Abstract
Global disruption of transient receptor potential-melastatin-like 7 (Trpm7) in mice results in embryonic lethality before embryonic day 7. Using tamoxifen-inducible disruption of Trpm7 and multiple Cre recombinase lines, we show that Trpm7 deletion before and during organogenesis results in severe tissue-specific developmental defects. We find that Trpm7 is essential for kidney development from metanephric mesenchyme but not ureteric bud. Disruption of neural crest Trpm7 at early stages results in loss of pigment cells and dorsal root ganglion neurons. In contrast, late disruption of brain-specific Trpm7 after embryonic day 10.5 does not alter normal brain development. We developed induced pluripotent stem cells and neural stem (NS) cells in which Trpm7 disruption could be induced. Trpm7−/− NS cells retained the capacities of self-renewal and differentiation into neurons and astrocytes. During in vitro differentiation of induced pluripotent stem cells to NS cells, Trpm7 disruption prevents the formation of the NS cell monolayer. The in vivo and in vitro results demonstrate a temporal requirement for the Trpm7 channel kinase during embryogenesis.
Keywords: gastrulation, magnesium, melanocytes, chanzyme, TRPM6
Within the large family of transient receptor potential (TRP) proteins, ubiquitously expressed TRPM7 functions as both a divalent-permeant ion channel and serine/threonine kinase (1–3). As a channel, TRPM7 is permeant to monovalent and many divalent ions (4), is inhibited by internal Mg2+ (5) and by phosphatidylinositol 4,5-bisphosphate hydrolysis (2), and is potentiated at low pH (6). Global Trpm7 disruption in mice leads to early embryonic lethality, whereas T cell-specific disruption results in developmental block of thymocytes at the double-negative stage and a progressive depletion of thymic medullary cells (7). Trpm7 mutations in zebrafish cause embryonic lethality, abnormal skeletogenesis, kidney stone formation, and albinism (8). In Xenopus laevis toads, Trpm7 disruption results in embryonic lethality during gastrulation (9). These pleiotropic effects suggest that TRPM7 is important during development, although the role of the ion channel in these various tissues remains obscure. To explore this question, we took advantage of genetic tools that allow tissue-specific and time-specific inactivation of the Trpm7 gene. Here, we show, via inducible disruption at different embryonic stages and postnatal ages, that loss of TRPM7 has the most profound effects on organs that develop earliest in embryogenesis, specifically before embryonic day (E) 7–E9.
Results
TRPM7 Is Essential Only for Early Stages (E7–E9) of Embryogenesis.
A tamoxifen (TM)-inducible (Cre-ER) transgenic line (10) was bred to Trpm7fl/fl mice to allow global Trpm7 disruption in Trpm7fl/fl (Cre-ER) embryos or adult mice. A single TM dose administered at early (E7–E9) gestation to pregnant females induced Trpm7 disruption in Trpm7fl/fl (Cre-ER) embryos and resulted in embryonic lethality within 48–72 h (Fig. 1A), with dying Trpm7fl/fl (Cre-ER) embryos exhibiting abnormal morphology and body patterning (Fig. 1B). In crosses between male Trpm7fl/fl (Cre-ER) and female Trpm7fl/fl mice, the absence of live Trpm7fl/fl (Cre-ER) embryos/pups in litters from dams treated early with TM confirmed embryonic lethality at E7–E9 (Fig. S1A). However, when TM injection was performed at E14.5, live Trpm7fl/fl (Cre-ER) pups were born in numbers consistent with expected Mendelian inheritance. In parallel injections, no dead Trpm7fl/fl (Cre-ER) embryos were found in utero 48 h after injection (Fig. S1A). Therefore, we conclude that disruption of Trpm7 at E7–E9, but not at E14.5, results in embryonic death.
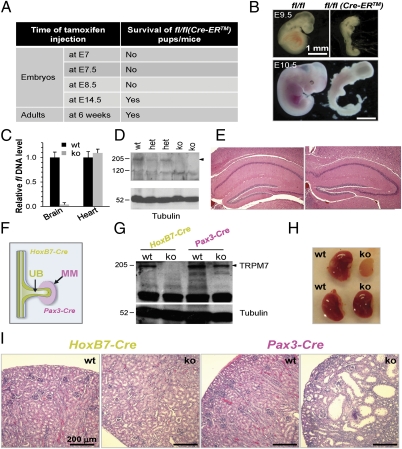
Fig. 1.
