Abstract
Dominant mutations or DNA amplification of tyrosine kinases are rare among the oncogenic alterations implicated in prostate cancer. We demonstrate that castration-resistant prostate cancer (CRPC) in men exhibits increased tyrosine phosphorylation, raising the question of whether enhanced tyrosine kinase activity is observed in prostate cancer in the absence of specific tyrosine kinase mutation or DNA amplification. We generated a mouse model of prostate cancer progression using commonly perturbed non-tyrosine kinase oncogenes and pathways and detected a significant up-regulation of tyrosine phosphorylation at the carcinoma stage. Phosphotyrosine peptide enrichment and quantitative mass spectrometry identified oncogene-specific tyrosine kinase signatures, including activation of EGFR, ephrin type-A receptor 2 (EPHA2), and JAK2. Kinase:substrate relationship analysis of the phosphopeptides also revealed ABL1 and SRC tyrosine kinase activation. The observation of elevated tyrosine kinase signaling in advanced prostate cancer and identification of specific tyrosine kinase pathways from genetically defined tumor models point to unique therapeutic approaches using tyrosine kinase inhibitors for advanced prostate cancer.
Keywords: AKT, androgen receptor, ERG, K-RAS, bioinformatics
The future of effective cancer treatment is based on the emerging concept of personalized therapy, which requires detailed analysis of the oncogenic lesions that drive disease. One prominent oncogenic change seen in many cancers is somatic-activating mutations of tyrosine kinases, including BCR-ABL in chronic myelogenous leukemia (CML), mast/stem cell growth factor receptor (SCFR or KIT) in gastrointestinal stromal tumors (GIST), and EGFR in lung cancer (1–3). The dependency on tyrosine kinase activation in these tumors has led to successful clinical treatment with tyrosine kinase inhibitors (4–6). In prostate cancer, great progress has been made in identifying the genetic determinants of disease progression such as increased expression of androgen receptor (AR) and myelocytomatosis oncogene cellular homolog (MYC), phosphatase and tensin homologue deleted on chromosome 10 (PTEN) deletion, and erythroblast transformation specific (ETS) family gene fusions (7–11). However, recent large-scale cancer genome studies show that activating somatic mutations or DNA amplification of tyrosine kinase genes are rare in prostate cancer (8). This reveals why clinical administration of tyrosine kinase inhibitors for the treatment of advanced prostate cancer has been less effective and strongly implies that a more complete understanding of the tyrosine kinases that contribute to this disease is warranted (12, 13).
Despite the paucity of activating somatic mutations in tyrosine kinases, recent evidence suggests that tyrosine kinase phosphorylation in prostate cancer contributes to disease progression. In androgen-depleted conditions, tyrosine kinase, non-receptor, 2 (TNK2 or ACK1), SRC, and erythroblastic leukemia viral oncogene homolog 2 [ERBB2 (HER-2/neu)] tyrosine kinase activity can restore AR function in prostate cancer cells (14–17). Increased expression of the tyrosine kinase SRC and AR can synergistically drive frank carcinoma of the mouse prostate (18). This relationship results in robust activation of SRC tyrosine kinase and MAPK signaling (18). SRC activity was also observed in a subset of castration-resistant prostate cancer (CRPC) patients, which correlated with lower overall survival and increased metastatic disease (19). These data support the idea that tyrosine kinase activity may play a prominent role in prostate cancer progression in the absence of activating mutations.
Nearly 50% of tyrosine kinases are thought to contribute to human cancers, yet tyrosine phosphorylation represents less than 1% of the phosphoproteome (20). Sensitive and specific methods capable of enriching tyrosine phosphorylated peptides via antibody binding followed by quantitative mass spectrometry (MS) identification has become useful for the elucidation of tyrosine kinase signaling pathways, nodes, and negative feedback mechanisms in different cancer types (21–23). The ability to sensitively characterize pathway alterations in the presence of activated tyrosine kinases or tyrosine kinase inhibitors can allow for the identification of new potential drug targets (21, 24). We use this approach to identify and characterize tyrosine kinase signaling networks in transformed tissues that do not express mutated tyrosine kinases.
Global tyrosine phosphorylation in clinical prostate cancer samples was measured by immunohistochemistry (IHC) and showed a substantial increase in tyrosine phosphorylation in late-stage disease. To study this in a controlled manner, we evaluated tyrosine phosphorylation in a mouse model of prostate cancer progression using oncogenes common to prostate tumorigenesis and observed robust tyrosine phosphorylation in the advanced tumor phenotypes. Unbiased phosphotyrosine proteomics was used to investigate the specific tyrosine kinase signaling pathways activated by each of the nontyrosine kinase oncogenes. Analysis of the tyrosine phosphoproteome of these tumors revealed oncogene-specific tyrosine kinase activation including EGFR, ephrin type A receptor 2 (EPHA2), JAK2, ABL1, and SRC.
