neural network parameters (SI Appendix, Supplement B).
Discrete Characters.
Hidden Networks Treated as Discrete Characters.
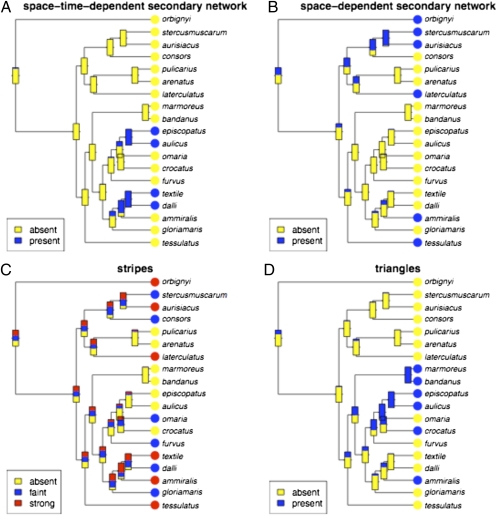
We can treat the presence or absence of a hidden neural network as a binary discrete character. Then, the presence or absence of this character can be mapped onto the phylogeny by using parsimony and maximum-likelihood reconstruction for discrete characters. The two methods give identical results. The presence of hidden networks was restricted to small subclades of the full clade. The presence/absence of hidden networks (Fig. 6 A and B show the presence of a space–time-dependent hidden network and space-dependent hidden network, respectively) showed strong phylogenetic clustering. Relatively few transitions from simple models (i.e., no hidden networks) to complex models (i.e., containing a hidden network) were needed for either character. For the space–time-dependent network, species in two small clades (C. episcopatus/C. aulicus and C. textile/C. dalli) are complex. For the space-dependent hidden network, a complex pattern is more dispersed in the phylogeny.
Fig. 6.
Maximum-likelihood estimates of selected discrete characters. The relative simplicity of the inferred evolution of pattern complexity is in striking contrast to what can be inferred about the evolution of specific features of the patterns when they are described as discrete characters, as illustrated in A–D.
Discrete phenotypic characters.
Other discrete characters were also mapped for comparison with the results for hidden networks. We mapped several discrete phenotypic characters on the phylogeny (SI Appendix, Supplement B). Cone shape is fairly scattered but shows some uniformity in small clades. Strikingly, prey preference shows extremely high conservation [as was clear in the discussion of Nam et al. (3)] compared with shell pattern characters. Each major clade is almost completely restricted to a certain prey, and the entire pattern is explained by the minimum possible number of transitions.
Fig. 6 shows the distributions of stripes and triangles in this group and the maximum-likelihood assignment of ancestral states. The presence and absence of stripes, in particular, is scattered throughout the phylogeny, indicating that they are evolutionarily labile, although triangle presence/absence shows some correlation with large clades. These observations are confirmed by standard parsimony statistics and their comparison with randomized-tip null models; presence/absence of stripes, despite these being visually striking patterns used in identification, appear to lack significant phylogenetic signal in that they do not show significantly more congruence with the phylogeny than is expected under the null model in which character states have been randomly shuffled among the phylogeny tips.
Inference of Ancestral Shell Patterns.
We used a Brownian motion model to estimate parameter values in the species ancestral to the living species. We then ran the neural-network model with these estimated parameter values to predict the pigmentation patterns in the ancestral species. Those patterns are shown at the nodes in Fig. 4. Ancestral states for each parameter common to all species were estimated by using maximum-likelihood estimation on the tree inferred from DNA sequences, modeling the evolution of each parameter as an independent Brownian motion process (15, 16). Two other available methods—generalized least-squares and phylogenetically independent contrasts—gave similar estimates.
For the additional parameters used in the hidden networks, ancestral character estimation was performed as follows. Phylogenetically independent contrasts were applied to reconstruct the ancestral states of the hidden networks because it works from the tips downward, and so, unlike maximum likelihood, can be used when parameters for hidden networks are not available in the rest of the clade.
The ancestral shell patterns are shown in Fig. 4. Each estimate has an associated variance and confidence intervals. To test the robustness of the ancestral patterns to uncertainty in parameter estimates, we randomly generated sets of parameters from the distribution of each parameter and generated ancestral patterns from each set. We found that some ancestral patterns are quite robust to uncertainty in estimated parameters whereas others are not. Fig. 7 shows two examples of each kind. The ancestral patterns for nodes 25 and 29 are quite similar for different sets of estimated parameters, whereas those for nodes 27 and 31 differ greatly among sets of estimated parameters, although various detailed similarities can still be detected even among these shells because of the underlying similarity of neural network parameters.
Fig. 7.
Possible realizations of ancestors. For each neural network parameter at each node, five values were drawn from their estimated distribution by using the predicted uncertainty around the maximum-likelihood estimate. Each set of parameter values for a node was then input into the neural-network model to produce a depiction of the possible ancestor. Some shell patterns appear to occupy larger regions of parameter space, and are thus more robust to perturbation. However, similarities can still be discerned even in shells that appear quite different at first glance.
