Abstract
Helicases are ubiquitous enzymes that unwind double-stranded DNA (dsDNA) to reveal single-stranded DNA (ssDNA) during essential processes such as replication, transcription, or repair. The Escherichia coli RecQ protein is a 3′ to 5′ helicase, which functions in the processes of homologous recombination and replication fork restart. Here, we analyzed the relationship between ATP hydrolysis by RecQ and its translocation on ssDNA. We monitored a single round of RecQ translocation on ssDNA by measuring the rates of inorganic phosphate release during translocation, and the dissociation of RecQ from ssDNA. We find that RecQ translocates with a rate of 16( ± 4) nucleotides/s and moves on average only 36( ± 2) nucleotides before dissociating. Fitting to an n-step kinetic model suggests that the helicase displays a nonuniform translocation mechanism in which it moves approximately five nucleotides rapidly before undergoing a rate-limiting kinetic slow step. Unexpectedly, RecQ requires a length of 34( ± 3) nucleotides to bind and translocate on ssDNA. This large site size suggests that several monomers are required to bind DNA prior to translocation. Energetically, the RecQ helicase couples the hydrolysis of one ATP molecule to the translocation of more than one nucleotide (1.6 ± 0.3). Thus, our data show that RecQ translocates on ssDNA by efficiently coupling the hydrolysis of one ATP molecule into structural alterations that result in movement of approximately two nucleotides, presumably by an inchworm mechanism. These attributes are consistent with the function of RecQ in recombination and replication.
Keywords: DNA motor, DNA repair, DNA unwinding, recombination
A diverse group of enzymes, the helicases play a prominent role in replication, repair, and recombination by unwinding double-stranded DNA (dsDNA). Moreover, these motor enzymes function by translocating on single-stranded DNA (ssDNA)—a process that can be as important in vivo as unwinding dsDNA. Important cellular processes that are carried out by translocating enzymes include removal of secondary structure from ssDNA, movement of Holliday junctions, and displacement of bound proteins from ssDNA (1).
A 3′ to 5′ helicase from E. coli, RecQ is the founding member of the RecQ-family of helicases (2). These enzymes belong to helicase superfamily-2 (SF2), but are more closely homologous to one another (3–5). Members of this family include Bloom (BLM), Werner (WRN), RECQ1, RECQ4, and RECQ5 from humans, Sgs1 from Saccharomyces cerevisiae, and Rqh1 from Schizosaccharomyces pombe (6). These proteins play important roles in DNA recombination and repair in their respective organisms. The function of BLM, WRN, and RECQ4 proteins are of particular interest because mutations in these helicases lead to Bloom’s, Werner’s, and Rothmund-Thomson syndromes, respectively. These genetic disorders are characterized by genomic instability and an increased incidence of cancer (6).
Recent studies of helicases are demonstrating the importance of translocation in lieu of, or in addition to, helicase activity. Helicases involved in recombination, such as Srs2 from S. cerevisiae, BLM, and RECQ5 negatively regulate recombination by acting as translocases to remove Rad51 protein from ssDNA (7–10), demonstrating a major biological function for translocation on ssDNA. A further example of an essential cellular function for translocation is provided by the DExH/D family of proteins. This family comprises a group of RNA helicases that are essential for RNA metabolism. The DExH/D helicases can unwind duplex RNA (11, 12), but equally importantly, these enzymes remodel both RNA structures and protein-ssRNA complexes (13).
The mechanisms of dsDNA unwinding and ssDNA translocation by RecQ are not well understood. In vitro, RecQ can unwind a variety of dsDNA substrates that mimic intermediates formed during homologous recombination (14, 15). RecQ can unwind plasmid-length dsDNA in the presence of SSB, which sequesters the unwound ssDNA strands (16). DNA unwinding by RecQ is cooperative with respect to ATP concentration, with a Hill coefficient of about 3, indicating that at least three monomers of RecQ comprise the active unwinding unit. However, from these steady-state experiments, it was not possible to determine the energetics of DNA unwinding relative to ATP hydrolysis.
Several translocation mechanisms for helicases have been proposed: Two popular mechanisms are the inchworm and the Brownian ratchet (1, 17, 18) . The SF1 helicases Bacillus stearothermophilus PcrA and E. coli UvrD move using an “inchworm” mechanism (18–21). The motor domains of these enzymes are comprised of two DNA binding sites. The binding of ATP induces a change in the position of the binding sites relative to one another, resulting in a movement wherein the trailing domain moves towards the leading one. When bound to DNA, the hydrolysis of ATP followed by release of ADP and phosphate causes coordinated domain opening and translocation of the helicase. These conformational changes, coupled to cycles of ATP hydrolysis and release of ADP and phosphate, result in a stepping movement of one nucleotide along the DNA lattice. In contrast, kinetic studies of NS3h helicase from Hepatitis C virus were interpreted to show that this enzyme unwinds DNA using a Brownian motor mechanism (17, 22). In this model, ATP hydrolysis does not drive sequential steps of translocation; rather, when ATP is bound, movement is random. Directional movement results from an asymmetric energy landscape, which arises from binding of the helicase to ssDNA and the associated cycles of nucleotide cofactor binding and hydrolysis. Unlike PcrA and UvrD, NS3h has one binding site for DNA. When the helicase binds ATP, the affinity for ssDNA is decreased. In this state, NS3h can move randomly, forward or backward, on the DNA lattice. Upon hydrolysis of the ATP, the helicase has a higher affinity for ssDNA and is locked into its current position. In contrast to the inchworm model, in this model ATP hydrolysis is not tightly coupled to translocation and multiple ATP molecules are hydrolyzed, inefficiently, before the helicase can take a step forward (1, 17).