Results
Tyrosine Phosphorylation Is Increased in Clinical Castration-Resistant Prostate Cancer Samples.
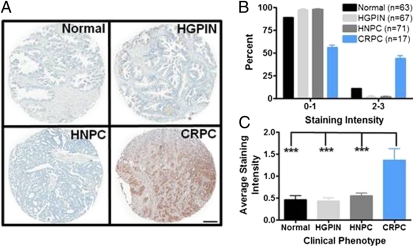
We performed IHC staining of prostate cancer tissue microarrays with the tyrosine phosphorylation-specific antibody 4G10 to evaluate phosphotyrosine expression during disease progression. CRPC (androgen independent) exhibited a robust increase in phosphotyrosine staining intensity compared with benign prostate, the precursor lesion high-grade prostatic intraepithelial neoplasia (HGPIN), or hormone naïve (androgen dependent) prostate cancer (HNPC) (Fig. 1A). Analysis of these tissue microarray samples indicated that 44% of CRPC specimens stain for phosphotyrosine at moderate to high levels (staining intensity 2–3), whereas only 11% of normal, 2% of HGPIN, and 2% of HNPC tissues stain at this intensity (Fig. 1B). Further, the average staining intensity of all of the CRPC tissue samples was significantly increased by over twofold compared with the other clinical phenotypes (Fig. 1C). These data reveal that tyrosine phosphorylation is present and elevated in CRPC and raise the notion that systemic treatment of patients with this disease may induce this response.
Fig. 1.
Robust phosphotyrosine expression is observed in castration-resistant prostate cancer (CRPC) specimens. (A) Representative image of immunohistochemical staining using the phosphotyrosine-specific antibody, 4G10, of prostate specimens ranging from normal to CRPC. Tissue spots from patients with CRPC show high levels of phosphotyrosine expression in the epithelial compartment. (B) Increased tyrosine phosphorylation is observed in CRPC, with nearly 50% of the patients displaying high-intensity staining (2, 3) compared with normal, HGPIN, or HNPC tissues. (C) Average staining intensity of all of the tissues clearly show a significant increase of tyrosine phosphorylation in CRPC patients. HGPIN, high-grade prostatic intraepithelial neoplasia; HNPC, hormone naïve prostate cancer; HRPC, hormone refractory prostate cancer. ***P < 0.001, one-way ANOVA. (Scale bar, 200 μm.)
Tyrosine Phosphorylation Is Robust in Mouse Models of Advanced Prostate Cancer.
The observation of increased tyrosine phosphorylation in late-stage prostate cancer specimens raises the question of whether tyrosine kinase activity is evident in prostate cancer models that do not express mutated or amplified tyrosine kinases. We recapitulated different stages of prostate cancer ranging from prostate intraepithelial neoplasia (PIN) to adenocarcinoma using the prostate in vivo regeneration model system (25, 26). We chose four of the most commonly perturbed oncogenes in prostate cancer, both in androgen-dependent and -independent states: activated AKT (myristoylated AKT, resembling PTEN deletion, ∼40–70% of prostate cancers), AR amplification (∼20–60% of prostate cancers), ERG rearrangements (∼40–70% of prostate cancers), and activated K-RAS (K-RASG12V, resembling RAS/RAF pathway activation, observed in ∼40–50% of prostate cancers) (7, 8, 11, 27–30).
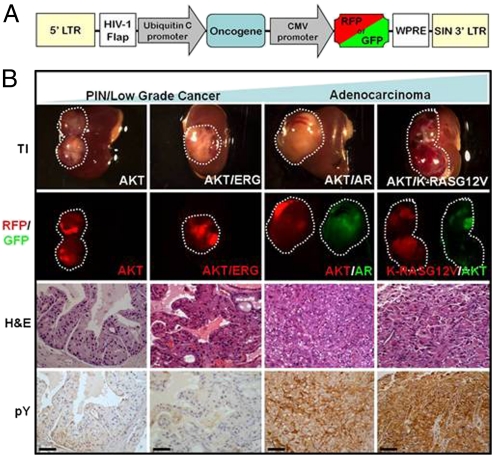
We infected total mouse prostate cells with AKT alone or in combination with each respective oncogene using a lentiviral vector delivery system (Fig. 2A) and evaluated the histological phenotype of the resulting tumors after 12 wk. These tumors displayed histological characteristics of PIN (AKT), well differentiated and less aggressive cancer (AKT/ERG), or adenocarcinoma (AKT/AR and AKT/K-RASG12V) (Fig. 2B). IHC and Western blot analysis confirmed ectopic expression of each oncogene (Fig. S1 A and B). IHC staining and Western blot analyses displayed a gradient of phosphotyrosine expression in these tumors ranging from low to undetectable levels of tyrosine phosphorylation in the normal and indolent lesions (mouse prostate, AKT, or AKT/ERG) to very high levels in the more advanced tumors (AKT/AR and AKT/K-RASG12V) (Fig. 2B and Fig. S2 A and B).
