Abstract
ATP8A2 is a P4-ATPase (“flippase”) located in membranes of retinal photoreceptors, brain cells, and testis, where it mediates transport of aminophospholipids toward the cytoplasmic leaflet. It has long been an enigma whether the mechanism of P4-ATPases resembles that of the well-characterized cation-transporting P-type ATPases, and it is unknown whether the flippases interact directly with the lipid and with counterions. Our results demonstrate that ATP8A2 forms a phosphoenzyme intermediate at the conserved aspartate (Asp416) in the P-type ATPase signature sequence and exists in E1P and E2P forms similar to the archetypical P-type ATPases. Using the properties of the phosphoenzyme, the partial reaction steps of the transport cycle were examined, and the roles of conserved residues Asp196, Glu198, Lys873, and Asn874 in the transport mechanism were elucidated. The former two residues in the A-domain T/D-G-E-S/T motif are involved in catalysis of E2P dephosphorylation, the glutamate being essential. Transported aminophospholipids activate the dephosphorylation similar to K+ activation of dephosphorylation in Na+,K+-ATPase. Lys873 mutants (particularly K873A and K873E) display a markedly reduced sensitivity to aminophospholipids. Hence, Lys873, located in transmembrane segment M5 at a “hot spot” for cation binding in Ca2+-ATPase and Na+,K+-ATPase, appears to participate directly in aminophospholipid binding or to mediate a crucial interaction within the ATP8A2-CDC50 complex. By contrast, Lys865 is unimportant for aminophospholipid sensitivity. Binding of Na+, H+, K+, Cl-, or Ca2+ to the E1 form as a counterion is not required for activation of phosphorylation from ATP. Therefore, phospholipids could be the only substrate transported by ATP8A2.
Keywords: lipid transport mechanism, mutagenesis, phospholipid flippase, phosphatidylserine, membrane asymmetry
P-type ATPases are a large family of membrane pumps believed to be transiently phosphorylated at the conserved aspartate residue of the DKTGT motif during the catalytic cycle. An important challenge is to understand how these proteins couple the utilization of ATP with transport of substances across the membrane. Among the most well-characterized P-type ATPases are the sarcoplasmic reticulum Ca2+-ATPase and the Na+,K+-ATPase. During the reaction cycle of these ATPases, the intermediates E1, E1P, E2P, and E2 (P representing phosphorylation) are formed sequentially, involving large movements of the three cytoplasmic domains, N (nucleotide binding), P (phosphorylation), and A (“actuator”), that are coupled through a linker region with the ion translocation occurring in the membrane domain M (1–3). For many other P-type ATPases, much less is known regarding the transport mechanism due to lack of structural and biochemical data, and even the existence of a phosphorylated intermediate is a conjecture based on sequence homology.
P4-ATPases constitute a subfamily of P-type ATPases implicated in the transport of aminophospholipids (4, 5). Using ATP as an energy source, the P4-ATPases “flip” aminophospholipids from the exoplasmic to the cytoplasmic leaflet of biological membranes, thus determining the curvature of the phospholipid bilayer (6, 7). In humans, there are 14 members of this family, which are expressed in various cell types (8). Mutations in many of these ATPases are linked to severe human disorders (8–11). The amino acid sequence homology suggests that the overall structure and domain topology of P4-ATPases is similar to that of the catalytic subunits of the Ca2+-ATPase and Na+,K+-ATPase, consisting of cytoplasmic N, P, and A domains and a membrane domain made up of ten transmembrane helices M1–M10. However, there is little information available on the actual mechanism of phospholipid transport. How is ATP hydrolysis coupled with the flipping of lipids? Do flippases form a phosphoenzyme existing in two major conformations, E1P and E2P? Is lipid flipping toward the cytoplasmic leaflet associated with dephosphorylation of E2P, like the transport of K+ from the exoplasmic to the cytoplasmic side by the Na+,K+-ATPase? Is the formation of the phosphoenzyme of flippases activated by a specific substrate being transported, as established for the phosphoenzyme intermediates of Ca2+-ATPase and Na+,K+-ATPase, which depend on the binding of Ca2+ and Na+, respectively?
ATP8A2 is a member of the mammalian P4-ATPase family, and we have shown that it transports the lipids phosphatidylserine (PS) and phosphatidylethanolamine (PE), but not phosphatidylcholine (PC), from the luminal to the cytoplasmic surface of photoreceptor outer segment discs (12). ATP8A2 exists in complex with its “β-subunit” CDC50A in photoreceptors, which is essential for function (12, 13). ATP8A2 is expressed in the retina, throughout the brain, as well as in testis, and mutations in ATP8A2 have recently been shown to cause severe mental retardation and other neurological problems in humans (9). The expression and purification of ATP8A2 (4) allows detailed mechanistic studies of this flippase, and here we have characterized the phosphoenzyme and its dependence on ions and the transported lipids. We have studied the consequences of key mutations in ATP8A2 located in the A, P, and M domains, and we have identified Lys873 of putative transmembrane segment M5 as an essential residue involved in PS-activated dephosphorylation, possibly as a PS interacting residue.
Results
ATP8A2 Phosphoenzyme.