In this paper, the pre-steady-state kinetics of translocation by RecQ on ssDNA are defined. We measured the rates of ATP hydrolysis by RecQ by tracking the phosphate released during a single round of translocation on oligonucleotides of increasing length. In parallel, we measure the lifetimes of RecQ-ssDNA complexes. The data were analyzed using an n-step model for enzymes translocating on ssDNA (20). We established parameters for translocation including the rate, processivity, kinetic step size, and coupling efficiency. Both a kinetic mechanism and a translocation model are proposed for RecQ and are discussed in the context of RecQ-family helicase function.
Results
Fluorescent Assays Measure Different Kinetic Aspects of Translocation
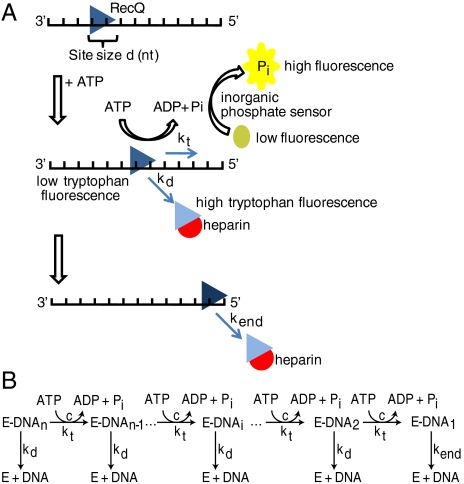
To monitor translocation on ssDNA, we took advantage of two fluorescence methods (Fig. 1A): (i) measuring the amount of ATP hydrolyzed in one round of translocation by RecQ on ssDNA by using a fluorescent sensor for inorganic phosphate (Pi) (23, 24); and (ii) measuring the lifetime of RecQ-ssDNA complexes by monitoring the dissociation kinetics via changes in the intrinsic tryptophan fluorescence (23, 24). The fluorescent sensor for Pi is phosphate binding protein (PBP) labeled with a coumarin fluorophore (MDCC) as described previously (25, 26).
Fig. 1.
Kinetic methods used to study translocation of RecQ on ssDNA. (A) RecQ (triangle) with site size d (in nucleotides) is prebound to ssDNA. Addition of ATP initiates translocation. Each kinetic step is coupled to hydrolysis of ATP, described by the coupling efficiency, c. By measuring the phosphate released using the fluorescent phosphate-binding protein, the coupling coefficient and rate of translocation are determined. Additionally, the quenching of tryptophan fluorescence was monitored to determine kinetic lifetimes. (B) Schematic of the n-step kinetic model describing translocation. The helicase (E) is initially prebound to ssDNA (DNA). The enzyme translocates along the lattice in distinct, measurable kinetic steps (E-DNAi, the ith species), hydrolyzing ATP until reaching the end.
In a Single Round of Translocation on ssDNA, RecQ Displays a Rapid Phase of ATP Hydrolysis Followed by a Slower Phase
Both the rate of translocation and the efficiency of coupling to ATP hydrolysis by RecQ on ssDNA were monitored using MDCC-PBP. To interpret the results, we used an n-step kinetic model defined previously to describe translocation by a helicase on ssDNA (24) (Fig. 1B). To ensure that the reaction reflected just a single round of translocation by RecQ, we used heparin to sequester the free RecQ protein. RecQ was initially incubated with ssDNA, and then rapidly mixed with ATP and heparin using a stopped-flow apparatus. RecQ is expected to translocate, dissociate from the ssDNA, and then bind to the heparin, resulting in only one round of translocation. In Fig. 2A, kinetic traces from the phosphate-release assay are shown for ssDNA of increasing length. We observed an initial rapid phase of phosphate release, followed by a substantially slower steady-state increase in phosphate production. As the length of ssDNA was increased, the duration and amplitude of the rapid phase increased as well. Only a slight amount of phosphate (1 PiRecQ) was produced when an oligonucleotide of 30 nucleotides (nt) was used (Fig. 2A). We inferred that the rapid phase of phosphate release is due to translocation on ssDNA, whereas the slow steady-state rate of phosphate release is from RecQ that transiently dissociates from the heparin, and subsequently rebinds the ssDNA (Fig. S1A). Heparin alone does not stimulate ATP hydrolysis by RecQ (Fig. S1B). Mixing of RecQ with a solution of dT70, heparin, and ATP results in the disappearance of the rapid phase of phosphate release, whereas a low level steady-state phase is still detectable (Fig. S1B). These control experiments support our interpretation that the initial rapid phase is due to translocation and ATP hydrolysis by RecQ, whereas the slow phase is due to infrequent rebinding of RecQ to ssDNA. A similar effect was also observed for experiments monitoring translocation of UvrD on ssDNA (23).