Fig. 2.
Phosphotyrosine expression is increased during prostate cancer progression. (A) Lentiviral vector diagram displaying the organization of oncogene and fluorescent marker expression used in these tumors. (B) Gross and histological morphology of each tumor type after 12-wk engraftment in SCID mice using the prostate regeneration protocol. Fluorescence corresponds to expression of a particular oncogene. IHC staining of progressive mouse tumor phenotypes reveals an increasing gradient of phosphotyrosine expression with more aggressive tumors expressing higher levels than indolent tumors. TI, transillumination; H&E, hematoxylin and eosin; pY, phosphotyrosine. (Scale bars, 50 μm.)
Phosphoproteomic Profiling Identifies Oncogene-Dependent Tyrosine Phosphorylation of Kinases and Phosphatases.
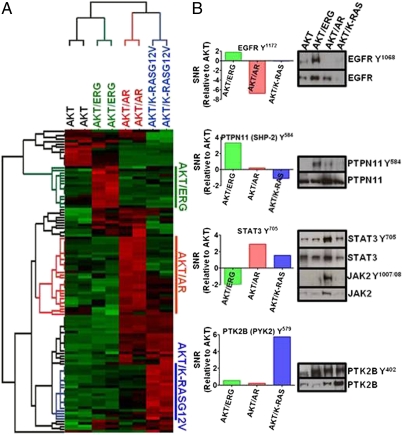
We enriched for tyrosine phosphorylated peptides and performed quantitative label-free MS to identify phosphopeptides that contribute to this increased tyrosine phosphorylation (21, 31). We identified 139 phosphopeptides corresponding to 102 proteins (Dataset S1). Statistical analysis (ANOVA, 0.2 cutoff) revealed differential phosphorylation of 116 phosphopeptides corresponding to 87 proteins across all of the tumor phenotypes. Unsupervised hierarchical clustering analysis identified unique and overlapping patterns of tyrosine phosphorylated peptides for each tumor type, with an increased abundance of tyrosine phosphorylation events observed in the more advanced tumors (AKT/AR and AKT/K-RASG12V) (Fig. 3A and Fig. S3). These data demonstrate oncogene-specific signatures of phosphotyrosine activation across the spectrum of prostate cancer progression.
Fig. 3.
Unique phosphotyrosine signatures are observed in a mouse model of prostate cancer progression. (A) Heatmap representing unique clusters of tyrosine phosphorylation for each mouse tumor phenotype. Each row corresponds to a unique phosphopeptide. Red, hyperphosphorylation; green, hypophosphorylation for each phosphopeptide. (B) Specific tyrosine kinases are observed in an oncogene-specific fashion. Signal-to-noise ratio (SNR) (relative to AKT) was calculated for each phosphorylation event and plotted. Positive SNR confirms elevation of that particular phosphorylation event. Western blotting validates indicated oncogene-specific phosphorylation results.
From the MS data, the activation sites of several tyrosine kinases and protein phosphatases were identified in the specific tumor types (Table 1 and Figs. S3 and S4) (32). Consistent with these findings, Western blotting confirmed high levels of a second EGFR phosphorylation site (Y1068) and PTPN11 (SHP-2) Y584 in AKT/ERG tumors (Fig. 3B). Activation of the JAK/STAT pathway was also revealed in AKT/AR tumors as high levels of phosphorylation of STAT3 Y705 were observed. Western blotting confirmed activation of the upstream kinase JAK2Y1007/08 and STAT3 Y705 in this tumor type (Fig. 3B). We additionally identified an increase in phosphorylation of PTK2B/PYK2/FAK2 Y579 and Y849 in AKT/K-RASG12V tumors and Western blot confirmed the phosphorylation of the activation site Y402 of PTK2B (Fig. 3B). Together, these data demonstrate that each combination of prostate cancer oncogenes generates distinct patterns of tyrosine kinase and phosphatase activity.
Table 1.
