Abstract
Maintenance of skeletal muscle structure and function requires efficient and precise metabolic control. Autophagy plays a key role in metabolic homeostasis of diverse tissues by recycling cellular constituents, particularly under conditions of caloric restriction, thereby normalizing cellular metabolism. Here we show that histone deacetylases (HDACs) 1 and 2 control skeletal muscle homeostasis and autophagy flux in mice. Skeletal muscle-specific deletion of both HDAC1 and HDAC2 results in perinatal lethality of a subset of mice, accompanied by mitochondrial abnormalities and sarcomere degeneration. Mutant mice that survive the first day of life develop a progressive myopathy characterized by muscle degeneration and regeneration, and abnormal metabolism resulting from a blockade to autophagy. HDAC1 and HDAC2 regulate skeletal muscle autophagy by mediating the induction of autophagic gene expression and the formation of autophagosomes, such that myofibers of mice lacking these HDACs accumulate toxic autophagic intermediates. Strikingly, feeding HDAC1/2 mutant mice a high-fat diet from the weaning age releases the block in autophagy and prevents myopathy in adult mice. These findings reveal an unprecedented and essential role for HDAC1 and HDAC2 in maintenance of skeletal muscle structure and function and show that, at least in some pathological conditions, myopathy may be mitigated by dietary modifications.
Keywords: autophagosome formation, muscle disease, muscle metabolism, epigenetic regulation
Cellular homeostasis is maintained by a balance between protein biosynthesis and degradation. Macroautophagy (herein referred to as autophagy) is a catabolic pathway responsible for the degradation of various cellular constituents or deleterious cellular components. The process involves formation of vesicles, called autophagosomes, that capture and deliver proteins to lysosomes for degradation (1). The resulting breakdown products are recycled and used to support cellular metabolism (2). Genetic studies with mice mutant for genes involved in autophagy substantiated the importance of basal autophagy for organelle turnover and cellular homeostasis (3–11). Various stress conditions, such as fasting, exercise, hypoxia, oxidative stress, and pathogen infection, trigger autophagy as an adaptive response to normalize cellular metabolism (12, 13).
Deregulation of autophagy has been implicated in cancer (14, 15), neurodegenerative disorders (4, 5, 10), and muscular diseases (16, 17). Epigenetic factors have been implicated in regulating autophagy during various pathological conditions (12). Alteration of the acetylation status of histones or other proteins through histone deacetylases (HDACs) is a key mechanism that controls gene transcription and protein function. Removal of acetyl groups from histone tails by HDACs promotes transcriptional repression by allowing chromatin compaction (18). HDACs also modulate the activity of a variety of transcription factors and large macromolecular complexes involved in diverse cellular processes (19). Although the roles of HDACs have been extensively studied in vitro using HDAC inhibitors, much remains to be learned about the functions of these enzymes in vivo (18). Mice lacking HDAC genes in different tissues have revealed highly specific functions for individual HDAC isoforms during development and adulthood (18). Global deletion of HDAC1 or HDAC2 in mice causes early lethality (20), necessitating conditional gene deletion to uncover the functions of these enzymes in adult tissues. Indeed, tissue-specific deletion of HDAC1 and HDAC2 has uncovered important roles of these enzymes in diverse cellular processes (20–22).
In the present study, we show that HDAC1 and HDAC2 control muscle homeostasis and autophagy flux in skeletal muscle. Approximately 40% of double knockout (dKO) mice lacking both HDAC1 and HDAC2 in skeletal muscle die within the first day of life; the remaining dKO mice develop a progressive myopathy, preceded by impaired autophagy flux, which correlates with a shift toward oxidative metabolism. HDAC1 and HDAC2 regulate autophagy flux, at least in part, by allowing the transcriptional induction of genes involved in autophagosome formation. Remarkably, feeding pregnant mice a high-fat diet (HFD) prevents perinatal mortality of dKO mice, whereas feeding adult dKO mice with HFD disallows the onset of myopathy and permits basal autophagy flux. This study defines a previously unrecognized role for HDAC1 and HDAC2 in regulating autophagy and muscle homeostasis and suggests that certain lethal myopathies might be mitigated by dietary modifications.
Results
Deletion of HDACs 1 and 2 Causes a Progressive Myopathy.