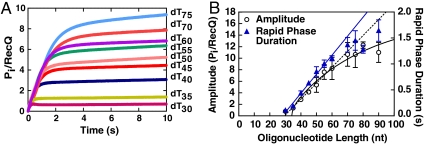
Fig. 2.
In a single round of translocation, RecQ moves more than one nucleotide per molecule of ATP hydrolyzed. (A) Kinetic traces for the release of phosphate by RecQ for oligonucleotides of varying lengths. Each trace was fit to Eq. 3 to give an amplitude and duration for phosphate release. (B) Plot of the amplitude (black circles) and duration (blue triangles) for the rapid phase in (A) versus DNA length. The dashed black line is a linear fit of the amplitude data up to 60 nt; the solid black line is the best fit of the data to Eq. 8. The solid blue line is a linear fit of the duration data up to 60 nt.
The kinetic traces for phosphate release were fit to Eq. 3, and the amplitude, A, of the rapid phase was plotted for each DNA length (Fig. 2B). Above a length of 30 nucleotides, the amplitude increases linearly with length until 60 nucleotides, whereupon the points noticeably begin to deviate from a line. The concentration of ssDNA is saturating with respect to the concentration of RecQ: increasing the concentration of the shortest oligonucleotide that resulted in an appreciable rapid phase of phosphate release (dT35) showed no change in the amplitude of phosphate release (Fig. S2). Thus, we interpret the deviation of the observed amplitude from a straight line to mean that, above 60 nucleotides, the length of the ssDNA is greater than the processivity of RecQ. On these longer substrates, RecQ does not always reach the end of the lattice, and dissociates from internal DNA sites. Because the amplitude for lengths below 60 nucleotides increases linearly, we assume that RecQ reaches the end of these molecules, and then dissociates. The linear portion of the data in Fig. 2B was fit to a line and a slope of 0.28 ± 0.02 Pi/[RecQ]/nucleotide was determined (Fig. 2B, black dashed line). The reciprocal of the slope is an estimate of the coupling efficiency per nucleotide, c/m, of translocation by RecQ (see Table 1 for definitions). However, one also needs to correct for the fact that RecQ binds randomly to the oligonucleotide substrate rather than starting at an end (27). Thus, on average, only half of the length of each oligonucleotide is available for translocation. With this assumption, the slope is multiplied by 2, resulting in 0.56 ± 0.04 ATP molecules hydrolyzed to move one nucleotide. This approximate estimate indicates that RecQ moves approximately two nucleotides for every ATP molecule hydrolyzed.
Table 1.
Kinetic parameters for translocation of RecQ on ssDNA
| Kinetic Parameter | Definition | Units | Dissociation Data | Pi Release Data |
| kt | Forward stepping rate | steps/s | 3.0 ± 0.6 | - |
| kend | Dissociation rate from DNA end | s-1 | 6 ± 1 | - |
| r | Ratio of binding DNA internally vs. end | - | 7.1 ± 0.7 | - |
| m | Kinetic step size | nt/step | 5.3 ± 0.9 | - |
| kd | Dissociation rate (0.5 mg/mL heparin) | s-1 | 0.75 ± 0.01 | - |
| kd | Dissociation rate (no heparin) | s-1 | 0.43 ± 0.04 | - |
| m•kt | Translocation rate | nt/s | 16 ± 4 | 14.0 ± 0.4 |
| d | Binding site size | nt | 34 ± 3 | 34 ± 0.7 |
| c | Coupling efficiency per step | ADP/step | - | 3.4 ± 0.4 |
| c/m | Coupling efficiency per nucleotide | ADP/nt | - | 0.6 ± 0.2 |
| m/c | Nucleotides translocated per ATP hydrolyzed | nt/ATP | - | 1.6 ± 0.3 |
| P | Processivity (0.5 mg/mL heparin) | - | 0.952 ± 0.005 | |
| P | Processivity (no heparin) | - | 0.972 ± 0.002 | - |
| Nav = 1/(1-P) | Average distance (0.5 mg/mL heparin) | nt | 21 ± 2 | - |
| Nav = 1/(1-P) | Average distance (no heparin) | nt | 36 ± 2 | - |
The parameters kt, kend, and r were determined by fitting the data in Fig. 4A to Eq. 6. The values are reported as an average of the three datasets with standard deviation. The dissociation rate constant from internal ssDNA sites was determined from experiment with poly dT using Eq. 4 (Fig. S5). The values for the binding site size, d, and kinetic step size, m, were determined from linear least squares fitting to the data in Fig. 4B. The coupling efficiency, c, and site size, d, in the “Pi release” data column were determined from the fit of Eq. 8 to the amplitudes of the phosphate-release data in Fig. 2B.