Oncogene-specific phosphoactivation of tyrosine kinases and phosphatases
| Oncogene combination | Tyrosine kinase (phosphoresidue) | Tyrosine phosphatase (phosphoresidue) | Functional significance* |
| AKT/ERG | EGFR (Y1172) | PTPN11 (Y584) | Enzymatic activation |
| PTPRA (Y825) | Enzymatic activation | ||
| INPPL1 (Y1136) | Unknown | ||
| AKT/AR | JAK2 (Y1007/08)† | PTPN11 (Y62) | Enzymatic activation |
| AKT/KRASG12V | EPHA2 (Y595) | Enzymatic activation | |
| LYN (Y508) | Enzymatic inhibition | ||
| PTK2B (Y579) | Unknown | ||
| PTK2B (Y849) | Unknown | ||
| EPHA2 (Y773) | Unknown | ||
| AKT/ERG | FER (Y402) | Alters cell motility | |
| AKT/AR | |||
| AKT/KRASG12V | |||
All measured phosphorylation events are relative to AKT-only lesions.
*Source: Phosphosite (http://www.phosphosite.org).
†JAK2 was not identified by MS, but inferred on the basis of high STAT3 Y705 phosphorylation observed in AKT/AR tumors.
Bioinformatic Inference of Tyrosine Kinase Activity Reveals Enrichment of Dasatinib Targets in AKT/AR Tumors.
In addition to direct observation of phosphorylated tyrosine kinases and phosphatases by MS, we sought to use the phosphotyrosine peptide data to infer kinase activities specific to each tumor type. We predicted the activated kinases directly upstream for each observed phosphorylation site using known relationships from PhosphoSite (32), kinase motifs from PhosphoMotif Finder (33) and Phosida (34), and predictions from NetworKin (35). We then performed an enrichment analysis of kinase-associated phosphorylation targets (Materials and Methods) to determine which kinase activities were predicted to be highly active in each tumor type.
Using this unbiased bioinformatic approach, we identified a statistically significant enrichment of the EGFR kinase substrate (D|E)pY in AKT/ERG but not in AKT/AR or AKT/K-RASG12V tumors (Fig. S5 and Dataset S2). Notably, this bioinformatic prediction was in direct agreement with our phosphoproteomic and Western blot data (Fig. 3B). Inference of kinase activity in AKT/K-RASG12V tumors further revealed an enrichment of ERK1/2 and MEK1/2 substrates, consistent with direct activation of MAPK signaling by the K-RASG12V oncogene (Fig. 4B and Fig. S4 and Dataset S2) (36).
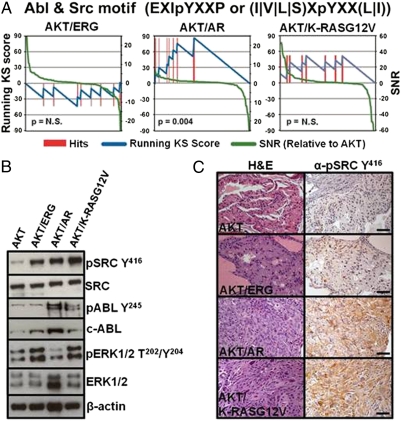
Fig. 4.
Bioinformatic analysis reveals enrichment of dasatinib tyrosine kinase targets in AKT/AR tumors. (A) Enrichment analysis of tyrosine phosphosite motifs reveals enrichment of phosphosubstrates of the tyrosine kinases ABL and SRC, targets of the tyrosine kinase inhibitor dasatinib, in AKT/AR tumors. No significant enrichment of these phosphopeptides were observed in either AKT/ERG or AKT/K-RASG12V tumors. Enrichment scores for all kinase motifs are shown in Dataset S2. (B) Western blot and (C) IHC staining for the activated kinases ABL, SRC, or ERK1/2 reveal tumor-specific activation of these kinases. (Scale bars, 50 μm.)
Evaluating kinase activity from AKT/AR phosphopeptides revealed statistically significant enrichment of two motifs associated with ABL1 and SRC kinases [EXIpYXXP and (I|V|L|S)XpYXX(L|I), respectively] (37). Because these kinases are both targets of the tyrosine kinase inhibitor, dasatinib, we combined these motifs into a “dasatinib target” group and found enrichment of predicted ABL1 and SRC substrates in AKT/AR tumors (Fig. 4A and Dataset S2). AKT/K-RASG12V and AKT/ERG tumors demonstrated modest and no enrichment of these motifs, respectively. Western blotting and IHC validated this bioinformatic prediction, as both SRC Y416 and ABL1 Y245 were highly phosphorylated only in the AKT/AR tumor type, whereas SRC Y416 but not ABL1 Y245 were phosphorylated in AKT/ERG and AKT/K-RASG12V tumors (Fig. 4 B and C). This result demonstrates that substrate-based bioinformatic approaches for inferring kinase activity can reveal oncogene-specific tyrosine kinase activation not originally identified directly by phospho-MS.