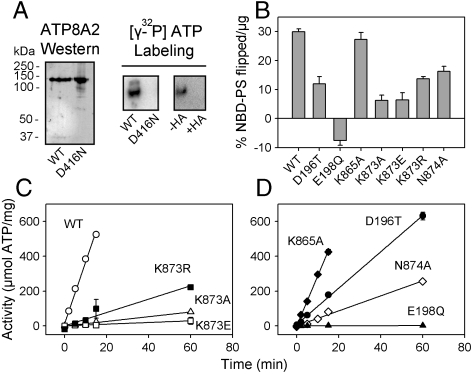
In the presence of [γ-32P]ATP and Mg2+, purified ATP8A2 reconstituted into PC lipid vesicles formed an acid-stable phosphorylated intermediate detected as an intensely 32P-labeled band by phosphorimaging following SDS-PAGE at pH 6.0 (Fig. 1A, Center). Incubation with hydroxylamine prior to SDS-PAGE abolished labeling, indicating that the phosphoenzyme intermediate is an acyl phosphate (Fig. 1A, Right). The apparent Mg2+ affinity for activation of phosphorylation was similar to that of Na+,K+-ATPase (Fig. S1A). To verify that the phosphorylation site of ATP8A2 is Asp416 in the DKTGT P-type ATPase signature sequence, we carried out a similar phosphorylation analysis of the mutant D416N. D416N was expressed at a 10-fold lower level compared to wild type. Nevertheless, when using an amount of D416N similar to wild type, no 32P incorporation was observed (Fig. 1A, Left and Center).
Fig. 1.
ATP8A2 phosphoenzyme and overall function of wild-type ATP8A2 and mutants. (A) Western blot of PC reconstituted wild type and D416N mutant with ATP8A2-specific antibody and analysis of 32P-labeling by SDS-PAGE at pH 6.0 following treatment of [γ-32P]ATP-incubated enzyme with (+) or without (−) hydroxylamine (HA). (B) Transport of NBD-PS measured using the fluorescence-dithionite assay. A positive value for NBD-PS transport indicates transport from the exoplasmic to the cytoplasmic leaflet, whereas a negative value indicates transport in the opposite direction (see further in Fig. S3B). (C and D) ATPase activity of 90PC:10PS reconstituted enzyme. In B, C, and D, the activity is calculated relative to the amount of ATP8A2 protein present. See additional information including statistical analysis in Table S1.
Expression and Overall Function of Wild-Type ATP8A2 and the DGET and M5 Mutants.
We examined the functional consequences of ATP8A2 mutations D196T, E198Q, K865A, K873A, K873E, K873R, and N874A. Because the glutamate of the A-domain motif TGES in Ca2+-ATPase is a key residue in catalysis of dephosphorylation of the E2P phosphoenzyme intermediate (1, 14), we selected the corresponding ATP8A2 glutamate for analysis by replacement with glutamine (E198Q). In all P4-ATPases, the residue corresponding to threonine in the TGES motif is an aspartate (DGET motif, see Fig. S2). To study the functional impact of this characteristic feature we analyzed the mutant D196T. Furthermore, we selected the highly conserved Lys873 and Asn874 in the middle of M5 for analysis, because M5 of Ca2+-ATPase and Na+,K+-ATPase is pivotal in the ion translocating events (1–3, 15–18). Lys873 was replaced with alanine, arginine, and glutamate (K873A, K873R, K873E), and Asn874 was replaced with alanine (N874A). For comparison, Lys865 located near the cytoplasmic border of M5 was also mutated (K865A). These mutants were generally expressed at levels rather similar to the wild type (Fig. S3A). Each mutant and the wild type were purified and reconstituted into vesicles consisting of either PC or a mixture of 90% PC and 10% PS (“90PC:10PS”). Both wild type and mutants were largely free of any contaminant proteins, and the PC and 90PC:10PS preparations contained roughly the same concentration of ATP8A2 (Fig. S3A). Comparable levels of CDC50A copurified with wild type and mutants. Previous studies have indicated that ATP8A2 reconstituted by this method is oriented such that 70% is accessible to trypsin digestion and ATP (ATP binding domain facing outward) (4). Hence, in assays of the reconstituted vesicles 70% of the enzyme is expected to be active, and the outer leaflet corresponds functionally to the cytoplasmic leaflet of the cell membrane.
Reconstitution was also carried out in PC together with NBD-labeled PS (NBD-PS, 1-oleoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoserine), which allowed for the determination of flippase activity using fluorescence spectroscopy (Fig. 1B and Table S1). ATP or the nonhydrolyzable AMPPNP was added to the reconstituted enzyme, and flipping of NBD-PS was allowed to occur for 2.5 min at 23 °C, followed by dilution to stop the ATP utilization and addition of dithionite to bleach NBD-PS in the outer leaflet, thereby allowing a precise determination of % NBD-PS contained in the inner leaflet (4). The difference in inner leaflet % NBD-PS for AMPPNP versus ATP represents the ATP energized transport to the outer leaflet corresponding to the cytoplasmic side (see examples of traces in Fig. S3B). D196T, K873R, and N874A were found to transport NBD-PS at about one-third to one-half of the level transported by wild type, and K873A and K873E about one-fifth of wild type. K865A transported NBD-PS at a similar level as wild type. E198Q seemed to catalyze a small net transport of NBD-PS in the opposite direction, from the outer to the inner leaflet (Fig. 1B and Fig. S3B, see further explanation in legend to Fig. S3).
ATP8A2 wild type and mutants reconstituted in 90PC:10PS were assayed for ATPase activity at 37 °C. As seen in Fig. 1 C and D, ATP utilization was linear over time. Compared with wild type, K873A and K873E displayed approximately 30- and 70-fold lower specific ATPase activity (relative to amount of ATP8A2 protein present), respectively. D196T was reduced by 3-fold, whereas K873R and N874A were reduced by 10-fold. K865A exhibited ATPase activity similar to wild type. ATPase activity was not detectable for E198Q or D416N (Fig. 1D and Table S1).