To determine the rate of translocation, the reciprocal of the observed kinetic rate constant for phosphate production from the rapid phase of each kinetic trace was plotted versus DNA length (Fig. 2B, blue triangles). The reciprocal of the observed rate constant is related to the kinetic lifetime of the complex responsible for the rapid phase of phosphate release. The duration of this phase increases with the length of the oligonucleotide, consistent with the idea that RecQ is translocating to the end of the ssDNA and then dissociating. However, again, the duration of the rapid phase is linear only up to a length of 60 nucleotides, again implying that lengths above 60 nucleotides exceed the processive distance for translocation by RecQ, and that RecQ begins to dissociate from an internal site prior to reaching an end. A linear fit to the data between 35 and 60 nucleotides gives a slope of 0.037 ± 0.001 seconds/nucleotide (Fig. 2B, blue line). Again, we assume binding of RecQ occurs, on average, in the middle of each oligonucleotide. Thus, from the slope of the line and our assumption, we estimate a translocation rate of 14 ± 0.4 nt/s for RecQ on ssDNA.
The Lifetime of RecQ-ssDNA Complexes Reveals Slower Dissociation from Internal ssDNA Binding Sites than from ssDNA Ends.
Deduction of a comprehensive kinetic model for RecQ function requires an evaluation of many kinetic states (24). To both reduce the number of fitting parameters and increase the accuracy of the model, we independently determined the rate constant for dissociation of RecQ from internal ssDNA binding sites, kd. Poly dT was used because of its large size (approximately 4,000 nt), ensuring a high probability of dissociation from an internal site relative to dissociation from an end, and to permit direct comparisons to the dT-containing oligonucleotides used herein. We observed that the native tryptophan fluorescence of RecQ was quenched upon addition of poly dT (Fig. S3) and took advantage of this property to measure the rate of RecQ dissociation from both poly dT and the oligonucleotides that were used in the phosphate-release assays.
RecQ was prebound to the ssDNA, and then rapidly mixed with ATP and heparin: dissociation of RecQ resulted in an increase in fluorescence (Fig. 3A). As the length of DNA was increased, the duration of the dissociation trace increased as well (Fig. 3A). The fluorescence increase is due to the dissociation of RecQ from ssDNA and not from binding of the helicase to heparin: experiments in which RecQ was mixed with heparin show no change in tryptophan fluorescence (Fig. S4). Each kinetic trace for dissociation of RecQ from ssDNA was fit to a single exponential function to obtain an observed rate constant for dissociation, kobs, and amplitude. Plotting the reciprocal of kobs versus the length of the oligonucleotides showed that the characteristic relaxation time of the RecQ-ssDNA complexes increased linearly up to 60 nucleotides (Fig. 3B) and then began to plateau; this result coincided almost exactly with the data obtained from the phosphate-release assay (Fig. 2B).
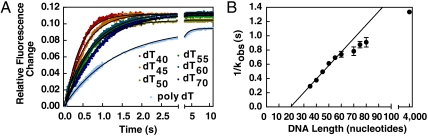
Fig. 3.
The dwell time of RecQ on ssDNA increases with oligonucleotide length. (A) Kinetic traces for the dissociation of RecQ-ssDNA complexes, measured by fluorescence, for oligonucleotides of varying lengths. The traces were each fit to Eq. 4 (black lines). The amplitudes are the same for all substrates. (B) The reciprocal of the rate constants determined from (A) are plotted as a function of DNA length. The solid line is a fit of the initial linear region (up to 60 nt); the slope is 0.019 ± 0.001 s/nt.
For poly dT, RecQ dissociated much more slowly than from the other DNA substrates, indicating that the length of poly dT is much greater than the processivity of RecQ. The lifetime of the RecQ-ssDNA complexes increases with DNA length because kobs becomes more dependent on the dissociation rate constant from internal DNA sites, kd, which is slower than the dissociation rate constant from DNA ends, kend (Table 1). The data for dT35-dT60 were fit to a line to obtain a slope of 0.019 ± 0.001 s/nt. The inferred translocation rate is the inverse of the slope corrected for the random binding of RecQ to ssDNA. Thus, the inverse of the slope was divided by two to obtain an estimate of approximately 26 nt/s for the translocation rate. Global fitting of all kinetic traces, described below, yielded a value of 16 ± 4 nt/s, which is a more robust measure of the translocation rate (Table 1).