Assembly of Oncogene-Specific Tyrosine Kinase Signaling Networks from Phosphoproteomic Data and Public Databases.
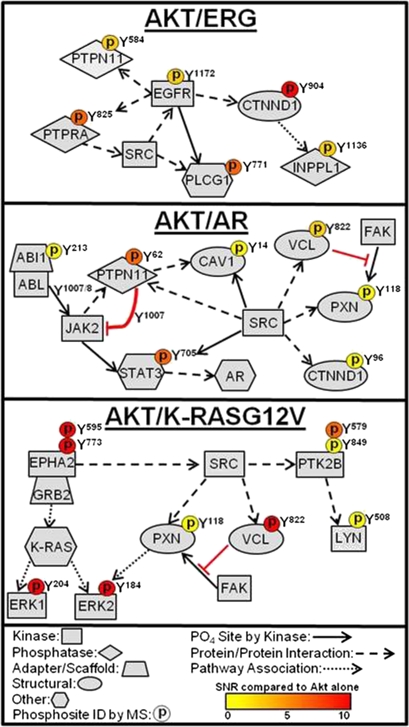
We next sought to combine our phosphopeptide and bioinformatics data with information from public databases of protein–protein interactions (Human Protein Reference Database, HPRD) and posttranslational modifications (Phosphosite) to manually construct tyrosine kinase signaling networks for each oncogene combination. In AKT/ERG tumors, identification of the EGFR substrate Y771 of phospholipase C, gamma 1 (PLCG1), and EGFR interacting proteins catenin, delta 1 (p120 catenin, CTNND1), PTPN11, and PTPRA, suggest strong association and activation of the EGFR tyrosine kinase pathway (Fig. 5). In AKT/AR tumors, detection of elevated SRC and ABL1 activity prompted us to investigate other substrates and binding partners of these kinases within our phosphoproteomic data. The identification of SRC and ABL1 substrates Y705 of STAT3, Y14 of caveolin-1 (CAV-1), and Y1007/1008 of JAK2 with binding partners vinculin (VCL) Y822, paxillin (PXN) Y118, CTNND1 Y96, and PTPN11 Y62, suggest that, along with JAK2, these kinases act in concert toward the development of AKT/AR tumors (Fig. 5). The identification of the activation site of EPHA2 Y595 and downstream effectors ERK1 Y204 and ERK2 Y184 reveals strong MAPK activation in AKT/K-RASG12V tumors (Fig. 5). Further, the identification of VCL Y822 and PXN Y118 in AKT/AR and AKT/K-RASG12V tumors suggests that regulation of focal adhesions may be important for motility and survival in these tumors. The phosphorylation of PXN at Y118 by focal adhesion kinase (FAK) increases cell motility and survival, which are characteristic features of cells that have undergone an epithelial-to-mesenchymal transition (EMT) (38). The possibility of an EMT phenotype would be consistent with previous tumor phenotypes where SRC activation was observed (18). The manual curation of phosphotyrosine networks suggest novel associations of tyrosine kinase signaling with defined oncogenic insults in prostate cancer.
Fig. 5.
Curation of phosphoproteomic profiling and bioinformatics delineates distinct tyrosine kinase signaling pathways in an oncogene-specific manner. Selected substrate and interaction pathways from each tyrosine kinase were generated from a combination of our phosphoproteomics dataset and the HPRD and Phosphosite databases. An elevated phosphorylation event identified by MS is indicated by a phosphorylation residue depicted above the protein and color coded. Solid arrow, protein is a direct substrate of the upstream kinase at that site. Dashed arrow, protein interacts directly with the upstream kinase/protein. Dotted arrow, protein is found within the pathway of the upstream kinase/protein.
Discussion
Many studies have linked the aberrant activation of tyrosine kinases by somatic mutation or DNA amplification to a wide array of cancers (39, 40). We demonstrate oncogene-specific signatures of global phosphotyrosine activity without ectopic expression of mutant tyrosine kinases in a mouse model of prostate cancer progression. The activation of tyrosine kinase signaling suggests the presence of alternative mechanisms regulating tyrosine kinase activity not related to activating mutations (18, 21, 22). These include but are not limited to loss of negative feedback mechanisms (e.g., increased or decreased phosphatase activity), transcriptional up-regulation of kinases, or increased stabilization of tyrosine kinases through decreased protein degradation (22, 41, 42). Our data suggest that some of these mechanisms may control tyrosine kinase signaling in our mouse model of prostate cancer.