Dephosphorylation and Interaction with Aminophospholipids.
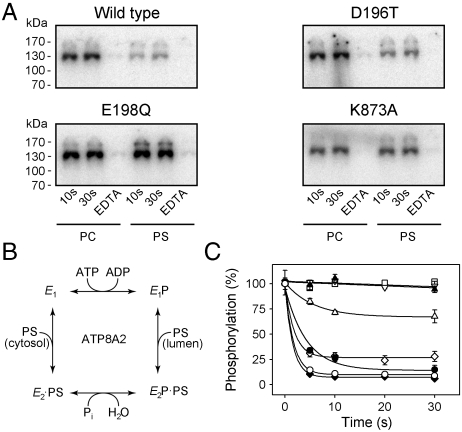
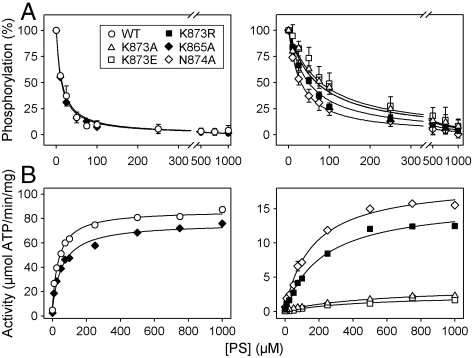
Like wild-type ATP8A2, the DGET and M5 mutants formed a phosphoenzyme from [γ-32P]ATP (see examples of phosphorylation gels in Fig. 2A). The phosphorylation stoichiometry was for most mutants rather similar to that of the wild type (Table S2). The apparent affinity for ATP determined for a selected group of mutants was also wild type-like (Fig. S1B, K0.5 ∼ 0.1 μM, where K0.5 is the ligand concentration giving half maximal effect). Following chelation of Mg2+ with EDTA, no phosphorylation was observed (Fig. 2A). Interestingly, E198Q and K873A exhibited phosphorylation levels in 90PC:10PS of approximately the same magnitude as in PC, whereas the wild type and D196T displayed a much lower steady-state level of phosphorylation in 90PC:10PS than in PC (Fig. 2A), as did native ATP8A2 purified from photoreceptor outer segments (Fig. S3C). This consequence of the presence of PS can be explained by assuming that PS activates the dephosphorylation in analogy with the K+-induced dephosphorylation of Na+,K+-ATPase, as illustrated in the reaction scheme in Fig. 2B. To further address this issue, PC reconstituted wild type and mutants solubilized in CHAPS detergent (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid) were phosphorylated followed by addition of PS (Fig. 2C). In control experiments in which PC was added instead of PS, the phosphoenzyme levels remained almost maximal during 30 s for wild type as well as mutants. Upon addition of PS, the wild type dephosphorylated rapidly, reaching a level less than 10% of the initial level in PC within 5 s. D196T dephosphorylated somewhat slower than wild type, whereas K865A was indistinguishable from wild type. N874A dephosphorylated to a level close to 25% that of the wild type. In contrast, the level of phosphoenzyme remained constant at 100% for E198Q even after 30 s incubation with PS, and K873A was only weakly reactive to PS, displaying relatively slow kinetics of dephosphorylation, reaching a constant phosphoenzyme level as high as 70%. This led us to employ the phosphorylation assay to determine the apparent affinity for PS as well as PE by measuring the dephosphorylation at various concentrations of the added aminophospholipid (Fig. 3A, Fig. S1C and D, and Table S2). The wild type exhibited a K0.5 of approximately 12 μM for PS and approximately 140 μM for PE, in agreement with the previously demonstrated selectivity for PS versus PE (4, 12). K865A was rather similar to wild type. Importantly, K873A and K873E displayed marked reductions (7- and 8-fold, respectively) of the apparent affinity for PS, whereas the PS affinities of K873R and N874A were more moderately reduced (4- and 3-fold, respectively). Using the ATPase activity assay, K873A and K873E also displayed a conspicuous 13- to 16-fold reduction of the apparent affinity for activation by PS, and these mutants were completely insensitive to PE within the concentration range tested. K873R and N874A displayed an intermediate reduction in apparent affinity for PS, and little or no ATPase activation by PE was observed. K865A was again wild type-like (Fig. 3B, Fig. S1D, and Table S2). Similar studies with D196T showed a wild type-like or even enhanced apparent affinity for PS (Fig. S1C). These results indicate that Lys873 could be a critical lipid interacting residue and that the positive charge of the side chain is important.
Fig. 2.
Dephosphorylation induced by phosphatidylserine. (A) Wild-type ATP8A2 and mutants reconstituted in PC or 90PC:10PS (“PS”) were phosphorylated for 10 s or 30 s with [γ-32P]ATP in SPM (for background, EDTA was added). Equal amounts of protein reconstituted in PC and 90PC:10PS were loaded on the gel. (B) Proposed reaction scheme showing various enzyme intermediates and their interaction with nucleotides and phosphatidylserine (PS). (C) Following 10 s phosphorylation of PC reconstituted enzyme with [γ-32P]ATP in SPM containing CHAPS, PS dissolved in CHAPS was added at a final concentration of 55 μM, and quenching was performed at the indicated time intervals for wild type (open circles), D196T (filled circles), E198Q (filled triangle), K865A (filled diamond), K873A (open triangle pointing up), and N874A (open diamond). The time course with PC added instead of PS is shown for wild type (open triangle pointing down) and K873A (open square).
Fig. 3.