In addition, we could determine the translocation processivity of RecQ from the values of the forward and dissociation rate constants (Eq. 1) to be 0.952 ± 0.005; this parameter is the probability that RecQ will translocate with an additional kinetic step on ssDNA. Consequently, on average, RecQ translocates 21 ± 2 nucleotides before dissociating from ssDNA (Eq. 2). This value, however, is calculated in the presence of heparin, which we determined actually promotes dissociation of RecQ from ssDNA (Fig. S5A). To determine the unperturbed processivity, we measure kd as a function of heparin concentration, and extrapolated the values to zero heparin (Fig. S5B). In the absence of heparin, P = 0.972 ± 0.002, and RecQ translocates on average 36 ± 2 nucleotides before dissociating (Table 1).
RecQ Couples the Hydrolysis of One ATP Molecule to the Movement of Approximately Two Nucleotides During Translocation.
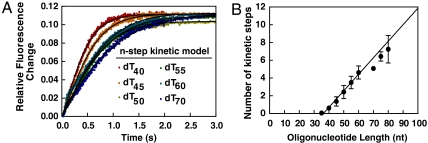
To determine the kinetic parameters for translocation by RecQ on ssDNA, we used the n-step kinetic model defined previously (24). Using this formalism, the translocation of a helicase on, and dissociation from, DNA are given by Eq. 6. As detailed in the Supporting Information, three sets of traces for dissociation of RecQ from dT35-dT80 were globally fit to Eq. 6. The results (Fig. 4A, black curves) gave values for the global translocation parameters: kt, the forward stepping rate constant; kend, the dissociation rate constant; and r, the probability of RecQ binding ssDNA internally versus an end (Table 1), as well as the number of kinetic steps, n, RecQ takes for each length of DNA (see Fig. 4B). The rate constant for dissociation of RecQ from internal ssDNA sites, kd, is the measured dissociation rate constant for the RecQ-poly dT complex, and it was held constant throughout the fit. The kinetic step size, m, was determined by plotting the number of kinetic steps versus the oligonucleotide length and then fitting Eq. 7 to the data (Fig. 4B). We observed that above 60 nucleotides the number of steps RecQ takes on longer DNA substrates deviates from a straight line. As a result, we only fit the first six points (dT35-dT60) to Eq. 7 to determine the kinetic step size; RecQ has a kinetic step size, m, of 5.3 ± 0.9 nucleotides•step-1 and a site size, d, of 34 ± 3 nucleotides.
Fig. 4.
Kinetic modeling of translocation by RecQ. (A) The kinetic traces from Fig. 3 were globally fit to Eq. 6. Shown are the best fits to the n-step kinetic model (black line) using the parameters summarized in Table 1. (B) Plot of the maximum number of kinetic steps as a function of oligonucleotide length. The data are linear with respect to the DNA lengths up to 60 nucleotides. The solid line is a fit to Eq. 7. Fitting of Eq. 7 to three datasets yields a kinetic step size of 5.3 ± 0.9 nt per kinetic step, and a binding site size of 34 ± 3 nt.
We used the parameters that were obtained from fitting of Eq. 6 to the dissociation traces to calculate the relationship of ATP hydrolysis to translocation for RecQ. In the above analysis of the phosphate-release data, we assumed that the processivity of RecQ on the shorter oligonucleotides (dT35-dT60) was unity. This simplification assumes that, for shorter oligonucleotides, RecQ will always reach the DNA end, which may underestimate the coupling efficiency. To confirm our qualitative, but intuitive, analysis of the coupling efficiency and to more accurately calculate the number of ATP molecules coupled to each kinetic step, we used Eq. 8 to fit the amplitudes of the kinetic traces for the phosphate-release data in Fig. 2B (black line) (23). The values r and m in Eq. 8 were fixed as the values determined from analysis of the dissociation experiments, and the parameters for the site size (d), and the coupling efficiency (c) were obtained by fitting as detailed in the Supporting Information. Our analysis of the phosphate-release data shows that RecQ has a binding site size of 34 ± 1 nucleotides and a coupling efficiency of 3.4 ± 0.4 ATP per kinetic step (Table 1); these parameters are in good agreement with those obtained from the independent dissociation data. Taking the value of the coupling efficiency (c) with the value of the kinetic step size (m), we calculate that RecQ uses 0.6 ± 0.2 ATP molecules per nucleotide stepped, or that approximately two nucleotides are traversed for every one ATP molecule hydrolyzed, consistent with the value determined above from the linear fit of the data in Fig. 2B
Discussion
Little is known about the mechanism of translocation for RecQ helicases in comparison to SF1 and other SF2 helicases. As such, we analyzed the kinetics of a single round of translocation of RecQ on ssDNA by monitoring the dissociation of RecQ from oligonucleotides of defined sizes and measuring the associated ATP hydrolysis (Table 1). RecQ translocates on ssDNA with a rate of 16 ± 4 nucleotides per second and a processivity (P) of 0.972 ± 0.005, which means that RecQ translocates 36 ± 2 nucleotides on average before dissociating from ssDNA. This value is consistent with steady-state analysis of ATP hydrolysis, which also indicates that the enzyme travels approximately 19 ± 6 nucleotides before dissociating from ssDNA (28). Finally, we determined that RecQ advances more than one nucleotide (1.6 ± 0.3) per ATP hydrolyzed. Because our kinetic analysis demonstrates that RecQ tightly couples ATP hydrolysis to movement on DNA, with 0.6 molecules of ATP hydrolyzed per nucleotide moved, we suggest that the enzyme translocates via an inchworm mechanism. Had the mechanism for translocation been that of a Brownian Ratchet, one would have expected the ratio of ATP hydrolyzed to nucleotides stepped to be greater than one (1).