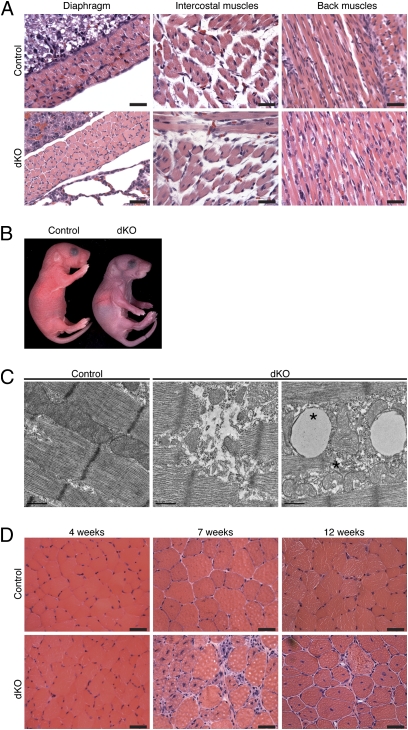
We generated mice lacking HDAC1 or HDAC2 in skeletal muscle by crossing HDAC1loxP/loxP or HDAC2loxP/loxP mice (20) to myogenin-Cre transgenic mice, which express Cre recombinase exclusively in skeletal muscle (23). The transgene of the myogenin-Cre mouse line consists of the mouse myogenin promoter and the skeletal muscle-specific enhancer of the mouse Mef2c gene, which are expressed only in the skeletal muscle lineage from embryonic day (E) 8.5 to adulthood (24, 25). Efficient deletion of Hdac1 and Hdac2 genes from skeletal muscle of HDAC1fl/fl; HDAC2fl/fl; myogenin-Cre mice (hereafter referred to as dKO mice) at postnatal day (P) 1 was confirmed by real-time RT-PCR analysis (Fig. S1). Mice lacking up to three alleles of Hdac1 and Hdac2 were viable and showed normal skeletal muscle architecture by histological analysis (Fig. S2). dKO mice were seen at expected Mendelian ratios at E18.5, but a significant reduction of approximately 40% between the number of expected and observed dKO mice was detected 10 d after birth (Fig. S3). Although no apparent skeletal muscle abnormalities were detected by histological analysis at P1 (Fig. 1A), dying dKO mice appeared cyanotic (Fig. 1B), suggesting that death was due to impaired function of respiratory muscles. Electron microscopy of P1 diaphragm muscle revealed ultrastructural abnormalities, including degeneration of sarcomeres and misshapen mitochondria, in dKO myofibers of the subset of affected mice(Fig. 1C).
Fig. 1.
dKO mice die perinatally or develop a progressive myopathy. (A) H&E sections of various muscles of dKO mice and control mice, a few hours after birth. (Scale bars, 40 μm.) (B) Picture of dKO and control mice a few hours after birth. (C) Electron microscopic pictures of diaphragm isolated from 1-d-old control and dKO mice, showing ultrastructural abnormalities, including swollen and cup-shaped mitochondria (asterisk). (Scale bars, 0.5 μm.) (D) H&E sections of TA muscle from control and dKO mice at indicated ages. (Scale bars, 40 μm.)
dKO mice that survived the first day of life developed a progressive myopathy in the tibialis anterior (TA) and extensor digitorum longus muscles, starting from 7 wk of age, characterized by myofiber degeneration, centronuclear myofibers, and inflammatory cell infiltration (Fig. 1D). At 4 wk of age, before histological onset of myopathy, no Evans blue dye uptake was seen in dKO myofibers (Fig. S4), and no signs of muscle damage were evident in dKO mice, as measured by creatine kinase assay (Fig. S5).
Deletion of HDAC1 and HDAC2 Blocks Autophagy Flux.
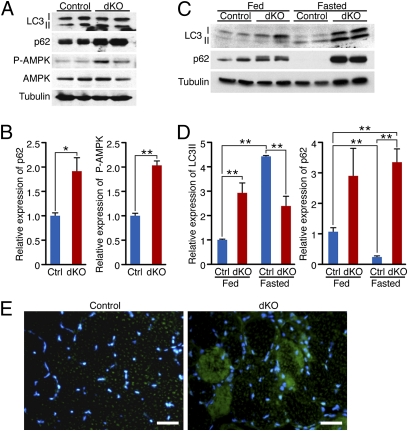
The phenotype of dKO mice partially resembles prior descriptions of mice deficient in autophagy (7, 26–28). Autophagy flux can be monitored by detection of the modification of endogenous LC3 (microtubule-associated protein1 light chain 3) using standard immunoblotting procedures (29, 30). LC3 exists in two forms: the free mature form LC3-I and the membrane-bound lipidated LC3-II form, which is a marker of autophagosome formation. p62 is a protein sequestered in autophagosomes and lost when autophagosomes fuse with lysosomes. Thus, an increase in p62 abundance indicates inhibition of autophagy (31–33). As early as a few hours after birth, no difference was detected in the conversion of LC3-I to LC3-II between dKO and HDAC1fl/fl; HDAC2fl/fl (hereafter referred to as control) mice; however, significant accumulation of p62 protein was seen in muscle from dKO mice, suggesting a block in autophagy flux only a few hours after birth (Fig. 2 A and B).