TM-induced and tissue-specific Trpm7 disruption reveals a spatiotemporal requirement of the channel kinase for embryogenesis. (A) Trpm7 is temporally required for embryogenesis. TM-induced global Trpm7 disruption at E7–E9, but not at later stages (>E14.5 or adults), was consistently lethal. After timed breeding [male Trpm7fl/fl (Cre-ER) mice with female Trpm7fl/fl mice], TM was injected i.p. into pregnant females at E7, E7.5, E8.5, and E14.5. TM was also injected into 6-wk-old Trpm7fl/fl (Cre-ER) adult mice to induce Trpm7 disruption. (B) Representative images of grossly deformed, nonviable embryos at E9.5 (Upper) and E10.5 (Lower). Trpm7 disruption was induced at E7–E9. Detailed results are described in Fig. S1. (C) Real-time PCR genotyping of genomic DNA from Trpm7fl/fl [Nestin-Cre (ko)] and Trpm7fl/fl littermate control (wt) mice indicates depletion of the fl allele (the loxP-flanked exon 17) in ko brain but not heart. Quantification of the fl level in ko mice was normalized to wt. (D) Depletion of TRPM7 protein in the ko brain. Brain lysates were prepared from wt, het, and ko mice and analyzed by Western blotting. The arrowhead indicates TRPM7. (E) Normal brain histology of Nestin-Cre Trpm7 ko mice. Representative H&E-stained hippocampal sections from 12-wk-old wt and ko mice. (F) Diagram showing the differential expression pattern of the HoxB7-Cre and Pax3-Cre transgenes at E11.5: Hox B7-Cre was expressed in the UB, whereas Pax-3 Cre was expressed in the MM. (G) Western blot analysis of TRPM7 from kidney lysates in wt and HoxB7-Cre sections from P12 or Pax-3 Cre Trpm7 ko mice. (H) Images of P12 kidneys (wt vs. ko) mediated by Pax3-Cre (Upper) and HoxB7-Cre (Lower). (I) H&E staining of sections from P12 kidneys (wt vs. ko). The HoxB7-Cre ko kidney histology was normal, whereas the Pax3-Cre ko kidney contained cysts and a reduced number of glomeruli.
We also injected 6-wk-old Trpm7fl/fl (Cre-ER) adult mice with TM to generate adult Trpm7 global knockout (ko) mice. All mice were grossly normal 1–2 mo after injection (Fig. 1B). Tail DNA genotyping for the fl allele (the loxP-flanked Trpm7 exon 17) confirmed global Trpm7 disruption in these Trpm7fl/fl (Cre-ER) adult mice, as shown by emergence of the null allele and loss of the fl allele in tail DNA (Fig. S1B). To study the effect of global Trpm7 disruption in a tissue-specific manner, three matched daily TM injections (25–50 μg/g of body weight) in 4-wk-old Trpm7fl/fl control and Trpm7fl/fl (Cre-ER) mice were used. Trpm7 mRNA levels were measured, and histology was analyzed from tissues collected 4 wk after TM injection in Trpm7fl/fl control and Trpm7fl/fl (Cre-ER) KO mice. Exon 17-containing Trpm7 mRNA was nearly absent in kidney and heart and was significantly reduced in the ko liver (Fig. S1C), confirming TM-induced Trpm7 disruption. These organs displayed normal histology as shown by H&E staining (Fig. S1D). No morphological or obvious behavioral abnormalities were found in TM-induced Trpm7 ko adult mice, in contrast to the embryonic lethality caused by complete early Trpm7 deletion. Thus, at least in the few months following Trpm7 deletion in adults, loss of TRPM7 in adult mice does not result in major phenotypic differences.
Nestin-Cre–Mediated Trpm7 Disruption Does Affect Brain Development.
After neural induction and neurulation, ectoderm gives rise to neural tube (E9.5), whose epithelium contains the neural stem (NS) cells that are progenitors of all neural lineages. NS cells express Nestin and are poised to differentiate into neurons and astrocytes in vitro. NS cells can also be obtained by in vitro differentiation of ES cells, and such generated NS cells are capable of self-renewal and further differentiation into neurons and astrocytes (11). Nestin-Cre Trpm7 ko mice were generated by breeding Trpm7+/fl (Nestin-Cre) male mice with Trpm7fl/fl female mice. A normal Mendelian ratio was observed in pups (18 ko from 69 pups in 8 litters; 26% vs. 25% expected). As expected, the fl allele mRNA was absent in brain but not heart, and TRPM7 protein was substantially lower in brain lysates of the Nestin-Cre Trpm7 ko mice (Fig. 1 C and D). However, Trpm7fl/fl (Nestin-Cre) (or Nestin-Cre Trpm7 ko) mice were normal in overall appearance, size, reproductive ability, behavior, and brain histology (Fig. 1E) and survived to adulthood. Because the Nestin promoter/enhancer that drives the Nestin-Cre transgene begins to be expressed at E10.5, we conclude that TRPM7 is not required for mouse brain development after E10.5.
Spatiotemporal Requirement of TRPM7 for Kidney Development.
During gastrulation, the ectoderm, mesoderm, and endoderm germ layers are derived from pluripotent ES cells. During kidney development, nephrons are derived from metanephric mesenchyme (MM), whereas collecting ducts develop from ureteric bud (UB). At E10.5, reciprocal inductive interactions between the UB and MM drive kidney morphogenesis (12). The MM induces the outgrowth of the UB, which invades the MM by E11.5, inducing condensation of MM cells around UB tips and formation of the condensed cap. During mesenchyme-to-epithelia transformation, the cap mesenchymal cells form spherical renal vesicles that develop into comma-shaped bodies, S-shaped bodies, and, eventually, mature nephrons (12). Wnt, Notch, TGF-β, and receptor tyrosine kinase pathways are all known to be important during nephrogenesis (13, 14).