Apparent affinities of wild type and mutants for PS. (A) Following phosphorylation of PC reconstituted enzyme at 0 °C with [γ-32P]ATP in SPM containing CHAPS, PS dissolved in CHAPS was added at the indicated concentrations, and dephosphorylation was terminated 5 s later. K0.5 values for activation by PS (apparent PS affinities) were as follows: Wild type (12 μM), K865A (11 μM), K873A (88 μM), K873E (99 μM), K873R (46 μM), N874A (31 μM). (B) ATPase activity of wild type and mutants in the presence of CHAPS and PC with the indicated concentrations of PS. K0.5 values for activation by PS were as follows: Wild type (38 μM), K865A (58 μM), K873A (600 μM), K873E (490 μM), K873R (230 μM), N874A (170 μM). Refer to Table S2 for statistical analysis.
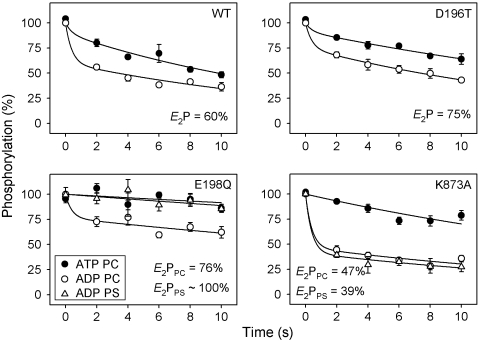
Distribution of the Phosphoenzyme Between E1P and E2P Forms.
For Ca2+-ATPase and Na+,K+-ATPase, it is well known that the phosphoenzyme resides in two major conformational states, E1P and E2P. E1P is the phosphoenzyme intermediate formed by phosphorylation with ATP and is able to donate the phosphoryl group back to ADP. Following the conformational change to E2P, the phosphoenzyme intermediate is insensitive to ADP, and dephosphorylation can only occur by reaction with water, liberating Pi. The structural basis is that in E2P the TGES motif becomes inserted in the catalytic site, thereby partly occupying the space previously taken up by ADP in the E1P form (1). To examine whether the phosphoenzyme of ATP8A2 exists in similar ADP-sensitive and -insensitive states, the dephosphorylation upon addition of either ATP or ADP to phosphoenzyme formed in the presence of PC was followed for selected mutants (Fig. 4, circles). Two exponential decay phases could be readily distinguished from the ADP dephosphorylation time course, a rapid component corresponding to the reaction of accumulated E1P with ADP and a slower phase, which may represent the hydrolysis of E2P and/or back conversion of E2P to E1P followed by reaction with ADP. The initial steady-state E1P and E2P fractions were estimated by fitting a biexponential decay function to the data (Fig. 4 and Table S1). For wild type, the phosphoenzyme distribution corresponded to 60% E2P, whereas somewhat more E2P was accumulated for D196T and E198Q but less for K873A. For wild type and D196T in 90PC:10PS, the low steady-state phosphorylation level and rapid PS-induced dephosphorylation precluded a similar determination of the E2P fraction, but for E198Q and K873A in 90PC:10PS this analysis was possible. For E198Q, the E2P fraction of the phosphoenzyme increased to approximately 100% in the presence of PS, whereas for K873A the presence of PS had little effect on the distribution of the phosphoenzyme between E1P and E2P (Fig. 4, triangles).
Fig. 4.
Sensitivity of the phosphoenzyme to ADP. Following phosphorylation with [γ-32P]ATP in SPM at 0 °C, enzyme reconstituted in PC (circles) or 90PC:10PS (triangles) was treated with excess unlabeled ATP (filled symbols) or ADP (open symbols) for the indicated times. The amplitude of the rapid phase of the ADP-induced decay reflects E1P, whereas the slow phase corresponds to E2P (E2P fraction indicated in panels). Refer to Table S1 for statistical analysis.
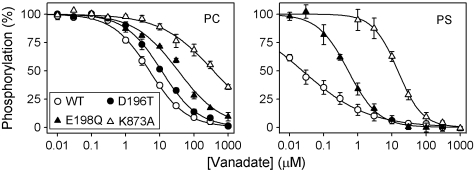
Vanadate Binding.
Vanadate is known as an analog of the penta-coordinated transition state of E2P dephosphorylation in P-type ATPases, which binds specifically to the E2 form (19). Because vanadate is also a potent inhibitor of ATP8A2 (12), we studied the changes in apparent vanadate affinity of ATP8A2 induced by the mutations and binding of PS for selected mutants, using a phosphorylation assay that takes advantage of the competition between vanadate and ATP (see SI Materials and Methods). Wild type and mutants displayed distinct vanadate affinities that depended on the presence of PS (Fig. 5 and Table S1). In PC, the wild type exhibited a K0.5 value for vanadate binding of approximately 5 μM, whereas D196T and E198Q showed approximately 2- and 7-fold lower apparent affinities, respectively. PC reconstituted K873A displayed a marked approximately 80-fold reduction of vanadate affinity relative to wild type. In 90PC:10PS the low phosphorylation levels of wild type and D196T precluded an accurate determination of vanadate affinity by this method, but it was nevertheless clear that a considerable affinity increase occurred as a consequence of the interaction with PS (shown for wild type in Fig. 5, approximately 200-fold affinity increase). For E198Q and K873A, the apparent affinities for vanadate were, respectively, 70- and approximately 25-fold higher than the corresponding affinities determined in the absence of PS. Hence, the presence of PS increased the vanadate affinity to a lesser extent in K873A as compared with E198Q and wild type.
Fig. 5.