Fig. 2.
HDAC1 and HDAC2 regulate autophagy flux in skeletal muscle. (A) Western blot showing autophagy markers (LC3 I-II and p62), phosphorylated AMPK (P-AMPK), and total AMPK in skeletal muscle of neonatal control and dKO mice. Tubulin was used as loading control. (B) Densitometric analysis of Western blot showing p62 and P-AMPK expression in dKO relative to control (Ctrl) mice. Values were normalized to tubulin. n = 4. *P < 0.05; **P < 0.005. (C) Western blot showing LC3 I-II and p62 in skeletal muscle of 4-wk-old control and dKO mice, fed or fasted for 24 h. Tubulin was used as loading control. (D) Densitometric analysis of Western blot showing LC3 II and p62 expression in dKO relative to Ctrl mice. Values were normalized to tubulin. n = 6. **P < 0.005. (E) Immunostaining of histological sections of TA muscle for p62 (green) and nuclei (blue) at 7 wk of age. (Scale bars, 40 μm.)
Because autophagy supplies energy during the early postnatal period of starvation (27, 34), we assessed the energy status of P1 skeletal muscle by measuring the activated form of AMP-activated protein kinase (AMPK), which functions as an energy sensor (35, 36). AMPK is activated by phosphorylation of the α-subunit after an increase in the intracellular AMP/ATP ratio, and it is up-regulated in mice with mutations in autophagy genes (27). Consistent with a block in autophagy flux, we found a significant increase of the phosphorylated form of AMPK in P1 dKO skeletal muscle compared with controls (Fig. 2 A and B). We analyzed the autophagy pathway at baseline and upon induction in response to 24 h of fasting, at 4 wk of age, before the histological onset of the myopathy. Upon fasting, as expected, there was a decrease of LC3-I and an increase of LC3-II in control mice, whereas in dKO mice there was an increase of both LC3-I and LC3-II, showing a defect in autophagy flux (Fig. 2 C and D). Strikingly, p62 protein robustly accumulated in dKO muscle, both at baseline and upon fasting (Fig. 2 C and D), indicative of a block in autophagy under both conditions. Accumulation of p62 protein was confirmed in dKO TA by immunostaining at 7 wk of age (Fig. 2E).
Because a block in the proteasome pathway can lead to an accumulation of p62 protein, we assessed proteasome activity in vivo in control and dKO mice. Proteasome activity was detected by coelectroporating a reporter plasmid, Ub-G76V-YFP (37), which is normally degraded by the proteasome and quickly accumulates if proteasome activity is impaired (7), and dsRED plasmid, for monitoring transfection efficiency, in TA muscle of 4-wk-old control and dKO mice. Twelve days later, we could not detect any YFP signal in the transfected TA, indicating that proteasome activity was not impaired in dKO mice (Fig. S6). As a positive control, we detected strong YFP signal colocalizing in the dsRED-positive myofibers in mice treated with the proteasome inhibitor MG262 (38) (Fig. S6).
HDAC1 and HDAC2 Modulate Autophagosome Formation in Skeletal Muscle.
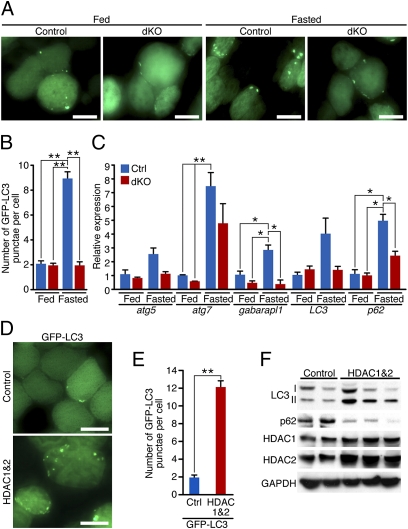
We monitored steady-state levels of autophagosomes by electroporating GFP-LC3 (7, 38, 39) into TA of control and dKO mice at 4 wk of age. One week later, we induced autophagy by fasting mice for 24 h. Localization of GFP-LC3-I is cytoplasmic and results in a diffuse fluorescence signal, whereas GFP-LC3-II associates with autophagosome membranes and is seen as punctate (39). An increased number of autophagosomes labeled by GFP-LC3 punctae was observed in control mice in response to fasting, but not in dKO mice (Fig. 3 A and B).
Fig. 3.