The HoxB7-Cre and Pax3-Cre transgenes are expressed at E11.5 during kidney development, but HoxB7-Cre is exclusively expressed in the UB (15), whereas Pax3-Cre is expressed in the MM (16) (Fig. 1F). After birth, at P12, TRPM7 protein was readily detectable in control kidneys but barely detectable in kidneys from HoxB7-Cre ko mice. In contrast, P12 kidneys from Pax3-Cre ko mice had TRPM7 protein levels that were only slightly lower than wt (Fig. 1G). This suggests that TRPM7 expression at P12 is predominantly in cells of UB rather than MM origin. However, P12 HoxB7-Cre ko kidney size (Fig. 1H) and histology (Fig. 1I, Left) were normal, whereas P12 Pax3-Cre ko kidneys were less than half the size of those of littermate controls (Fig. 1H), indicative of an important function of TRPM7 in the MM. In P12 Pax3-Cre ko kidneys, there were far fewer glomeruli and many large renal cysts were apparent (Fig. 1I, Right), indicating defective nephrogenesis.
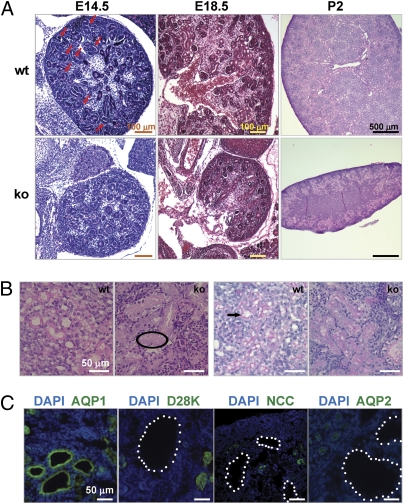
We then examined the Pax3-Cre Trpm7 ko kidney histology earlier in development. The ko kidneys were smaller than those of controls at E14.5 and E18.5 (Fig. 2A). Comma- and S-shaped structures, which reflect the transition from renal vesicles to segmented nephrons, were abundant in the control kidney at E14.5. In contrast, the ko kidney had only round and spherical tubular structures (Fig. 2A, Left). At E18.5, when numerous glomeruli were present in control kidneys (15.8 ± 2.8 per section, n = 4), very few glomeruli (1.7 ± 0.5 per section, n = 4) were found in ko kidneys (Fig. 2A, Center). By postnatal day (P) 2, renal tubules were dilated but no cysts were observed (Fig. 2A, Right). Eosinophilic material consistent with protein was present in the lumen of dilated renal tubules (Fig. 2B, Left) and the basement membrane was disrupted around dilated tubules of the ko kidney, compared with the well-organized basement membrane of controls (Fig. 2B, Right). Small cysts first emerged at P4 in ko kidneys. Immunostaining with markers specific for proximal tubules [aquaporin 1 (AQP1)], connecting tubules (Calbindin D28K), distal convoluted tubules [NaCl cotransporter (NCC)], and collecting ducts (AQP2) showed that the renal cysts were exclusive to the proximal tubules (Fig. 2C). In summary, the distinct kidney phenotypes resulting from the two differentially expressed transgenes show that MM TRPM7, but not UB TRPM7, is essential for nephrogenesis. This indicates that there is a spatiotemporal requirement for TRPM7 during kidney development.
Fig. 2.
Pax3-Cre–mediated Trpm7 disruption in kidney resulted in early defective nephrogenesis and a progressive kidney failure phenotype. (A) H&E staining of kidney sections at E14.5, E18.5, and P2 from Pax3-Cre Trpm7 ko mice. Spherical renal vesicles were present in E14.5 ko embryonic kidney, but comma- and S-shaped bodies were clearly absent (compared with wt control; arrows). There were reduced glomeruli in ko embryonic kidney at E18.5 and dilated tubules in the renal cortex of ko kidney at P2. (B) High-magnification images of dilated renal tubules in P2 ko kidney. (Left) Note even distribution of eosinophilic acellular material the tubular lumen of ko (example circled). (Right) Note well-organized basement membrane in wt kidney (arrow), and damaged ones in ko kidney (diffuse purple in periodic acid–Schiff stain). (C) Proximal tubular cysts first appeared at P4 in ko kidney. The cysts were associated with proximal tubules (AQP1+) but not connecting tubules (D28K+), distal convoluted tubules (NCC+), or collecting ducts (AQP2+). Cysts are highlighted by white dots. Nuclei were stained by DAPI.
Temporal Requirement for TRPM7 During Development of Neural Crest-Derived Pigment Cells.