Tyrosine phosphorylation of the protein tyrosine phosphatase PTPN11 may contribute to the phosphotyrosine signatures observed in our tumors. Activity of this phosphatase is often associated with increased signaling activity (43, 44). This phosphatase was highly phosphorylated on Y62 and Y584 in AKT/AR and AKT/ERG tumors, respectively. In EGFR-expressing fibroblasts, epidermal growth factor (EGF) stimulation resulted in Y584 phosphorylation of PTPN11 leading to RAS/ERK pathway activation (45). This supports our findings that Y584 of PTPN11 is highly phosphorylated in AKT/ERG tumors and suggests receptor tyrosine kinase pathway-mediated activation of PTPN11. PTPN11 inhibition leads to decreased xenograft growth of lung and prostate tumors and reduced activity of numerous tyrosine kinases, including SRC (46). PTPN11 Y62/63 activation results in tyrosine dephosphorylation of the inactive site of SRC Y530 by regulation of the Csk regulator PAG/Cbp, indicating that SRC activity in AKT/AR tumors may be dependent on PTPN11 activation (43, 46).
Transcriptional up-regulation of tyrosine kinases may also enhance tyrosine kinase activity as suggested by the phosphorylation of EPHA2 at Y595 in the AKT/K-RASG12V tumors. EPHA2 was shown to be a transcriptional target of the RAS–MAPK pathway and ligand-stimulated EPHA2 negatively regulates RAS activity (47). Constitutive activation of RAS through mutation bypasses the negative regulation of EPHA2 and results in increased MAPK activation, which is in direct agreement with our phosphoproteomic data. RAS activation may reveal why high expression levels of EPHA2 protein are observed in breast and prostate cancer and supports further clinical investigation of the connection between RAS mutation and EPHA2 status in these diseases (48, 49).
Tyrosine kinase activation offers therapeutic opportunities following the emerging successes of tyrosine kinase inhibitor therapies (5, 50). Our observation of SRC activity supports previous work that this kinase synergizes with other genes, including AR, to contribute to prostate adenocarcinoma (18, 51). SRC has also been shown to interact with the intracellular region of ERBB2 (HER-2), supporting the notion that SRC may be an important node for targeted therapy in advanced prostate cancer (17, 52). In support of these data, the SRC and ABL1 tyrosine kinase inhibitor dasatinib in combination with docetaxel is currently in phase III clinical trials for advanced prostate cancer and has shown modest phase I/II trial results in overall patient survival (53). Due to the heterogeneity of prostate cancer, this modest effect may be a result of the general administration of dasatinib without stratification of patients on the basis of SRC and ABL1 activity.
Strong activation of the EGFR pathway was observed in AKT/ERG-expressing mouse prostate tumors. Roughly half of all prostate cancer patients display the TMPRSS2-ERG translocation, a gene rearrangement fusing the androgen-regulated promoter of TMPRSS2 with the ETS transcription factor ERG, which is considered to be a marker for prostate cancer progression from PIN to adenocarcinoma (54). The product of the TMPRSS2-ERG translocation was shown to interact with the enzyme poly (ADP ribose) polymerase 1 (PARP1), and inhibition of this enzyme abrogates growth of prostate cancer xenografts that ectopically express ERG (55). PARP1 inhibition represents a promising treatment option for patients with TMPRSS2-ERG translocations. Our data suggest that EGFR activity level is another candidate target in patients with TMPRSS2-ERG translocations. This result is in agreement with recent reports of SPINK1+/ETS− prostate cancers where SPINK1-mediated growth occurs via EGFR signaling, demonstrating alternative pathways to activate EGFR (56). It will be important to further evaluate the relationship between EGFR activity and ERG clinically.
Our data suggest the molecular stratification of patients to target prostate cancer with tyrosine kinase inhibitors even in tumors without obvious tyrosine kinase mutations. Future work will extend this approach to prostate cancer patients to match tyrosine kinase inhibitor therapies with signaling activation patterns for targeted treatment of this disease.
Materials and Methods
Clinical Prostate Tissue Microarrays, Lentiviral Vector Construction, Prostate Regeneration and Prostate Epithelial Viral Infections, and Western Blot and Immunohistochemistry can be found in SI Materials and Methods.
Quantitative Analysis of Phosphotyrosine Peptides by Mass Spectrometry.