HDAC1 and HDAC2 regulate autophagosome formation in skeletal muscle. (A) Representative fields of GFP-LC3-electroporated TA muscle of adult control and dKO mice. Eight days after the electroporation, mice were fasted for 24 h. (Scale bars, 40 μm.) (B) Counts of GFP-LC3 punctae per myofiber in control (Ctrl) or dKO mice, fed or fasted for 24 h. n = 4–7 (≈400 fibers each). Values are mean ± SEM. **P < 0.005. (C) Real-time RT-PCR analysis, at 4 wk of age, for atg5, atg7, gaparapl1, LC3, and p62 mRNA of control (Ctrl) and dKO mice, fed or fasted for 48 h. Data are expressed as mean ± SEM. n = 4. *P < 0.05. **P < 0.005. (D) Representative fields of TA muscles 1 wk after coelectroporation of GFP-LC3 and either control or a combination of HDAC1 and HDAC2 expression plasmids. (Scale bars, 40 μm.) (E) Counts of GFP-LC3 punctae per myofiber in muscles coelectroporated with GFP-LC3 and either control or a combination of HDAC1 and HDAC2 expression plasmids. n = 6–8 (≈400 fibers each). Values are mean ± SEM. **P < 0.005. (F) Western blot analysis for autophagy markers LC3 I-II and p62, HDAC1, and HDAC2 in TA muscles 8 d after electroporation of either a control plasmid or a combination of HDAC1 and HDAC2 expressing plasmids.
To investigate whether deletion of HDAC1 and HDAC2 in skeletal muscle affects autophagy induction at the transcriptional level, we measured transcripts of genes involved in autophagy flux at baseline and in response to 48 h of fasting. For all of the genes analyzed—atg5, atg7, gabarapl1, LC3, and p62—expression was induced upon fasting in control mice, but to a lesser extent in dKO mice (Fig. 3C).
HDAC1 and HDAC2 Overexpression Is Sufficient to Promote Autophagy Flux.
To investigate whether the ectopic expression of HDAC1 and HDAC2 in skeletal muscle was sufficient to increase the number of autophagosomes and to promote autophagy flux, we coelectroporated TA muscles from WT mice with GFP-LC3 plasmid and either a control plasmid or a combination of HDAC1 and HDAC2 expression plasmids. After 8 d, we analyzed the presence of autophagosomes by fluorescence. Muscles coelectroporated with HDAC1 and HDAC2 showed a significantly higher number of GFP-LC3 punctae compared with those coelectroporated with a control plasmid (Fig. 3 D and E). Importantly, Western blot analysis revealed an increase of lipidated LC3-II and a decrease in p62 protein levels in muscles that overexpressed HDAC1 and HDAC2 compared with those electroporated with a control plasmid (Fig. 3F), indicating that HDAC1 and HDAC2 overexpression promotes autophagy flux.
dKO Mice Develop a Metabolic Disorder with the Histological Onset of Myopathy.
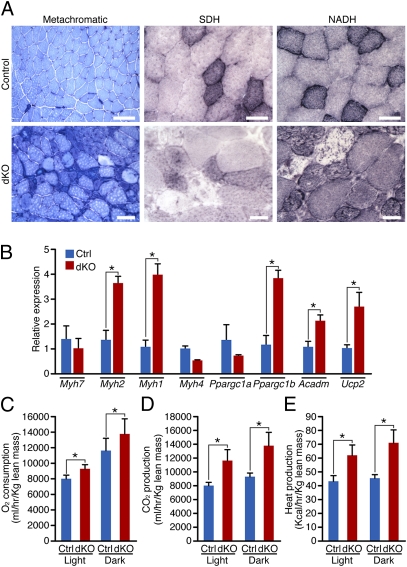
Gene array analysis performed on 4-wk-old TA muscles showed that only 18 transcripts out of 45,282 detected were significantly up-regulated in dKO mice (Fig. S7). Gene ontology analysis using the DAVID bioinformatics browser (40) indicated that the most significantly enriched up-regulated genes in dKO mice participate in metabolism (Fig. S8). On the basis of these findings, we compared the metabolic state of skeletal muscle of control and dKO mice. At 4 wk of age, histological analysis and enzymatic staining of TA muscle did not reveal any major differences between control and dKO mice at baseline (Fig. S9A). Expression of different myosin heavy chain (Myh) genes and genes important for metabolic regulation, such as Ppargc1a and Ppargc1b, was analyzed at baseline and in fasting conditions, in which the requirements for the protective role mediated by autophagy are more pronounced. No significant differences were detected in the expression of Myh or Ppargc1a genes between control and dKO mice, whereas fasting induced a significant reduction in the expression of Ppargc1b only in control mice (Fig. S9B). In contrast, at 7 wk of age, when the histological onset of the myopathy was observed, metachromatic staining of the TA muscle revealed an increase in oxidative fibers (darker blue) in dKO mice compared with control mice; succinic dehydrogenase (SDH) and NADH staining highlighted compromised activity of mitochondria in a subset of myofibers (Fig. 4A). Real-time RT-PCR analysis showed up-regulation of Myh2, Myh1, Ppargc1b, Acadm (a protein essential for converting fatty acids to energy in mitochondria), and Ucp2 at baseline, in dKO compared with control mice (Fig. 4B).