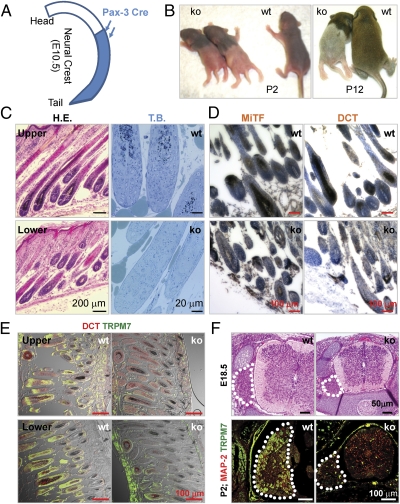
Pax3 is endogenously expressed in neural crest (NC) cells (17). The expression of Cre recombinase in Pax3-Cre mice largely recapitulates the normal Pax3 expression pattern, but Cre expression at E10.5 is detected in NC tissue in the lower but not upper trunk (18) (Fig. 3A). This Cre expression pattern resulted in normal agouti fur in the upper trunk but white fur in the lower trunk in Pax3-Cre Trpm7 ko mice (Fig. 3B). Similarly, pigment cells were present in hair follicles of the upper but not the lower trunk of P2 dorsal skin sections (Fig. 3C, Left). Toluidine blue staining, which yields excellent contrast between melanin and background, revealed that pigment cells were absent in the lower trunk of ko mice (Fig. 3C, Right). Immunohistochemical labeling with antibodies against microphthalmia-associated transcription factor and dopachrome tautomerase (DCT) revealed drastic reductions of pigment cells in the dorsal lumbar skin sections of P2 Pax3-Cre Trpm7 ko mice (Fig. 3D). The Pax3-Cre transgenic line exemplifies the point that promoter-driven Cre does not follow the exact endogenous pattern of the native gene because of differences in expression levels. However, by E14.5, Cre expression was detected in skin pigment cells throughout the embryo. Indeed, TRPM7 was depleted in hair follicles of both the upper and lower trunk of P2 ko mice, but DCT+ cells and pigment in hair follicles were reduced only in the lower trunk (Fig. 3E). These results indicate that NC Trpm7 deletion at E10.5 disrupts normal development of pigment cells but its deletion at E14.5 in skin pigment cells does not affect cell survival or normal melanogenesis. Finally, tyrosinase (Tyr) promoter Cre-mediated Notch ko mice were prematurely gray (19) because Notch is required for maintenance of melanocyte stem cells. However, we found that Tyr-Cre Trpm7 ko mice, expressing at E10.5 (20), exhibited normal hair color throughout their life span, indicating that TRPM7 is not required for maintenance of these stem cells.
Fig. 3.
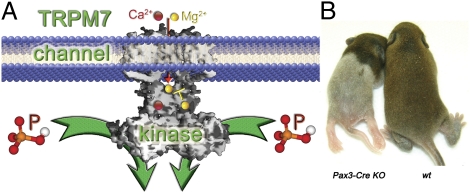
Trpm7 is temporally required for development of NC-derived pigment cells and DRG sensory neurons. (A) Diagram showing unique pattern of Pax3-Cre expression in E10.5 NC. The Cre transgene does not faithfully follow the endogenous Pax3 expression pattern and was not expressed in the upper (cervical and part of the thoracic) region of the NC (18). (B) Unique phenotype of Pax-3 Cre Trpm7 ko mice (P2 or P12): loss of pigment only in the lower trunk and paresis of only the hind legs. (C) Absence of pigment cells only in the lower trunk of the ko mice. (Left) H&E (H.E.) staining shows that pigment cells are present in the upper but not lower trunk in dorsal skin from a P12 ko mouse. (Right) Toluidine blue (T.B.) staining of P2 dorsal lumbar skin sections shows loss of pigment cells in the lower trunk of the ko mouse compared with wt littermate controls. (D) Immunohistochemistry of P2 dorsal lumbar skin sections confirmed the absence of microphthalmia-associated transcription factor (MiTF+) (Left) or DCT+ (Right) pigment cells in the lower trunk of ko mice. (E) Merged immunofluorescence and differential interference contrast (DIC) images show that Trpm7 was disrupted in both the upper and lower trunk of a P2 ko mouse, but the lower trunk had few DCT+ pigment cells and lacked pigment in comparison to the upper trunk. (F) (Upper) H&E staining of E18.5 lumbar spinal cord and DRG. (Lower) Loss of TRPM7 in large-diameter dorsal root ganglion (DRG) sensory neurons in ko mice (immunofluorescence, P2 lumbar DRG).
TRPM7 Is Temporally Required for Development of NC-Derived Dorsal Root Ganglion Neurons.
NC progenitors also give rise to dorsal root ganglion (DRG) sensory neurons. Consistent with the spatiotemporal timing of Pax3-Cre expression, Pax3-Cre Trpm7 ko mouse hind legs were paralyzed, although the forelimbs appeared normal; thus, the mice dragged their hind legs in a kneeling posture (Fig. 3B). E18 ko lumbar DRG sections appeared to have normal gray/white matter distributions, but cross-sectional areas were consistently smaller than in wt (Fig. 3F, Upper). TRPM7 was depleted in ko lumbar DRG with concurrent loss of TRPM7+ large-diameter sensory neurons (Fig. 3F, Lower). The ko DRG contained exclusively small neurons that stained for the neuronal marker microtubule-associated protein 2 (MAP2) (Fig. 3F, Lower). These results provide additional evidence for the temporal requirement of TRPM7 in NC-derived lineages: TRPM7 is essential for NC progenitors but becomes dispensable once the progenitors are committed.
Generation of Trpm7fl/fl (Cre-ER)-Induced Pluripotent Stem and NS Cells.
Induced pluripotent stem (iPS) cells provide a powerful tool to understand cellular and molecular mechanisms of neurogenesis. As an alternative to ES cells, iPS cells can be generated by reprogramming somatic cells via retroviral expression of a combination of factors, such as Oct4, Sox2, Klf4, Myc, and Nanog (21, 22). We used both in vivo and in vitro strategies to define a cellular basis for the temporal requirement of the TRPM7 channel kinase during embryogenesis.