UvrD and PcrA helicases, both SF1 helicases, couple the hydrolysis of one ATP molecule to step 1 nucleotide (19, 20, 23). Structural and biochemical analysis of the PcrA and UvrD proteins reveal that translocation occurs by the binding and relative motion of the N- and C-terminal core domains (domains 1A and 2A) (5). When the helicase domain is bound to ssDNA, a cleft between domain 1A and 2A serves as the site for ATP binding. Upon binding of the nucleotide cofactor, the cleft closes around the ATP molecule, and the nucleotides of ssDNA are flipped between binding pockets on the surface of the 1A and 2A domains. Hydrolysis results in the release of the nucleotide cofactor and opening of the cleft. Repeated cycles of binding and hydrolysis followed by opening and closing of the cleft lead to translocation in a motion similar to an inchworm. Although similar studies of the RecQ helicase are not available, the crystal structure for RecQ does support a similar translocation mechanism (29). RecQ contains both the N- and C-terminal core domains found in SF1 helicases. Furthermore, residues that have been shown to be essential for coupling ATP binding and hydrolysis to translocation are essential for the activity of RecQ (30). These data, together with the coupling efficiency for translocation, argue that RecQ uses a similar mechanism for translocation as SF1 helicases, but notably, with a step size of two nucleotides per ATP hydrolyzed. Interestingly, a kinetic analysis of a different SF2 helicase, RecG, led to the conclusion that RecG unwinds 3 base pairs per ATP hydrolyzed (31). Moreover, recent single-molecule analysis of the NS3 helicase led to the observation of individual steps that ranged from 1–3 bp in size, with 0.5 bp increments, during unwinding of an RNA duplex (32). Thus, it appears that the translocation step of at least three SF2 motors is larger than that of the prototypical SF1 motors. The likely explanation for RecQ is that the movement of the two RecA-domains is sufficiently large to span two nucleotides; the fact that only 1.6 nucleotides are translocated per ATP hydrolyzed implies that either the physical movement is always two nucleotides (or larger) but some events are nonproductive, or that RecQ can undergo a mixture of both 1- and 2-nucleotide steps due to variations in the domain movements that accompany translocation.
Our kinetic model suggests that the affinity of RecQ for ssDNA cycles during translocation due to the binding and hydrolysis of ATP. Binding of ATP results in a high affinity of RecQ for ssDNA, while either the hydrolysis to ADP and phosphate, or release of the cofactor products equivalently decreases affinity, contributing to dissociation of RecQ from the DNA. Analysis of the binding of RecQ to ssDNA shows that either of the nonhydrolysable ATP analogues, adenosine 5′-(β,γ-imido) triphosphate (AMP-PNP) and adenosine 5′-O-(3-thio) triphosphate (ATPγS), increases the equilibrium binding affinity of RecQ for ssDNA relative to the affinity in the presence or absence of ADP, which is the same (27, 33). These data support a DNA binding affinity cycle similar to that of E. coli RecA, in which ATP binding increases the affinity of the protein for ssDNA, whereas hydrolysis leads to cofactor product states with lower affinity (33). The observed modulation of the ssDNA-binding affinity of RecQ suggests that dissociation of the protein most likely occurs after ATP hydrolysis. Thus, the dissociation rate constant, kd, likely measures dissociation of the non-ATP-bound forms of RecQ.
The simplest model suggested by the kinetic analyses is that RecQ takes three fast kinetic steps, where each kinetic step consumes one ATP molecule to translocate two nucleotides, followed by a kinetically slow step. A nonuniform translocation model was also proposed for NS3 and UvrD helicases (23, 34). In the case of NS3 protein, a large unwinding step size was observed to consist of smaller kinetic step sizes (32). For the translocation by UvrD, nonuniform stepping was also observed wherein the helicase took four fast kinetic steps to translocate four nucleotides (23). RecQ may utilize a mechanism similar to UvrD for pulling on ssDNA via one of the core domains during the fast kinetic steps, while the slow kinetic step comes about when the lagging core domain releases the bound ssDNA. Unfortunately, the basis for the nonuniform stepping of these helicases is not well understood (24); however, a recent analysis shows that static heterogeneity in the translocation rates of individual molecules leads to an overestimate in the step size determined from ensemble kinetic analysis (35). This interpretation would suggest that RecQ displays an intrinsic heterogeneity with regard to translocation, an interpretation that is in accord with single-molecule data (28).