Vanadate binding determined by inhibition of phosphorylation from [γ-32P]ATP. Enzyme reconstituted in PC or 90PC:10PS (“PS”) was incubated at 25 °C with the indicated concentration of orthovanadate in SPM containing CHAPS and subsequently phosphorylated with [γ-32P]ATP at 0 °C. The maximal phosphorylation obtained following incubation in the absence of vanadate was taken as 100%. Refer to Table S1 for K0.5 values and statistical analysis.
Dependence of Phosphorylation on Ions.
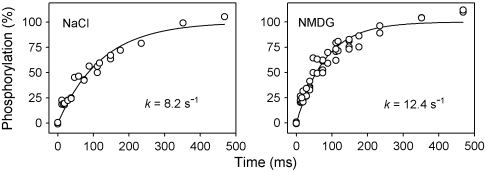
The studies of the phosphoenzyme reported above were carried out in the presence of 150 mM NaCl. In cation-transporting P-type ATPases like Na+,K+-ATPase and Ca2+-ATPase, the ion being translocated from the cytoplasmic side toward the exoplasmic side of the membrane activates the reaction of the E1 state with ATP leading to phosphoenzyme formation. To search for candidate cations or anions required to activate the phosphorylation of ATP8A2, wild-type ATP8A2 reconstituted in PC or 90PC:10PS was dialyzed against medium containing 150 mM N-methyl-D-glucamine (NMDG), a cationic sugar that provides for ionic strength but is unable to substitute for Na+ in the activation of the Na+,K+-ATPase. Although the dialysis reduced the concentrations of Na+ and K+ to less than 25 μM as determined by atomic absorption spectrometry, and Cl- levels were presumably equally low, the phosphorylation rate constant of PC reconstituted ATP8A2 determined in rapid-kinetic measurements was not reduced in the NMDG dialyzed sample (Fig. 6 and Table S3). Moreover, addition of various salts to this sample slightly reduced the steady-state phosphorylation levels in PC and only slightly increased the ATPase activity in 90PC:10PS. Removal of Ca2+ with EGTA had no significant effect either (Fig. S4A).
Fig. 6.
Ionic determinants of phosphorylation. Using a quenched-flow module, rapid kinetics of phosphorylation at 25 °C was studied with expressed ATP8A2 reconstituted in PC in SPM (“NaCl”) or NMDG. All data points are shown. The rate constants extracted from the data are indicated. Refer to Table S3 for statistical analysis.
An obvious candidate for ion countertransport and activation of phosphorylation would be H+. Hence, we examined the dependence of the phosphorylation on pH. There was no significant effect of pH on the phosphorylation rate constant of PC reconstituted enzyme in the range of pH 6.5 to pH 9.0 (Fig. S4B; compare also with pH 7.5 in Fig. 6 and Table S3), and over a broad range of pH values, no large differences were observed in the steady-state levels of phosphoenzyme in both PC and PS (Fig. S4B), indicating that H+ is not required for phosphorylation. This is in agreement with earlier work showing that the ATPase activity remains maximal at pH 9.0 (4).
The phosphorylation rate constants of D196T, E198Q, and K873A determined in PC at pH 7.5 in the presence of 150 mM NaCl were similar to that of the expressed wild-type ATP8A2 and the native ATP8A2 from photoreceptors reconstituted in PC (Fig. S5 and Table S3). The Ca2+-ATPase exhibited a 2-fold higher rate constant in medium of identical composition except for the addition of 100 μM Ca2+. Because of the strong inhibition of dephosphorylation in E198Q, rapid kinetic measurements of phosphorylation could be conducted with E198Q in 90PC:10PS, and the rate constant was found 1.6-fold reduced relative to that of E198Q in PC (Fig. S5 and Table S3).
Discussion
The present findings provide insight into the molecular mechanism of phospholipid transport in P4-ATPases, extending our understanding of ion transport to an important but relatively uncharacterized subfamily of mammalian P-type ATPases.
First of all, our results indicate that the phosphoenzyme intermediate of ATP8A2, like that of Ca2+-ATPase and Na+,K+-ATPase, is formed by phosphorylation of the aspartic acid of the DKTG signature sequence in the P domain and exists in ADP-sensitive E1P and ADP-insensitive E2P forms. Comparison of various crystal structures of the Ca2+-ATPase has revealed that transformation of E1P to E2P and dephosphorylation of E2P is accomplished by a rotation of the A domain, allowing the glutamate of the TGES motif to catalyze hydrolysis of the aspartyl phosphoryl bond by acting as a base extracting a proton from the attacking water molecule in the transition state complex (1, 14). Here we have found that replacement of the corresponding Glu198 in ATP8A2 with glutamine leads to block of E2P dephosphorylation with resulting accumulation of E2P (Figs. 2 and 4), in accordance with a similar mechanism for dephosphorylation in P4-ATPases.