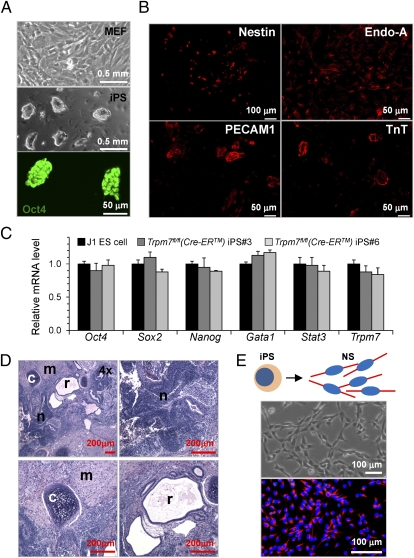
We established Trpm7fl/fl (Cre-ER) iPS cells in which Trpm7 could be disrupted by administration of TM. Mouse embryonic fibroblasts (MEFs) were isolated from E13.5 Trpm7fl/fl (Cre-ER) embryos and reprogrammed by retroviral expression of Oct4, Sox2, and Klf4 (Fig. 4A, Top). Multiple iPS clones maintained ES cell-like morphology (Fig. 4A, Middle) and expressed Oct4 (Fig. 4A, Bottom). Two independent iPS clones, iPS3 and iPS6, were used to avoid clonal diversity. In vitro differentiation assays showed that both iPS3 (Fig. 4B) and iPS6 (Fig. S2A) gave rise to all three germ layers: ectoderm (Nestin+ and NS cells), endoderm (cytokeratin Endo-A+), and mesoderm (PECAM1+ endothelial cells and Troponin T+ cardiomyocytes). Similar expression patterns of a panel of genes known to be expressed in ES cells (including Oct4, Sox2, and Nanog) were found in iPS3, iPS6, and a J1 ES cell line used to generate two ko mouse lines (Fig. 4C). We then tested the pluripotency of the Trpm7fl/fl (Cre-ER) iPS cells by assaying teratoma formation after iPS cell injection into immunocompromised mice. After iPS cell injection, teratomas formed from iPS3 (Fig. 4D) and iPS6 (Fig. S2B), with tissue resembling all three germ layers—ectoderm (neural epithelium), mesoderm (muscle and cartilage), and endoderm (respiratory duct)—demonstrating that these iPS cells are pluripotent. Trpm7fl/fl (Cre-ER) iPS cells were differentiated (Fig. 4E, Top) and formed a monolayer of NS cells. The Trpm7fl/fl (Cre-ER) NS cells exhibit bipolar morphology and form an adherent monolayer that is dramatically different from the morphology of the parental iPS cells (Fig. 4E, Middle). The cells in the monolayer stained positive for Nestin (Fig. 4E, Bottom), and thus consisted primarily of NS cells.
Fig. 4.
Trpm7fl/fl (Cre-ER) iPS cells and NS cells. (A) (Top) Isolated MEFs from E13.5 Trpm7fl/fl (Cre-ER) embryos. iPS cells were generated by reprogramming Trpm7fl/fl (Cre-ER) MEFs via retroviral expression of Oct4, Sox2, and Klf4. (Middle) ES cell-like morphology of iPS cells; phase contrast microscopy. (Bottom) Oct4 expression in iPS cells. (B) In vitro differentiation of iPS3 cells into ectodermal (Nestin+), endodermal (cytokeratin, Endo-A+), and mesodermal [PECAM1+, Troponin T+ (TnT+)] cells. (C) Real-time RT-PCR analysis of a panel of genes in two Trpm7fl/fl (Cre-ER) iPS clones (IPS3 and iPS6). Both clones exhibit a similar expression pattern as J1 ES cells. Expression was normalized to that of J1 ES cells. (D) Five weeks after injection of Trpm7fl/fl (Cre-ER) iPS3 in immunocompromised mice, all three germ layers were present in teratomas: neural epithelium (n; ectodermal), muscle and cartilage (m and c; mesodermal), and respiratory ducts (r; endodermal). Analysis of Trpm7fl/fl (Cre-ER) iPS6 is shown in Fig. S2. (E) Trpm7fl/fl (Cre-ER) NS cells were derived from pluripotent stem cells (29) (Materials and Methods). Monolayer of Trpm7fl/fl (Cre-ER) NS cells (Middle) is a nearly a homogeneous population of NS cells, as shown by immunofluorescence staining with the anti-Nestin antibody (red; Bottom). Nuclei were stained by DAPI (blue; Bottom).
Trpm7 Disruption in NS Cells Does Not Affect Self-Renewal and Differentiation.
Disruption of the fl Trpm7 allele by TM treatment of Trpm7fl/fl (Cre-ER) NS cells yielded Trpm7−/− NS cells (Fig. 5A). Consistently, no TRPM7 current was detected in Trpm7−/− NS cells (Fig. 5B). One common hypothesis is that TRPM7's function is to maintain relatively constant levels of global intracellular [Mg2+], because high levels of Mg2+ are required in cultured cells for survival (23). However, similar to lymphocytes from lck-Cre Trpm7 ko mice (7), total Mg2+ content was equivalent in Trpm7−/− and control (wt) NS cells (Fig. 5C). In addition, similar rates of proliferation were observed in Trpm7−/− and control NS cells, independent of extracellular Mg2+ (Fig. 5D). Trpm7−/− NS cells clearly displayed the capacity to differentiate into neurons (MAP2+, Fig. 6A) and astrocytes (GFAP+, Fig. 6B). Like wt NS cells, Trpm7−/− NS cells can be maintained indefinitely because they retain their ability to differentiate after >50 passages. In summary, loss of TRPM7 in NS cells did not affect the properties of self-renewal or differentiation (Fig. S3), consistent with the finding that Nestin-Cre Trpm7 ko mouse brain develops normally after Cre recombinase disruption in Nestin+ cells. Thus, our in vivo and in vitro results indicate that TRPM7 is dispensable for neurogenesis once NS cells have been established.