Fig. 4.
Altered skeletal muscle energy expenditure in dKO mice. (A) Histological sections of TA isolated from 7-wk-old control and dKO mice were analyzed by metachromatic ATPase, SDH, and NADH staining. (Scale bars, 100 μm for metachromatic ATPase, 40 μm for SDH and NADH staining.) (B) Real-time RT-PCR analysis of different Myh genes, Ppargc1a, Ppargc1b, Acadm, and Ucp2 in control (Ctrl) and dKO mice, at 7 wk of age. Data are expressed as mean ± SEM. n = 4. *P < 0.05. dKO mice show significantly higher (C) O2 consumption, (D) CO2 production, and (E) heat production than control mice, between 5 and 7 wk of age. *P < 0.05. Values are mean ± SEM. n = 7.
Suspecting an altered energy state in dKO mice, we assessed metabolic parameters of control and dKO mice, using metabolic chambers. dKO mice showed a significant increase in O2 production, CO2 consumption, and heat production, both in light and dark periods (Fig. 4 D–F), indicative of elevated energy expenditure. No significant differences were detected in physical activity, suggesting that heightened energy expenditure is not a consequence of increased voluntary movement or differences in body weight, lean mass, or food or water consumption between control and dKO mice (Fig. S10 A–E).
HFD Prevents Myopathy in dKO Mice.
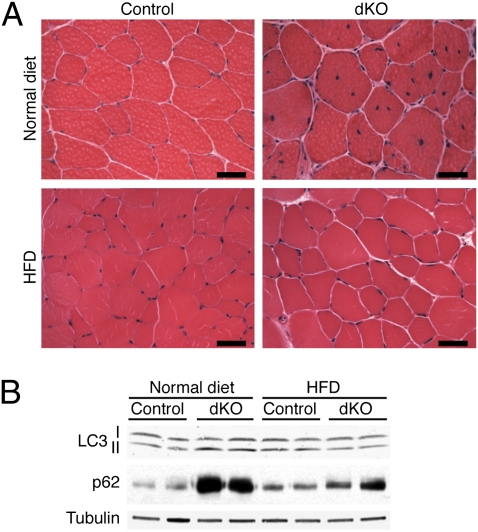
Because dKO mice exhibited higher energy expenditure and increased expression of genes involved in fatty acid metabolism, we challenged them with an HFD starting at weaning age. As previously reported (41), 8 wk of HFD was not sufficient to cause an increase in body weight in control or dKO female mice (Fig. S11), but surprisingly, 3-mo-old dKO mice showed none of the histological features of myopathy, such as muscle degeneration or centronuclear myofibers (Fig. 5A). Analysis of autophagy markers after 8 wk of normal chow or HFD showed no major differences in the lipidation status of LC3 (Fig. 5B). Strikingly, however, dKO mice on HFD did not accumulate p62 protein (Fig. 5B), nor did they show significant differences in the expression of Myh genes or Ppargc1b compared with control mice (Fig. S12A). HFD did not alter the expression of other HDACs in skeletal muscle, except for Hdac3, in both control and dKO mice (Fig. S12B).
Fig. 5.
HFD prevents the block of autophagy flux, myopathy, and perinatal mortality in dKO mice. (A) H&E sections of TA muscle of 3-mo-old control and dKO mice, fed with either normal chow or HFD for 8 wk. (Scale bars, 40 μm.) (B) Western blot analysis for autophagy markers LC3 I-II and p62 in control and dKO mice, fed with normal chow or HFD for 8 wk. Tubulin was used as loading control.
Because HFD was able to rescue the phenotype of adult dKO mice, we wondered whether feeding pregnant mice HFD could prevent the perinatal lethality observed in approximately 40% of dKO mice. Six pregnant mice were fed HFD, starting from the day after conception. We obtained a total of 45 pups, 19 of which were dKO, and none of them died perinatally. Ten days after birth, by genotyping, we confirmed no significant difference between the number of expected and obtained dKO mice (Fig. S13).