Discussion and Conclusions
We have taken a step in applying modern phylogenetic methods to understanding the development of complex phenotypic characters. The pigmentation patterns of Conus shells can be generated by a neural-network model that has a sound anatomical and physiological basis. The model parameters fitted to observed patterns show a substantial phylogenetic signal, indicating that the processes governing evolutionary change in shell patterns are, to some extent, gradual across the phylogeny. Our analyses have allowed us to estimate the shell pigmentation patterns of ancestral species, identify lineages in which one or more parameters have evolved rapidly, and measure the degree to which different parameters correlate with the phylogeny.
Our results are summarized in Fig. 4. This figure shows that pigmentation patterns in living species are well approximated by the neural-network model presented in this study. It also shows the inferred ancestral shell patterns. Often, recent ancestors of sister species show recognizable similarity to the pigmentation patterns in living species (e.g., nodes 26–27 and 31–32). Nodes more remote from the present often show ancestors that are generally similar to the living species (nodes 21, 22, 24, and 33). However, some ancestors are strikingly different from any of the living species in the group we analyzed. Interestingly, such patterns can be found in other living species. For example, the strong striping perpendicular to the axis of coiling of the shell found in node 37 is quite similar to that of Conus hirasei, Conus papuensis, or Conus mucronatus (17). Striping parallel to the axis of coiling of the shell, observed in other estimated ancestors, can also be found in living species, for example in some specimens of Conus hyaena and Conus generalis (ref. 17, pp. 354 and 392).
A unique feature of our results is that the inferred pigmentation patterns of ancestors may be quite different from the patterns of their descendants. The patterns generated by the neural-network model are not necessarily smooth functions of the parameter values. Instead, they can vary discontinuously when parameter values move into a different bifurcation region that produces qualitatively different patterns. The role of bifurcation boundaries in evolution was recognized in earlier studies of limb morphogenesis (18, 19). This feature of our results is quite different from what is usually found when inferring ancestral states of continuously variable characters. A well known limitation of methods for estimating ancestral states is that it is impossible for estimates to fall outside the range of the living species analyzed. This limitation does not apply to pigmentation patterns. Although the same averaging procedure is being used on each parameter of the neural-network model, it is possible, and even likely, that a set of estimated parameters will be in a region of parameter space not inhabited by any living species. In addition, the sensitivity of the neural network to perturbations means that small, gradual evolutionary shifts in one or a few parameters of the neural network can shift a shell from one pattern regime into an entirely dissimilar one.
We have necessarily made simplifying assumptions in our analysis to illustrate the overall logic of our method in a straightforward way. Although the DNA-based tree used in this study has strong statistical support, an important assumption is that the branch lengths inferred from the DNA sequence data are known without error, and that they have been accurately renormalized to an absolute time scale. A more formal analysis would begin with the raw DNA sequence alignment and fossil calibration points, and then integrate ancestral state estimates and parameters of evolutionary models, over the space of data-supported chronogram phylogenies (20).
A second assumption is that the set of parameter values for each species is unique and estimated without error. Given the number of parameters involved, a formal proof of uniqueness seems impossible; however, extensive experience with the numerical properties of the model suggests that each pattern is determined by a unique optimal (in the sense of a best fit to the observed pattern) set of parameters.
A third assumption is that the parameters evolved independently of one another on the phylogeny. That assumption is largely supported by our analysis of phylogenetically independent contrasts. Correlation in parameters could be accounted for by using a model of correlated Brownian motion on the phylogeny, but such a model was not needed for our analysis.
In estimating parameters of ancestral species and predicting their pigmentation patterns, we have not taken into account the range of parameters consistent with estimated values for living species. Parameter values estimated by using maximum likelihood and a Brownian motion model have associated confidence intervals that could make more than one qualitatively different pigmentation pattern for each ancestral species consistent with patterns in living species. Application of our method to a group of cone snails with a detailed fossil record—for example, those in southeastern North America (21)—might allow a more rigorous assessment of the accuracy of these techniques, and of what degree of uncertainty should be assigned to them. Usefully and remarkably, shell pigmentation patterns in fossil Conus can be visualized under UV light (21). Application of this technique to Conus fossils could provide a partial validation of our predicted ancestral patterns.
Our analysis is somewhat similar to that of Allen et al. (11), who examined spotted patterns in felids by using a morphogen-diffusion model of pattern formation. Allen et al. showed that there is little phylogenetic signal in the model parameters, indicating that spotting patterns in felids evolve convergently under ecological influences. One difference between their study (11) and the present one is that we found phylogenetic signal in most of the neural network parameters that produce shell pigmentation patterns. This allowed us to infer ancestral patterns and to identify lineages in which relatively rapid evolution of some parameters have taken place.