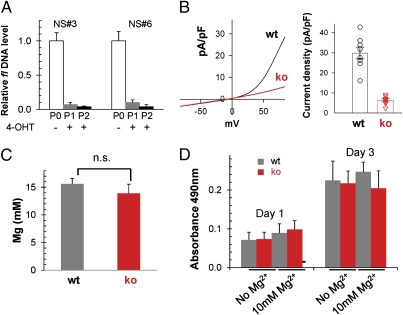
Fig. 5.
Trpm7−/− NS cells survive, renew, and have normal total magnesium content. (A) Genomic DNA real-time PCR shows that 2-μM TM treatment of Trpm7fl/fl (Cre-ER) NS cells for two passages (P1, P2) led to complete deletion of the fl allele, in comparison to untreated NS cells (P0), resulting in Trpm7−/− NS cells (ko). (B) Whole-cell patch-clamp of ko cells confirms the loss of TRPM7 current. (Left) Representative whole-cell currents. (Right) Pooled results of TRPM7 current density at +100 mV in wt (n = 8) and ko (n = 8) NS cells. (C) Measurement of total Mg2+ content in wt and ko NS cells. Total Mg2+ and K+ were measured via ICP-MS from 4 wt and 3 ko NS cell samples; [Mg2+] was calculated based on the Mg2+/K+ ratio. (D) Proliferation assay (MTS) shows no significant difference between wt and ko NS cells at either day 1 or day 3. Additionally, there is similar proliferation in Trpm7−/− and wt NS cells in the presence or absence of extracellular Mg2+ (10 mM).
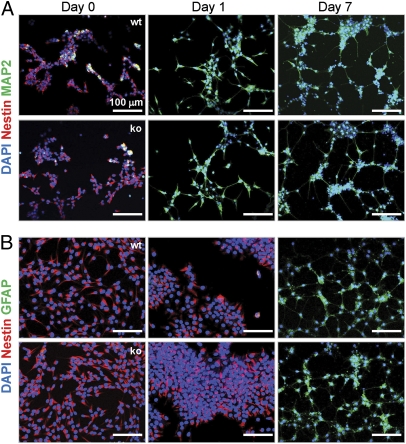
Fig. 6.
Trpm7−/− NS cells retain the potential to differentiate into neurons and astrocytes. Trpm7fl/fl (wt) or Trpm7−/− (ko) NS cells were seeded on chamber slides and then switched to differentiation medium for neurons (A) or astrocytes (B), as described in Materials and Methods. At days 0, 1, and 7, cells were fixed and double-stained with antibodies for Nestin or MAP2 (for neuron differentiation) or Nestin and GFAP (for astrocyte differentiation). Nuclei were counterstained with DAPI.
Trpm7 Disruption in iPS Cells Results in Cell Death.
TM treatment of the Trpm7fl/fl (Cre-ER) iPS cells (iPS3 and iPS6) resulted in Trpm7−/− iPS cells (Fig. 7A). Forty-eight hours after TM treatment, Trpm7−/− iPS cells were found to be floating in culture. When cells were passaged at 72 h posttreatment, the majority of the Trpm7−/− iPS cells had detached; control J1 ES cells treated similarly were not affected (Fig. 7B). In side-by-side experiments, 10 mM MgCl2 in the cell medium did not prevent loss of Trpm7−/− iPS cells (Fig. 7B, Bottom).
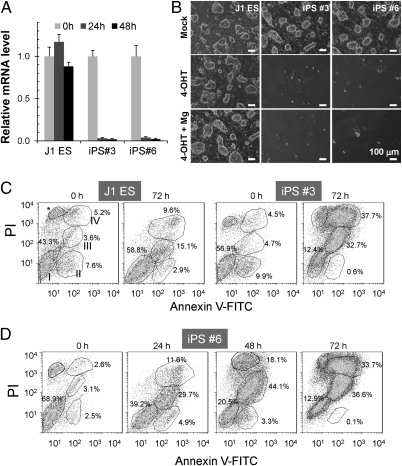
Fig. 7.