Discussion
Using a genetic approach to delete HDAC1 and HDAC2 specifically in skeletal muscle, this study reveals a previously unrecognized function of these enzymes in regulating autophagy flux and skeletal muscle homeostasis in vivo (Fig. 6). One copy of a WT allele of either Hdac1 or Hdac2 is sufficient for proper skeletal muscle development, suggesting a redundant function of these genes. When both HDAC1 and HDAC2 were deleted from skeletal muscle, approximately 40% of mice died perinatally, showing myofibrillar and mitochondrial abnormalities, accumulation of p62, and up-regulation of the activated form of AMPK, a marker of energy deficit. Mice mutant in autophagy genes show similar features (26, 27), highlighting the importance of this cellular mechanism for survival during the perinatal period of energy deprivation. dKO mice that survived the first day of life developed a progressive myopathy, starting from 7 wk of age, characterized by muscle degeneration and regeneration, similar to mice defective in autophagy (3, 4, 7–9). Importantly, before any histological evidence of myopathy, autophagosome induction and formation were impaired in dKO mice in response to fasting. Conversely, ectopic expression of HDAC1 and HDAC2 in skeletal muscle was sufficient to accumulate autophagosomes and promote autophagy flux.
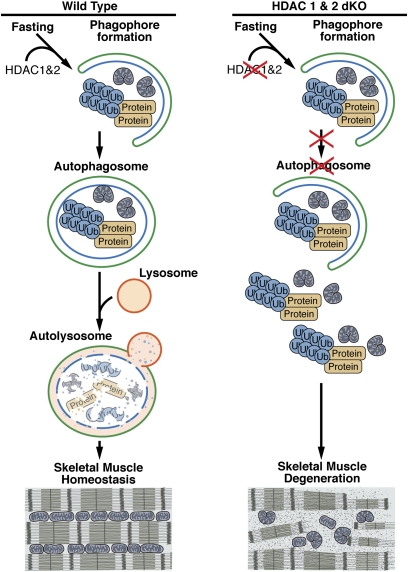
Fig. 6.
Model for the regulation of skeletal muscle homeostasis by HDAC1 and HDAC2. HDAC1 and HDAC2 promote autophagosome induction and formation. In the absence of HDAC1 and HDAC2 in skeletal muscle, autophagy flux is impaired, causing accumulation of protein aggregates and damaged mitochondria and eventually muscle degeneration.
Gene array analysis performed on skeletal muscle of 4-wk-old mice revealed only modest changes in gene expression between control and dKO mice (0.04% up-regulated and 0.08% down-regulated), suggesting that HDAC1 and HDAC2 do not function as global repressors of gene transcription in skeletal muscle. The specificity of deregulated genes in dKO mice suggested possible alterations in the metabolic status of skeletal muscle. Indeed, skeletal muscle of dKO mice exhibited a shift in metabolism toward a more oxidative state, and dKO mice showed increased energy expenditure, although the altered respiratory rate observed in dKO mice may be a consequence of abnormalities in the diaphragm.
Because the changes in metabolism of dKO mice appeared only after the histological onset of myopathy, except for the modulation of Ppargc1b expression after fasting, they are likely a consequence of compromised skeletal muscle homeostasis. However, there is a mechanistic and molecular link between autophagy and cellular metabolism (2, 34). Indeed, the breakdown products from autophagy are substrates for both new biosynthesis and energy production. Recently, a unique role of autophagy in regulating glucose metabolism after acute exercise has been shown (13). In addition, autophagy is critical for survival during fasting and for reprogramming of cell metabolism (34). After a period of starvation, newborn pups switch from the placenta, high in carbohydrates and amino acids, to milk, a high-fat and low-carbohydrate diet. Although 8 wk of HFD did not trigger autophagy in control mice, it is intriguing that dKO mice fed with HFD did not develop myopathy, did not accumulate autophagy intermediates, and showed no changes in the expression of genes involved in oxidative metabolism, despite similar expression of other Hdacs, compared with control mice.
HFD leads to an imbalance between energy intake and expenditure, and eventually obesity. One of the major effects of HFD in skeletal muscle is lipid accumulation. Multiple pathways may contribute to lipid accumulation in skeletal muscle upon HFD, such as increased fatty acid uptake into muscle from circulation and/or defective muscle mitochondrial metabolism (42). Conflicting data regarding the expression and activity of Ppargc1 genes, which are key regulators of mitochondrial biogenesis and fatty acid oxidation in skeletal muscle (43) have been reported (42, 44). In addition, HFD reduces insulin signaling and autophagy flux, with a consequent impairment of GLUT4 translocation to the plasma membrane (13, 45, 46). It is possible that HFD rescues the dKO phenotype by prolonging caloric availability and by diminishing the requirement of autophagy for skeletal muscle metabolism homeostasis. These findings further confirm a link between autophagy, metabolism, and cellular homeostasis.