The fact that flipping of PS and to a lesser extent PE is mediated by the purified complex of ATP8A2 with CDC50A provides a clear indication that no other protein is involved (4, 12). The present results demonstrate that PS and PE activate the dephosphorylation of ATP8A2 in analogy with the activating effect of K+ on the dephosphorylation of Na+,K+-ATPase, as depicted in the scheme of Fig. 2B. This is deduced both from the much lower steady-state level of phosphoenzyme seen with expressed or purified native wild-type ATP8A2 in 90PC:10PS as compared with the enzyme in PC, and from the dephosphorylation induced by addition of PS or PE (Figs. 2 and 3, and Fig. S1D). It is therefore conceivable that aminophospholipids interact with the ATP8A2-CDC50A complex in the E2P form to modify the conformation, such that the transition state complex in the dephosphorylation reaction is stabilized and the rate of dephosphorylation thereby enhanced. In accordance with this hypothesis, vanadate was found to bind much stronger in the presence of PS than only in PC (Fig. 5). Because vanadate binds to P-type ATPases in the E2 form and not in E1, a shift of the E1-E2 equilibrium in favor of E2 could also contribute to the increase of apparent affinity for vanadate induced by PS. Both of these mechanisms would be analogous to the effect of binding of K+ to the transport site of the Na+,K+-ATPase, suggesting the existence of a PS binding site on the ATP8A2-CDC50A complex. In support of the scheme in Fig. 2B we also found that for E198Q, PS shifted the distribution of the phosphoenzyme intermediate between E1P and E2P in favor of E2P (Fig. 4).
Our results pinpoint Lys873 as a strong candidate for a residue interacting with the transported aminophospholipids. Mutant K873A displayed substantially reduced ATPase and lipid transport activities. K873A was phosphorylated at a rate similar to wild type (Fig. S5), but displayed significantly slower kinetics of PS-activated dephosphorylation and higher steady-state levels of the phosphoenzyme in PS, relative to wild type (Fig. 2). Importantly, K873A and K873E exhibited a conspicuous reduction of the affinity for PS activation of dephosphorylation and ATPase activity, relative to wild type, and no response to PE within the concentration range tested (Fig. 3, Fig. S1D, and Table S2). K873A furthermore displayed a weaker affinity for vanadate than the wild type and the other mutants in PC as well as in 90PC:10PS, and although the affinity of K873A for vanadate was significantly higher in the presence of PS than in its absence, the difference was not as large as for E198Q and wild type, thus supporting a weaker binding of PS to K873A. In addition, PS did not increase the E2P/E1P ratio of K873A, in contrast to E198Q. Hence, several pieces of evidence indicate that Lys873 is a key player in the functional interactions with PS and PE. Because K873R showed less effect on aminophospholipid sensitivity than K873A, despite the potentially disturbing bulkiness of the arginine side chain, the positive charge of the Lys873 side chain seems important. Mutation of Asn874 also reduced PS affinity moderately, which might be an indirect effect of its proximity to Lys873 or reflect an involvement to some extent of Asn874 in aminophospholipid binding. By contrast, the alanine replacement of Lys865, also located in M5, but nearer the cytoplasmic boundary than Lys873, did not affect the sensitivity to aminophospholipid.
The alignment of amino acid sequences depicted in Fig. S2 reveals that Lys873 and Asn874 of ATP8A2 are located corresponding to the cation binding region in the middle of M5 in Na+,K+-ATPase and Ca2+-ATPase and are rather well conserved among P4-ATPases. Only ATP9A and ATP9B show a semiconservative replacement of the lysine with arginine. The Na+,K+-ATPase residue in the corresponding position is a serine, which contributes its side-chain oxygen to K+ binding (2, 3, 16, 18). In addition, the asparagine (corresponding to Asn874) next to the serine as well as a threonine and a glutamate in M5 also contribute to K+ binding in Na+,K+-ATPase (2, 3, 17, 18). Furthermore, the corresponding asparagine and glutamate of the Ca2+-ATPase provide ligands for Ca2+ binding (1, 15). It is also intriguing that the H+,K+-ATPase has a lysine at the position corresponding to Lys873 of ATP8A2. This lysine seems to help preventing the H+,K+-ATPase from transporting more than one proton and one K+ ion in each ATP hydrolysis cycle (20), in contrast to the 3Na+∶2K+∶1ATP stoichiometry of the Na+,K+-ATPase. In Na+,K+-ATPase, replacement of the corresponding serine with alanine reduces the affinity for extracellular K+ 30-fold, and the kinetics indicate a role as a gating residue (16). Furthermore, the same serine seems to contribute to Na+ binding in the E1 form (18, 21) in accordance with an alternating extracellular and cytoplasmic exposure during the transport cycle. Hence, by analogy it may be speculated that during the transport of aminophospholipid by ATP8A2 the lipid head group is bound from the exoplasmic leaflet to Lys873. Because the Lys873 mutations affected PE interaction with the E2P form of ATP8A2 even more than PS interaction, it is not likely that the lysine binds the carboxylate group specific to PS, but it rather interacts with the head group phosphate. Later in the cycle, following the dephosphorylation, Lys873 might be involved in the ejection of the lipid from ATP8A2 into the cytoplasmic leaflet by repelling the positive charge of the aminophospholipid head group common to both PS and PE. Because the removal of the phospholipid head group from the membrane-water interphase may be energetically more demanding than the movement of the hydrocarbon chains through the membrane, the interaction of the head group with a protein site is likely a crucial part of the flipping mechanism. If the CDC50 subunit plays an essential role in lipid flipping, the alternative to a direct interaction of Lys873 with the lipid head group could be a more indirect engagement in the flipping by tight interaction with CDC50.
Unlike the majority of the P-type ATPase subclasses, the P4-ATPases possess an aspartate at the position corresponding to the threonine of the TGES motif. Although our results show that Asp196 is less crucial for the dephosphorylation of E2P than Glu198, mutation D196T did slow dephosphorylation significantly in the presence of PS (Fig. 2C), leading to reduced ATPase activity (Fig. 1D). In the Ca2+-ATPase and Na+, K+-ATPase structures, the side chain of the threonine apparently stabilizes the TGES loop through a hydrogen bond formed with the backbone nitrogen of the glutamate, and replacement of the threonine with alanine markedly reduces the rate of dephosphorylation in Na+,K+-ATPase (22). The effect observed for the D196T mutation of ATP8A2 is similar although less dramatic, and it is therefore likely that in the P4-ATPases the aspartate side chain contributes to stabilize the catalytic assembly through additional bond formation(s) that might be under long-distance control by PS binding.