A total of 300–500 mg of frozen tumor mass was homogenized and sonicated in urea lysis buffer (20 mM Hepes pH 8.0, 9 M urea, 2.5 mM sodium pyrophosphate, 1.0 mM β-glycerophosphate, 1% N-octyl glycoside, 2 mM sodium orthovanadate). A total of 35 mg of total protein was used for phosphotyrosine peptide immunoprecipitation as previously described (21, 57, 58). Additional details can be found in SI Materials and Methods.
Prediction of Kinase-Substrate Relationships.
For each phosphopeptide, we predicted the potential upstream kinases using three types of data: (i) NetworKIN 2.0 kinase-substrate relationships (http://networkin.info/version_2_0/search.php), (ii) PhosphoSite kinase-substrate dataset (http://www.phosphosite.org/), and (iii) consensus kinase motifs culled from the Human Protein Reference Database's PhosphoMotif Finder (http://www.hprd.org/PhosphoMotif_finder) and Phosida (http://www.phosida.de/).
Enrichment Analysis of Kinase Activity.
Phosphotyrosine peptides were ranked by the signal-to-noise ratio observed for a given perturbation (e.g., AKT/AR tumors compared with AKT alone). Having annotated the phosphopeptides with their predicted upstream kinases, we calculated a Kolmogorov–Smirnov statistic against the expected distribution for each upstream kinase. The statistical significance of enrichment was then determined by permutation analysis. This approach is analogous to the normalized enrichment score of gene set enrichment analysis (59). The enrichment scores for all putative upstream kinases are shown in Dataset S2. Additional details can be found in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the O.N.W. laboratory for helpful comments and discussion on the manuscript. We thank Mireille Riedinger for purifying the 4G10 antibody used in MS studies. We thank the Tissue Procurement Core Laboratory at University of California, Los Angeles (UCLA) for assistance on tissue processing and H&E staining. J.M.D. is supported by the Department of Defense Prostate Cancer Research Program (PC101928). J.M.D., N.A.G., and D.A.S. are supported by the UCLA Tumor Biology Program, US Department of Health and Human Services, Ruth L. Kirschstein Institutional National Research Service Award T32 CA009056. T.S. is supported by California Institute for Regenerative Medicine Training Grant TG2-01169. A.S.G. is supported by a Career Development Award from the Specialized Program of Research Excellence (SPORE) in Prostate Cancer [Principal Investigator (PI), Robert Reiter]. J.H. is supported by UCLA SPORE in Prostate Cancer (PI, Robert Reiter), Department of Defense Prostate Cancer Research Program (PC101008), CalTech-UCLA Joint Center for Translational Medicine Program, and National Cancer Institute (1R01CA158627-01; PI, Leonard Marks). T.G.G. is partially supported by the CalTech-UCLA Joint Center for Translational Medicine. J.H. and O.N.W. are supported by a Prostate Cancer Foundation Challenge Award (to O.N.W., PI). O.N.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: MS2 spectra for all phosphopeptides reported in this paper have been deposited in the PRIDE database, http://www.ebi.ac.uk/pride/ (accession nos. 20879–20889).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120985109/-/DCSupplemental.
References
- 1.Nakahara M, et al. A novel gain-of-function mutation of c-kit gene in gastrointestinal stromal tumors. Gastroenterology. 1998;115:1090–1095. doi: 10.1016/s0016-5085(98)70079-4. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 3.Dienstmann R, Martinez P, Felip E. Personalizing therapy with targeted agents in non-small cell lung cancer. Oncotarget. 2011;2:165–177. doi: 10.18632/oncotarget.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim KS, et al. Predictors of the response to gefitinib in refractory non-small cell lung cancer. Clin Cancer Res. 2005;11:2244–2251. doi: 10.1158/1078-0432.CCR-04-2081. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich MC, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visakorpi T, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–531. [PubMed] [Google Scholar]
- 11.Yoshimoto M, et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet. 2006;169:128–137. doi: 10.1016/j.cancergencyto.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Pezaro C, et al. An open-label, single-arm phase two trial of gefitinib in patients with advanced or metastatic castration-resistant prostate cancer. Am J Clin Oncol. 2009;32:338–341. doi: 10.1097/COC.0b013e31818b946b. [DOI] [PubMed] [Google Scholar]
- 13.Curigliano G, et al. Absence of epidermal growth factor receptor gene mutations in patients with hormone refractory prostate cancer not responding to gefitinib. Prostate. 2007;67:603–604. doi: 10.1002/pros.20530. [DOI] [PubMed] [Google Scholar]
- 14.Guo Z, et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell. 2006;10:309–319. doi: 10.1016/j.ccr.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 15.Mahajan NP, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad Sci USA. 2007;104:8438–8443. doi: 10.1073/pnas.0700420104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Dasatinib inhibits site-specific tyrosine phosphorylation of androgen receptor by Ack1 and Src kinases. Oncogene. 2010;29:3208–3216. doi: 10.1038/onc.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craft N, Shostak Y, Carey M, Sawyers CL. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat Med. 1999;5:280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 18.Cai H, Babic I, Wei X, Huang J, Witte ON. Invasive prostate carcinoma driven by c-Src and androgen receptor synergy. Cancer Res. 2010;71:862–872. doi: 10.1158/0008-5472.CAN-10-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatarov O, et al. SRC family kinase activity is up-regulated in hormone-refractory prostate cancer. Clin Cancer Res. 2009;15:3540–3549. doi: 10.1158/1078-0432.CCR-08-1857. [DOI] [PubMed] [Google Scholar]