We found phylogenetic signal in the continuous parameters of the primary neural network and in the presence/absence of a hidden network, suggesting that the model reasonably approximates the developmental processes underlying pigmentation patterns in the Conus species we considered. In contrast, various features of the pigmentation patterns, such as the presence of stripes and dots, do not have significant phylogenetic signal (SI Appendix, Tables S2–S4). This is in agreement with the conclusion of Hendricks.*
Supplementary Material
Acknowledgments
The authors thank David Jablonski, John Huelsenbeck, Alan Kohn, Jonathan Hendricks, and Carole Hickman. We also acknowledge Hans Meinhardt for his encyclopedic treatment of morphogen-based lateral inhibition models, many of which provided the inspiration for our neural net models. N.J.M. was supported by National Science Foundation (NSF) Grant DEB-0919451, a Wang Fellowship, and a Tien Fellowship. J.E.V. and M.S. were supported in part by National Institutes of Health Grant R01-GM40282. G.O. was supported by NSF Grant DMS 0414039. B.E. was supported by NSF Grant DMS081713.
Footnotes
The authors declare no conflict of interest.
Data deposition: The computational parameters for the pattern formation model for each of the described species are available upon request.
*Hendricks JR, Geological Society of America Annual Meeting, November 2–5, 2003, Seattle, WA.
See Author Summary on page 1367.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119859109/-/DCSupplemental.
References
- 1.Campbell J, Ermentrout B, Oster G. A model for mollusk shell patterns based on neural activity. Veliger. 1986;28:369–388. [Google Scholar]
- 2.Boettiger A, Ermentrout B, Oster G. The neural origins of shell structure and pattern in aquatic mollusks. Proc Natl Acad Sci USA. 2009;106:6837–6842. doi: 10.1073/pnas.0810311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nam HH, Corneli PS, Watkins M, Olivera B, Bandyopadhyay P. Multiple genes elucidate the evolution of venomous snail-hunting Conus species. Mol Phylogenet Evol. 2009;53:645–652. doi: 10.1016/j.ympev.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Ferrante M, Migliore M, Ascoli GA. Feed-forward inhibition as a buffer of the neuronal input-output relation. Proc Natl Acad Sci USA. 2009;106:18004–18009. doi: 10.1073/pnas.0904784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermentrout B, Terman D. Mathematical Foundations of Neuroscience. New York: Springer; 2010. [Google Scholar]
- 6.Gutkin B, Pinto D, Ermentrout B. Mathematical neuroscience: From neurons to circuits to systems. J Physiol Paris. 2003;97:209–219. doi: 10.1016/j.jphysparis.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Kang K, Shelley M, Sompolinsky H. Mexican hats and pinwheels in visual cortex. Proc Natl Acad Sci USA. 2003;100:2848–2853. doi: 10.1073/pnas.0138051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meinhardt H, Klingler M. A model for pattern formation on the shells of molluscs. J Theor Biol. 1987;126:63–89. [Google Scholar]
- 9.Meinhardt H, Prusinkiewicz P, Fowler DR. The Algorithmic Beauty of Sea Shells. Berlin: Springer; 2003. [Google Scholar]
- 10.Murray JD. Mathematical Biology. 3rd Ed. New York: Springer; 2002. [Google Scholar]
- 11.Allen WL, Cuthill IC, Scott-Samuel NE, Baddeley R. Why the leopard got its spots: Relating pattern development to ecology in felids. Proc Biol Sci. 2010;278:1373–1380. doi: 10.1098/rspb.2010.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmon LJ, et al. Early bursts of body size and shape evolution are rare in comparative data. Evolution. 2010;64:2385–2396. doi: 10.1111/j.1558-5646.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 13.Maddison WP, Slatkin M. Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution. 1991;45:1184–1197. doi: 10.1111/j.1558-5646.1991.tb04385.x. [DOI] [PubMed] [Google Scholar]
- 14.Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 15.Paradis E. Analysis of Phylogenetics and Evolution with R. Use R! Series. Vol xii. New York: Springer; 2006. [Google Scholar]
- 16.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 17.Röckel D, Korn W, Kohn AJ. Manual of the Living Conidae: Indo-Pacific Region. Vol 1. Wiesbaden, Germany: Christa Hemmen; 1995. [Google Scholar]
- 18.Oster G, Alberch P, Murray J, Shubin N. Evolution and morphogenetic rules. The shape of the vertebrate limb in ontogeny and phylogeny. Evolution. 1988;42:862–884. doi: 10.1111/j.1558-5646.1988.tb02508.x. [DOI] [PubMed] [Google Scholar]
- 19.Oster G, Alberch P. Evolution and bifurcation of developmental programs. Evolution. 1982;36:444–459. doi: 10.1111/j.1558-5646.1982.tb05066.x. [DOI] [PubMed] [Google Scholar]
- 20.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendricks JR. The genus Conus (Mollusca: Neogastropoda) in the Plio-Pleistocene of the southeastern United States. Bull Am Paleontol. 2008;(375):1–180. [Google Scholar]