An explanation for the large site size (34 ± 3) of RecQ could be that a higher-ordered structure is required for efficient translocation. Several experimental observations suggest that three molecules, or more, of RecQ are needed for highest activity. First, ATP hydrolysis by RecQ is cooperative with respect to ATP concentration with a Hill coefficient of 2 (28). The unwinding of plasmid-length DNA is cooperative in ATP concentration as well, with a Hill coefficient of 3 (16). These data imply that at least two to three monomers of RecQ are involved in both the translocation on ssDNA and the unwinding of dsDNA. Second, RecQ is observed to bind to ssDNA with an equilibrium site size of 10 and a cooperativity of approximately 25 (36), implying that monomers interact with one another. Steady-state ATP hydrolysis assays on short oligonucleotides indicate that RecQ requires a length of at least 10 nucleotides to hydrolyze ATP (28). Assuming this site size reflects the requirement for a monomer of RecQ, then three monomers of RecQ would require 30 nucleotides to efficiently bind ssDNA prior to translocation. Based on structural and biochemical data, a “directed subunit–hand-off” model was proposed for translocation of the Rho transcription termination protein along RNA where each monomer advanced one step (one nucleotide, in this case) sequential in an asymmetric hexameric structure (37). Although the needed structural information for RecQ is currently insufficient to permit direct comparison, it is possible that a related sequential hand-off model is being used by RecQ multimers to advance along ssDNA in 2 nucleotide steps.
Interestingly, translocation by RecQ on ssDNA is slower and less processive than for other helicases. UvrD and PcrA translocate on ssDNA with rates of 193 nt/s and 233 nt/s, and processivities of 230 and 250 nucleotides, respectively (19, 20, 23). RecQ translocates with a 12-fold slower rate and 10-fold lower processivity on ssDNA. Thus, RecQ is not expected to be a rapid and processive helicase. Because the processivity for translocation is low, a protomer of RecQ is incapable of unwinding long regions of dsDNA in a single binding event. In seeming contradiction, RecQ can unwind plasmid-length dsDNA, but titrations revealed that SSB protein is required and the maximal rate of unwinding occurs at 1 RecQ monomer per 30 bp (16). This value is in agreement with our kinetic data for translocation, and suggests that while a RecQ protomer unwinds only a short region of dsDNA (approximately 30 bp), it can perform multiple rounds of binding and unwinding and/or many protomers can act autonomously to separate longer lengths of dsDNA, provided that SSB is present to trap the unwound strands of DNA and prevent the formation of nonproductive RecQ-ssDNA complexes.
Our analysis of the translocation by RecQ also illuminates the in vivo functions of this enzyme. RecQ functions at both the initial and terminal steps of genetic recombination (14, 38–40). At the initiation stage of DNA recombination, RecQ unwinds dsDNA and allows a RecJ, a 5′ to 3′ exonuclease to process the ssDNA, resulting in 3′ terminated overhang used for homologous pairing by the RecA protein (40). In addition, these two proteins also process ssDNA gaps at the sites of stalled replication forks (41). For homologous recombination, the minimal efficient processing segment (MEPS) is approximately 40 bp for the RecF-dependent pathway (42). This value is the minimum length required by RecA to carry out DNA strand exchange and, interestingly, is on the order of the processivity of RecQ. Thus, processing of a dsDNA break or ssDNA gap does not need to be extensive during recombination, reflecting the capability of RecQ to translocate and unwind only 30 base pairs per binding event; if more extensive unwinding tracts are needed, for example, to unwind through regions of DNA heterology, then multiple RecQ protomers can cooperate to unwind large regions of dsDNA (16, 28). In addition, RecQ, in conjunction with Topoisomerase III (Topo III), promotes catenation and decatenation of dsDNA, a conserved aspect of RecQ-family function (43, 44). This biochemical activity is important for dissolving double Holliday junctions during the last step of recombination (43, 44) and to separate the terminal regions of dsDNA during replication (45). To allow Topo III to catenate or decatenate DNA molecules, RecQ need only unwind a few base pairs to allow the type I topoisomerase to bind and function. Thus, the mechanism of unwinding by RecQ is well suited for its roles in vivo.
Materials and Methods
Proteins and DNA
RecQ was purified as described (14). Phosphate binding protein (PBP) was purified and labeled with 7-diethylamino-3-((((2-maleimidyl) ethyl)-amino) carbonyl) coumarin (MDCC) as described (25, 26). Oligonucleotides and poly dT were purified as in SI Text.
Equations and Data Analysis
The model and analysis was essentially as described (24). The equations used are listed below; parameters are defined in Table 1 or below. Analytical details are found in the SI Text:
 |
[1] |
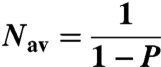 |
[2] |
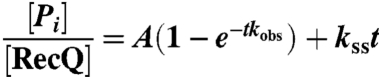 |
[3] |
where A is the amplitude, kobs is the observe rate constant, and kss the rate constant for steady-state ATP hydrolysis.
| [4] |
| [5] |
where kobs is the observed rate constant, kd is the intrinsic dissociation rate constant from internal DNA sites, kd,heparin is the dissociation rate constant in the presence of heparin.