Cell death after Trpm7 disruption in iPS cells. (A) TM induced rapid Trpm7 disruption in Trpm7fl/fl (Cre-ER) iPS cells. J1 ES cells and two iPS clones, iPS3 and iPS6 cells, were treated with 2 μM TM. (B) Of Trpm7fl/fl (Cre-ER) cells treated with TM, only a few individual iPS3 and iPS6 cells survived. MgCl2 (10 mM) in the culture medium did not rescue cells. J1 ES cells and controls were analyzed side-by-side. (C) TM-induced Trpm7 disruption in iPS cells resulted in death at 72 h. J1 ES and iPS 3 cells, before and after 72 h of TM treatment, were labeled with AxV-FITC and PI for flow cytometry analysis. Double-negative cells were viable (Group I), FITC+ PI− cells (Group II) are early apoptotic cells, and late apoptotic/necrotic cells can be separated into two distinct groups: FITC+ PIlo cells (Group III) and FITC+ PIhi cells (group IV). *Necrosis that is common for adherent cells treated with trypsin (analysis with Flowjo software). (D) Progressive cell death was triggered by Trpm7 disruption. iPS6 cells were treated with 2 μM TM for 0, 24, 48, and 72 h, respectively, and double-stained with AxV-FITC and PI for flow cytometry analysis. Dynamic distribution of the four subgroups of cell populations during the time course indicates progressive cell death induced by Trpm7 disruption.
In an attempt to determine whether TM-induced Trpm7 deletion precipitated necrotic or apoptotic cell death, iPS cells and J1 ES cells were labeled by Annexin V (AxV)-FITC and propidium iodide (PI) for flow cytometry analysis. As shown in Fig. 7C, TM-treated iPS3 cells were largely dead, as indicated by the high number of AxV+ and PI+ cells. We defined four subpopulations of FACS cells as follows: (i) live cells (AxV−PI−), (ii) early apoptotic (AxV+PI−), (iii) midstage apoptotic (AxV+PIlo), and (iv) late apoptotic/necrotic (AxV+PIhi). Early apoptotic cells were largely absent in iPS3 cells at 72 h after TM treatment (Fig. 7C), likely because the early apoptotic phase was rapid. Time course experiments were then conducted to determine whether dying iPS cells were, in fact, traversing through early apoptosis before reaching late apoptosis/necrosis. Fig. 7D shows that live iPS6 cells gradually decline over 72 h, whereas early apoptotic cells peak at 24 h, midstage apoptotic cells peak at 48 h, and apoptotic/necrotic cells peak at 72 h. Future studies will address whether ES cells follow the same pattern.
Discussion
We established a series of Trpm7 ko mice in which the channel kinase was disrupted at different embryonic stages and in different organs. Characterization of these mice revealed a temporal and spatial requirement of TRPM7 for embryogenesis and organogenesis. We also generated pluripotent iPS and neural precursor (NP) cells in which inducible TRPM7 depletion could be manipulated. This combination of in vivo and in vitro studies led us to identify key time points for TRPM7 regulation of organ formation.
In kidney, Trpm7 disruption in the UB vs. MM resulted in starkly contrasting outcomes. Given normal development in the UB lacking TRPM7, we surmise that TRPM7 is not essential for reciprocal UB/MM induction or the ensuing UB branching or nephron segmentation. In contrast, loss of TRPM7 in the MM resulted in abnormal nephrogenesis, manifested first in the failure of renal vesicles to progress to curved comma- and S-shaped structures. Progressively defective nephrogenesis was apparent from E18.5 to P12, which was typified by decreasing numbers of glomeruli, dilation of renal tubules at P2/3, and cyst formation after P4. Because we have not examined all potential time frames, our results do not indicate that TRPM7 is entirely dispensable for development of the UB-derived urinary system. Importantly, HoxB7-Cre expression is first detected in the UB at E11.5 (15), when the UB has already formed. Because both the UB and MM are derived from the intermediate mesoderm (IM) at E9.5, it remains to be seen whether Trpm7 disruption in the IM causes defects in the formation of the UB and MM.
It is surprising that Trpm7 disruption in iPS cells results in rapid cell death, because we found that many cell types do not require TRPM7 for survival (7). In this study, neurons and astrocytes from Nestin-Cre Trpm7−/− mice as well as melanocytes from Pax3-Cre and Tyr-Cre Trpm7−/− mice survived. Trpm7−/− MEF cells induced by TM, and even multipotent progenitors like Trpm7−/− NP cells, survived in vitro. It has been reported that TRPM7 is also required for maintenance of bone marrow-derived mesenchymal stem cells (24). The question is why TRPM7 is dispensable for survival of most terminally differentiated cells types and some multipotent stem cells, such as NP cells, but is required for pluripotent stem cells and some multipotent stem cells, such as mesenchymal stem cells. One worrisome potential artifact is that the combination of nonspecific TM effects (25), combined with the loss of TRPM7, results in rapid iPS cell death. Another possibility is that Trpm7 disruption leads to diminished expression of key genes required for pluripotent stem cell maintenance, such as OCT4, NANOG, and STAT3, or other growth regulatory genes, such as FGF13, FGF7, and midkine (7).