- 20.Del Rosario AM, White FM. Quantifying oncogenic phosphotyrosine signaling networks through systems biology. Curr Opin Genet Dev. 2010;20:23–30. doi: 10.1016/j.gde.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubbi L, et al. Global phosphoproteomics reveals crosstalk between Bcr-Abl and negative feedback mechanisms controlling Src signaling. Sci Signal. 2011;4:ra18. doi: 10.1126/scisignal.2001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun T, et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144:703–718. doi: 10.1016/j.cell.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Li J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawson DA, et al. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci USA. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bismar TA, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–485. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
- 28.Linja MJ, et al. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–3555. [PubMed] [Google Scholar]
- 29.Clark J, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 30.Brenner JC, Chinnaiyan AM. Translocations in epithelial cancers. Biochim Biophys Acta. 2009;1796:201–215. doi: 10.1016/j.bbcan.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 32.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 33.Amanchy R, et al. A curated compendium of phosphorylation motifs. Nat Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 34.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2011;39(Database issue):D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linding R, et al. NetworKIN: A resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36(Database issue):D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 37.Keshava Prasad TS, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subauste MC, et al. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. J Cell Biol. 2004;165:371–381. doi: 10.1083/jcb.200308011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 40.Olivier M, Taniere P. Somatic mutations in cancer prognosis and prediction: Lessons from TP53 and EGFR genes. Curr Opin Oncol. 2011;23:88–92. doi: 10.1097/CCO.0b013e3283412dfa. [DOI] [PubMed] [Google Scholar]
- 41.Bao J, Gur G, Yarden Y. Src promotes destruction of c-Cbl: implications for oncogenic synergy between Src and growth factor receptors. Proc Natl Acad Sci USA. 2003;100:2438–2443. doi: 10.1073/pnas.0437945100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasquale EB. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang SQ, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13:341–355. doi: 10.1016/s1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 44.Cunnick JM, et al. Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. J Biol Chem. 2002;277:9498–9504. doi: 10.1074/jbc.M110547200. [DOI] [PubMed] [Google Scholar]
- 45.Mitra S, Beach C, Feng GS, Plattner R. SHP-2 is a novel target of Abl kinases during cell proliferation. J Cell Sci. 2008;121:3335–3346. doi: 10.1242/jcs.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren Y, et al. Critical role of Shp2 in tumor growth involving regulation of c-Myc. Genes Cancer. 2010;1:994–1007. doi: 10.1177/1947601910395582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macrae M, et al. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8:111–118. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 48.Walker-Daniels J, et al. Overexpression of the EphA2 tyrosine kinase in prostate cancer. Prostate. 1999;41:275–280. doi: 10.1002/(sici)1097-0045(19991201)41:4<275::aid-pros8>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 49.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61:2301–2306. [PubMed] [Google Scholar]
- 50.Verweij J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: Randomised trial. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 51.Cai H, et al. Differential transformation capacity of Src family kinases during the initiation of prostate cancer. Proc Natl Acad Sci USA. 2011;108:6579–6584. doi: 10.1073/pnas.1103904108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- 53.Araujo JC, et al. Dasatinib combined with docetaxel for castration-resistant prostate cancer: Results from a phase 1-2 study. Cancer. 2011;118:63–71. doi: 10.1002/cncr.26204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brenner JC, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ateeq B, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Transl Med. 2011;3(72):72ra17. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skaggs BJ, et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci USA. 2006;103:19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimman A, et al. Activation of aortic endothelial cells by oxidized phospholipids: A phosphoproteomic analysis. J Proteome Res. 2010;9:2812–2824. doi: 10.1021/pr901194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.