Having realized the similarities between the aminophospholipid transport occurring in relation to dephosphorylation of ATP8A2 and the K+ transport by the Na+,K+-ATPase, one may also wonder how much the mechanism of phosphorylation of the E1 form by ATP resembles that of the archetypical cation-transporting P-type ATPases. Apparent ATP and Mg2+ affinities determined in the steady-state phosphorylation experiments were quite similar to the values reported for the Na+,K+-ATPase and Ca2+-ATPase under comparable conditions (cf. Fig. S1 and refs. 23 and 24). The rate constant of phosphorylation was somewhat lower for ATP8A2 (approximately 10 s-1) than the rate constants observed for Na+,K+-ATPase and Ca2+-ATPase (20–30 s-1; cf. Fig. S5 and ref. 22), but the difference is not dramatic. In accordance with the model in which the A domain does not interact functionally with the catalytic site of the P domain until the transformation of E1P to E2P, D196T and E198Q did not significantly affect the rate of phosphorylation by ATP. The less than 2-fold reduction of the phosphorylation rate found for E198Q in 90PC:10PS relative to PC may result from the shift in the E1-E2 equilibrium toward E2 induced by PS binding. A crucial question is whether binding of an ion to the E1 form of the P4-ATPase is required for activation of phosphorylation, but the present data do not support a critical role of Na+, K+, Ca2+, Cl-, or H+ ions. Hence, aminophospholipids could be the only species being transported by ATP8A2.
Concluding Remarks
The present work provides evidence that the basic catalytic assembly of P4-ATPases is similar to that of the cation-transporting P-type ATPases. Phospholipid seems to participate directly in the enzyme cycle by binding to E2P, thereby activating the dephosphorylation, whereas countertransport of ions binding to the E1 form does not seem to be a requirement in ATP8A2 (at least not Na+, K+, Ca2+, Cl-, or H+). The highly conserved Lys873 in M5 is critical for aminophospholipid interaction, possibly contributing to lipid binding during the transport. These studies form a crucial starting point for future work aiming to identify other important residues involved in phospholipid transport.
Materials and Methods
Experimental procedures are described in detail in SI Materials and Methods, and only a brief account is given here. Mutations were introduced into full-length bovine ATP8A2 with 1D4 tag and were verified by sequencing of the entire coding sequence. Wild-type ATP8A2 and mutants were coexpressed with CDC50A in transiently transfected HEK293T cells, purified, and reconstituted into lipid vesicles. The purification of ATP8A2 with its associated CDC50A protein was facilitated by the C-terminal nine amino acid 1D4 tag on ATP8A2 allowing immunoaffinity chromatography using the Rho 1D4 antibody (25). ATP8A2 protein concentrations were calculated based on Coomassie blue staining of SDS-PAGE gels using known amounts of bovine serum albumin as standard. Flippase and ATPase activities were determined as described (4, 12) at 23 °C and 37 °C, respectively. Studies of the phosphoenzyme formed from [γ-32P]ATP were carried out by adopting methods previously introduced for Ca2+-ATPase and Na+,K+-ATPase (14, 22, 26). A Bio-Logic QFM-5 quenched-flow module (Bio-Logic Science Instruments) was applied for rapid kinetic measurements of phosphorylation rate, which were conducted at 25 °C. All other phosphorylation and dephosphorylation experiments were performed at 0 °C using manual mixing or magnetic stirrer. The standard phosphorylation medium (SPM) used unless otherwise indicated consisted of 50 mM HEPES-NaOH (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 2 μM [γ-32P]ATP. When indicated, the concentration of ATP or MgCl2 was varied, or CHAPS was added at a concentration of 10 mM to solubilize protein and lipid. Following termination of the phosphorylation reaction with acid or SDS, SDS-PAGE was performed in 5.8% gels under acidic conditions (26), and the 32P-labeled radioactive band corresponding to the phosphorylated flippase was quantified by phosphorimaging. Some experiments were carried out following removal of alkali metal ions and Cl- from the reconstituted protein by dialysis against three changes of 1 l of NMDG [10 mM HEPES-Tris pH 7.5, 150 mM NMDG-Acetate, 1 mM Mg-Acetate, 1 mM DTT, and 10% (wt/vol) sucrose]. Experiments were generally performed at least twice. The average values are reported, except where it was judged relevant to show all the individual data points (stated in legend). Error bars and “±” indicate SEM values.
Supplementary Material
Acknowledgments.
The authors thank Janne Petersen and Lene Jacobsen, Department of Biomedicine, Aarhus University, for their expert technical assistance. J.A.C. has been a visiting scientist at this department during part of the work. J.A.C. is supported by a National Sciences and Engineering Council predoctoral studentship. This study was funded in part by grants from the Danish Medical Research Council, the Novo Nordisk Foundation (Fabrikant Vilhelm Pedersen og Hustrus Legat), and the Lundbeck Foundation, and grants from the Canadian Institutes for Health Research (MOP-106667) and the National Institutes of Health (EY02422).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108862109/-/DCSupplemental.