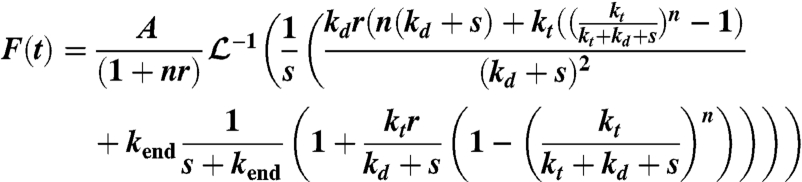 |
[6] |
where F(t) is the observed fluorescence; n, the number of kinetic step; and s, the Laplace variable.
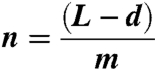 |
[7] |
where L is the DNA length in nucleotides, d is the binding site size for RecQ.
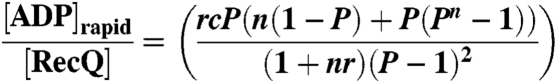 |
[8] |
where the processivity, P, in Eq. 8 is defined as 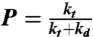 .
.
Equilibrium Binding, Dissociation, and Phosphate-Release Assays
Equilibrium binding was monitored using a fluorimeter (SLM-ISS) (26, 46). Rapid kinetic experiments used an SX.18MV-R stopped-flow reaction analyzer (Applied Photophysics); the final concentrations were 0.1 μM RecQ, 0.3 μM (molecules) oligonucleotide, 0.5 mM ATP, 0.5 mM Mg(OAc)2 and 0.5 mg/mL heparin, in 25 mM TrisOAc (pH 7.5) and 0.1 mM DTT. Phosphate-release assays contained 3 μM MDCC-PBP, and the buffer had a phosphate-mop consisting of 0.01 Units/mL of bacterial purine nucleoside phosphorylase and 100 μM 7-methylguanosine. Details are in SI Text.
Supplementary Material
Acknowledgments.
We thank I. Amitani, J. Graham, N. Handa, K. Morimatsu, T. Kim, and H.-Y. Lee for careful reading of the manuscripts. This work was supported by National Institutes of Health Grant GM41347 to S.C.K.; B.R. was supported in part by National Institutes of Health training Grant T32-GM007377.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119952109/-/DCSupplemental.
References
- 1.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: Mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama H, et al. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K-12: Identification of a new mutation (recQ1) that blocks the recF recombination pathway. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 4.Bennett RJ, Keck JL. Structure and function of RecQ DNA helicases. Crit Rev Biochem Mol Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 5.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 6.Chu WK, Hickson ID. RecQ helicases: Multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 7.Veaute X, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 8.Krejci L, et al. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J Biol Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- 9.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumont S, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 13.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 14.Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hishida T, et al. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 2004;18:1886–1897. doi: 10.1101/gad.1223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmon FG, Kowalczykowski SC. Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J Biol Chem. 2001;276:232–243. doi: 10.1074/jbc.M006555200. [DOI] [PubMed] [Google Scholar]
- 17.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 18.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 19.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 20.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin MK, Gurjar M, Patel SS. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat Struct Mol Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 23.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer CJ, Lohman TM. ATP-dependent translocation of proteins along single-stranded DNA: models and methods of analysis of pre-steady state kinetics. J Mol Biol. 2004;344:1265–1286. doi: 10.1016/j.jmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Brune M, et al. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37:10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 26.Johnson KA, editor. Kinetic Analysis of Macromolecules: A Practical Approach. New York: Oxford University Press; 2003. p. 256. [Google Scholar]
- 27.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 28.Rad B. Davis: University of California; 2010. Tracking the RecQ helicase on DNA: An insight into the mechanism of molecular motors. PhD thesis. [Google Scholar]
- 29.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33:6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Senac MM, Webb MR. Mechanism of translocation and kinetics of DNA unwinding by the helicase RecG. Biochemistry. 2005;44:16967–16976. doi: 10.1021/bi0512851. [DOI] [PubMed] [Google Scholar]
- 32.Cheng W, Arunajadai SG, Moffitt JR, Tinoco I, Jr, Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 34.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dou SX, Wang PY, Xu HQ, Xi XG. The DNA binding properties of the Escherichia coli RecQ helicase. J Biol Chem. 2004;279:6354–6363. doi: 10.1074/jbc.M311272200. [DOI] [PubMed] [Google Scholar]
- 37.Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 38.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 39.Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem. 2003;278:42668–42678. doi: 10.1074/jbc.M302994200. [DOI] [PubMed] [Google Scholar]
- 40.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E coli. Genes Dev. 2009;23:1234–1245. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 42.Shen P, Huang HV. Homologous recombination in Escherichia coli: Dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 44.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J Mol Biol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.