Recently it was shown that low-protein diet (LPD), through triggering autophagy, ameliorates myopathy in Col6a1−/− mice, a model of collagen VI muscular dystrophy (17). The rationale for feeding Col6a1−/− mice with LPD was that these mice displayed a delay in the induction of autophagy, contrary to dKO mice, which showed a block in autophagy flux even after 48 h of starvation. Because nutrient availability and metabolism are tightly linked and may promote disease, such as obesity and type 2 diabetes, further elucidation of the role of autophagy and its connection to metabolic pathways could provide insight for future clinical applications.
Although the present study reveals a heretofore unrecognized role of HDACs 1 and 2 in the control of autophagy, the precise targets for deacetylation by these HDACs that mediate this effect remain to be defined. Because HDACs repress gene transcription by deacetylating histones, it will be of interest to determine whether HDACs 1 and 2 act at the transcriptional level to modulate genes involved in the autophagy pathway. In addition, given that HDACs have been reported to deacetylate proteins other than histones (19), it is conceivable that HDACs 1 and 2 directly deacetylate autophagy proteins to regulate autophagosome induction.
HDAC inhibitors are used clinically as treatments for epilepsy, bipolar disorder, and cancer (47–49). Considering the broad use of HDAC inhibitors in clinical settings, and the role of HDACs 1 and 2 in regulating autophagy flux and skeletal muscle homeostasis, possible adverse consequences and side effects of administering HDAC inhibitors systemically should be considered.
Methods
Mice.
HDAC1 and HDAC2 conditional mutant mice, previously described (20), were crossed with myogenin-Cre transgenic mice, which express Cre recombinase under the control of the Mef2c skeletal muscle enhancer linked to the myogenin promoter (23). Mice were fed normal chow (no. 2016; Harlan Teklad Global) containing 12% kilocalories from fat or HFD (no. D12492; Research Diets) containing 60% kilocalories from fat.
Histology and Immunohistochemistry.
H&E staining, NADH staining, metachromatic ATPase staining, Evans blue dye uptake experiments, succinic dehydrogenase staining, and immunohistochemistry were performed as previously described (50–53). More information is provided in SI Methods. Electron microscopy was performed on P1 mouse diaphragms by the University of Texas Southwestern Electron Microscopy Core Facility.
Western Blot Analysis.
Western blot analyses were performed on TA muscles as previously described (52). More information is provided in SI Methods.
DNA Delivery by Electroporation.
Experiments were performed on 4-wk-old mice as previously described (50). When indicated, 8 d after the electroporation, some mice were fasted for 24 h, as previously described (38). For the ubiquitin-proteasome activity assay, TA muscles of 4-wk-old mice were cotransfected with dsRED and Ub-G76V-YFP plasmid, which is an ubiquitin-proteasome pathway activity reporter, as previously described (7). GFP-LC3 plasmid was received from J. A. Hill (54) (University of Texas Southwestern, Dallas, TX); Ub-G76V-YFP (37) (Addgene plasmid 11949) was purchased from Addgene. More information is provided in SI Methods.
RNA Isolation and RT-PCR.
RNA was isolated from TA muscles and RT-PCR was performed as previously described (50). Probe and primer information is given in SI Methods.
Microarray Analysis.
Total RNA was isolated from TA muscle of 4-wk-old mice (n = 3 control; n = 3 dKO mice). Microarray analysis was performed by the University of Texas Southwestern Microarray Core Facility using the Mouse Genome Illumina Mouse-6 V2 BeadChip. Genes that were up-regulated at least twofold were analyzed. More information is provided in SI Methods.
Metabolic Cage Studies.
Metabolic cage studies were performed by the University of Texas Southwestern Mouse Metabolic Phenotyping Core. Approximately 5-wk-old male dKO and control mice, receiving normal chow, were placed in TSE metabolic chambers for an initial 5-d acclimation period, followed immediately by a 4.5-d experimental period with data collection.
Statistical Analyses.
We performed statistical comparison of data, presented as mean ± SEM, using the two-tailed Student t test. We considered P < 0.05 to be statistically significant. A χ2 test was used to test whether the difference between the expected and observed dKO mice was statistically significant. An observed χ2 = 5.99 corresponds to P = 0.05; χ2 = 9.21 corresponds to P = 0.01.
Creatine Kinase Assay.
Creatine kinase levels were quantified from serum of 4-wk-old mice by using the enzyChrom creatine kinase assay kit from BioAssay Systems, according to the manufacturer's instructions.