References
- 1.Toyoshima C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim Biophys Acta. 2000;1793:941–946. doi: 10.1016/j.bbamcr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 3.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 4.Coleman JA, Kwok MC, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem. 2009;284:32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci USA. 2009;106:16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puts CF, Holthuis JC. Mechanism and significance of P4 ATPase-catalyzed lipid transport: Lessons from a Na+/K+-pump. Biochim Biophys Acta. 2009;1791:603–611. doi: 10.1016/j.bbalip.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Folmer DE, Elferink RP, Paulusma CC. P4 ATPases—Lipid flippases and their role in disease. Biochim Biophys Acta. 2009;1791:628–635. doi: 10.1016/j.bbalip.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Cacciagli P, et al. Disruption of the ATP8A2 gene in a patient with a t(10;13) de novo balanced translocation and a severe neurological phenotype. Eur J Hum Genet. 2010;18:1360–1363. doi: 10.1038/ejhg.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meguro M, et al. A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat Genet. 2001;28:19–20. doi: 10.1038/ng0501-19. [DOI] [PubMed] [Google Scholar]
- 11.Stapelbroek JM, et al. ATP8B1 is essential for maintaining normal hearing. Proc Natl Acad Sci USA. 2009;106:9709–9714. doi: 10.1073/pnas.0807919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman JA, Molday RS. Critical role of the β-subunit CDC50A in the stable expression, assembly, subcellular localization and lipid transport activity of the P4-ATPase ATP8A2. J Biol Chem. 2011;286:17205–17216. doi: 10.1074/jbc.M111.229419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Velden LM, et al. Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J Biol Chem. 2010;285:40088–40096. doi: 10.1074/jbc.M110.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clausen JD, Vilsen B, McIntosh DB, Einholm AP, Andersen JP. Glutamate-183 in the conserved TGES motif of domain A of sarcoplasmic reticulum Ca2+-ATPase assists in catalysis of E2/E2P partial reactions. Proc Natl Acad Sci USA. 2004;101:2776–2781. doi: 10.1073/pnas.0400122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke DM, Loo TW, Inesi G, MacLennan DH. Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Nature. 1989;339:476–478. doi: 10.1038/339476a0. [DOI] [PubMed] [Google Scholar]
- 16.Blostein R, Wilczynska A, Karlish SJ, Arguello JM, Lingrel JB. Evidence that Ser775 in the α subunit of the Na,K-ATPase is a residue in the cation binding pocket. J Biol Chem. 1997;272:24987–24993. doi: 10.1074/jbc.272.40.24987. [DOI] [PubMed] [Google Scholar]
- 17.Vilsen B. Mutant Glu781 → Ala of the rat kidney Na+,K+-ATPase displays low cation affinity and catalyzes ATP hydrolysis at a high rate in the absence of potassium ions. Biochemistry. 1995;34:1455–1463. doi: 10.1021/bi00004a041. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen PA, Nielsen JM, Rasmussen JH, Jorgensen PL. Contribution to Tl+, K+, and Na+ binding of Asn776, Ser775, Thr774, Thr772, and Tyr771 in cytoplasmic part of fifth transmembrane segment in α-subunit of renal Na,K-ATPase. Biochemistry. 1998;37:17818–17827. doi: 10.1021/bi981898w. [DOI] [PubMed] [Google Scholar]
- 19.Cantley LC, Jr, Cantley LG, Josephson L. A characterization of vanadate interactions with the (Na,K)-ATPase Mechanistic and regulatory implications. J Biol Chem. 1978;253:7361–7368. [PubMed] [Google Scholar]
- 20.Burnay M, Crambert G, Kharoubi-Hess S, Geering K, Horisberger JD. Electrogenicity of Na,K- and H,K-ATPase activity and presence of a positively charged amino acid in the fifth transmembrane segment. J Biol Chem. 2003;278:19237–19244. doi: 10.1074/jbc.M300946200. [DOI] [PubMed] [Google Scholar]
- 21.Einholm AP, Toustrup-Jensen MS, Holm R, Andersen JP, Vilsen B. The rapid-onset dystonia parkinsonism mutation D923N of the Na+,K+-ATPase α3 isoform disrupts Na+ interaction at the third Na+ site. J Biol Chem. 2010;285:26245–26254. doi: 10.1074/jbc.M110.123976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toustrup-Jensen M, Vilsen B. Importance of conserved Thr214 in domain A of the Na+,K+-ATPase for stabilization of the phosphoryl transition state complex in E2P dephosphorylation. J Biol Chem. 2003;278:11402–11410. doi: 10.1074/jbc.M212136200. [DOI] [PubMed] [Google Scholar]
- 23.McIntosh DB, Woolley DG, Vilsen B, Andersen JP. Mutagenesis of segment 487Phe-Ser-Arg-Asp-Arg-Lys492 of sarcoplasmic reticulum Ca2+-ATPase produces pumps defective in ATP binding. J Biol Chem. 1996;271:25778–25789. doi: 10.1074/jbc.271.42.25778. [DOI] [PubMed] [Google Scholar]
- 24.Vilsen B. A Glu329 → Gln variant of the α-subunit of the rat kidney Na+,K+-ATPase can sustain active transport of Na+ and K+ and Na+,K+-activated ATP hydrolysis with normal turnover number. FEBS Lett. 1993;333:44–50. doi: 10.1016/0014-5793(93)80372-2. [DOI] [PubMed] [Google Scholar]
- 25.Wong JP, Reboul E, Molday RS, Kast J. A carboxy-terminal affinity tag for the purification and mass spectrometric characterization of integral membrane proteins. J Proteome Res. 2009;8:2388–2396. doi: 10.1021/pr801008c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen JP, Vilsen B, Leberer E, MacLennan DH. Functional consequences of mutations in the β-strand sector of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1989;264:21018–21023. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.