Supplementary Material
Acknowledgments
We thank Rusty L. Montgomery for generating the HDAC1loxP/loxP and HDAC2loxP/loxP mice; Laura Brule for assistance at the University of Texas (UT) Southwestern Metabolic Phenotyping Core Facility; Christopher Gilpin and the UT Southwestern Electron Microscopy Core for electron microscopy advice and expertise; Beverly A. Rothermel for scientific advice; Nadia Rosenthal and Leslie Leinwand for comments on the manuscript; Cheryl Nolen for technical help; Jose Cabrera for graphics; Jennifer Brown for editorial assistance; and N. P. Dantuma for Addgene plasmid Ub-G76V-YFP. This work was supported by grants from the National Institutes of Health and by Grant I-0025 from the Robert A. Welch Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121159109/-/DCSupplemental.
References
- 1.Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: With an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 4.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 5.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 7.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Ebato C, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, Hara T. Intracellular quality control by autophagy: How does autophagy prevent neurodegeneration? Autophagy. 2006;2:302–304. doi: 10.4161/auto.2945. [DOI] [PubMed] [Google Scholar]
- 12.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He C, et al. Exercise-induced Bcl-2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012 doi: 10.1038/nature10758. 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raben N, et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grumati P, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 18.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 20.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. Redundant control of adipogenesis by histone deacetylases 1 and 2. J Biol Chem. 2010;285:14663–14670. doi: 10.1074/jbc.M109.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, et al. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci USA. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng TC, Wallace MC, Merlie JP, Olson EN. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science. 1993;261:215–218. doi: 10.1126/science.8392225. [DOI] [PubMed] [Google Scholar]
- 25.Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128:4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 32.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 33.Ichimura Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 34.Schiaffino S, Mammucari C, Sandri M. The role of autophagy in neonatal tissues: Just a response to amino acid starvation? Autophagy. 2008;4:727–730. doi: 10.4161/auto.6143. [DOI] [PubMed] [Google Scholar]
- 35.Hardie DG. Minireview: the AMP-activated protein kinase cascade: The key sensor of cellular energy status. Endocrinology. 2003;144:5179–5183. doi: 10.1210/en.2003-0982. [DOI] [PubMed] [Google Scholar]
- 36.Carling D. The AMP-activated protein kinase cascade—a unifying system for energy control. Trends Biochem Sci. 2004;29:18–24. doi: 10.1016/j.tibs.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Menéndez-Benito V, Verhoef LG, Masucci MG, Dantuma NP. Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum Mol Genet. 2005;14:2787–2799. doi: 10.1093/hmg/ddi312. [DOI] [PubMed] [Google Scholar]
- 38.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 41.Hwang LL, et al. Sex differences in high-fat diet-induced obesity, metabolic alterations and learning, and synaptic plasticity deficits in mice. Obesity (Silver Spring) 2010;18:463–469. doi: 10.1038/oby.2009.273. [DOI] [PubMed] [Google Scholar]
- 42.Turner N, et al. Excess lipid availability increases mitochondrial fatty acid oxidative capacity in muscle: evidence against a role for reduced fatty acid oxidation in lipid-induced insulin resistance in rodents. Diabetes. 2007;56:2085–2092. doi: 10.2337/db07-0093. [DOI] [PubMed] [Google Scholar]
- 43.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Sparks LM, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 45.Tremblay F, Lavigne C, Jacques H, Marette A. Defective insulin-induced GLUT4 translocation in skeletal muscle of high fat-fed rats is associated with alterations in both Akt/protein kinase B and atypical protein kinase C (zeta/lambda) activities. Diabetes. 2001;50:1901–1910. doi: 10.2337/diabetes.50.8.1901. [DOI] [PubMed] [Google Scholar]
- 46.Zierath JR, Houseknecht KL, Gnudi L, Kahn BB. High-fat feeding impairs insulin-stimulated GLUT4 recruitment via an early insulin-signaling defect. Diabetes. 1997;46:215–223. doi: 10.2337/diab.46.2.215. [DOI] [PubMed] [Google Scholar]
- 47.Kumagai T, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer. 2007;121:656–665. doi: 10.1002/ijc.22558. [DOI] [PubMed] [Google Scholar]
- 48.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phiel CJ, et al. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 50.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moresi V, et al. Tumor necrosis factor-alpha inhibition of skeletal muscle regeneration is mediated by a caspase-dependent stem cell response. Stem Cells. 2008;26:997–1008. doi: 10.1634/stemcells.2007-0493. [DOI] [PubMed] [Google Scholar]
- 52.Liu N, et al. Mice lacking microRNA 133a develop dynamin 2–dependent centronuclear myopathy. J Clin Invest. 2011;121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quiat D, et al. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc Natl Acad Sci USA. 2011;108:10196–10201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu H, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.