A requirement for TRPM7 to maintain pluripotent stem cells could provide an explanation of why Trpm7 disruption causes early embryonic lethality at the onset of gastrulation, similar to the phenotypes of Oct4, Nanog, and Sox2 ko mice. However, when gastrulation was bypassed using inducible Trpm7 disruption, unique time dependence was found in different tissues than could be explained by pluripotent stem cell maintenance. Among the progenitors of gastrulation, NP/NS cells are a population of ectodermal stem cells important for brain development, whereas mesenchymal stem cells are important for hematopoietic tissue and connective tissue development. The requirement of TRPM7 for maintenance of mesenchymal stem cells but not NP cells might explain TRPM7's temporal dependence. The strongest support for this model comes from our studies of NC-derived pigment cells in Trpm7 ko mice bred to different Cre transgenic mice. NC cells contribute to a cascade of multipotent progenitors that give rise to diverse cell types, including pigment cells. Pigment cells progress from ES/iPS cells to neural crest progenitor (NCP) cells and to early melanoblast stem cells, which later become niche-restricted in adults (26). These cells mature into melanocytes. Fortuitously, the unique Pax3-Cre transgenic Trpm7−/− mouse was illuminating. In this mouse, Cre was expressed in NCP cells of dorsal neural tube at E10.5 (lumbar but not cervical), whereas it was expressed in all melanoblasts at E14.5 (18). This pattern resulted in the unique pigment cell phenotype in the Pax3-Cre Trpm7 ko mice in which only the lower trunk failed to develop mature melanocytes. We conclude that Trpm7 disruption at E10.5 in NCP cells affects pigment cell development and that Trpm7 is dispensable for further development from melanoblasts at E14.5. Furthermore, we found that Tyr-Cre mediated Trpm7 ko mice exhibited normal hair color throughout their life span, indicating that TRPM7 is not required for maintenance of melanocyte stem cells. These results suggest that the development of pigment cells evolves sequentially via two intermediary progenitor cells, NCP and melanocyte stem cells, and that TRPM7 is required for maintenance of NCP cells but not melanocyte stem cells.
Nestin, an intermediate filament protein, is expressed in dividing cells of the nervous system during the early stages of development. Nestin-Cre Trpm7 ko brain developed normally, indicating that TRPM7 is no longer important to formation of the brain after ∼E10.5. Our in vitro studies of NP cells, which are similar to NS cells in that they give rise to neurons, astrocytes, and oligodendrocytes, clearly indicate that Trpm7−/− NP cells survive, self-renew, and differentiate in similar fashion to wt NP cells. These findings pinpoint a small window of time (from blastocyst to neural epithelium) during which TRPM7 is essential for neurogenesis.
In summary, TRPM7 is an important regulator of early embryogenesis and organogenesis. Our in vitro studies show a differential requirement for maintenance of pluripotent and multipotent stem cells. However, a detailed molecular mechanism is still lacking. The key to understanding TRPM7's biological function lies in identifying (i) the in vivo mechanism for TRPM7 channel gating and (ii) the native TRPM7 kinase substrates. Intracellular Mg2+ blocks TRPM7 with a Kd of ∼0.6 mM (5). Thus, the only known mechanisms for TRPM7 gating are depletion of intracellular Mg2+ levels below normal (∼0.4–0.8 mM) levels (5) or severe reduction of extracellular pH (to ∼5) (6). Despite the simplicity and appeal of these two mechanisms, we consistently find no evidence that cells are depleted of Mg2+ during development or that [Mg2+] is abnormally low in TRPM7 ko cells. Importantly, Mg2+ transporters (27, 28), Mg2+-permeant channels [e.g., cyclic nucleotide-gated, TRPV1, V2, M2, M6, as well as various transmitter-gated channels], and Mg2+ buffers appear sufficient to maintain intracellular Mg2+ homeostasis in the absence of TRPM7. A profound pH drop during normal early development has not been reported in mammals, to our knowledge, and, in any case, only conducts monovalent ions inwardly at low [pH]o (6). We hypothesize that after an unknown gating stimulus, localized entry of one of TRPM7's permeating divalent ions triggers activation of the kinase, which then leads to alteration of fundamental events critical to early development.
Materials and Methods
Animals.
Mice were maintained in the barrier facility at the Children's Hospital Boston. Trpm7fl/fl mice were generated previously (7). Timed breeding of Trpm7fl/fl (Cre-ER) male and Trpm7fl/fl female mice was used to isolate Trpm7fl/fl (Cre-ER) MEF cells. A CRE-ER, TM-inducible transgenic (10) was used for inducible Trpm7 disruption. The Pax3-Cre transgenic was obtained from Jonathan Epstein (University of Pennsylvania, Philadelphia, PA) (18), and the HoxB7-Cre transgenic was obtained from Carleton Bates (University of Pittsburgh, Pittsburgh, PA) (15). Mice were maintained on a mixed 129/SvEvTac and C57BL/6J background. Timed breeding at night and detection of mating plugs the next morning marked the embryonic stage as E0.5. Newborn pups were designated as P1. Mice were maintained in the barrier facility at the Children's Hospital Boston under approved institutional animal care and use committee protocols. For i.p. TM injections, 50 mg/mL TM was dissolved in ethanol and aliquoted at −20 °C. Before injection, frozen TM stock was warmed to 50 °C, diluted 1:5 in autoclaved sunflower seed oil, and emulsified. The injection volume was 50–100 μL, depending on mouse body weight. Injected females were 12–16 wk old and had previously delivered healthy litters. A Nestin-Cre transgenic mouse (003771; Jackson Laboratory) was used for conditional Trpm7 disruption in brain. Immunocompromised SCID mice (001303; Jackson Laboratory) were used for iPS cell injection for teratoma formation. All procedures were performed under approved protocols.
Genotyping and Genomic DNA Quantitative PCR.
DNA was prepared using a mouse-tail kit (Qiagen) for PCR genotyping. The primer sequences are as follows: Trpm7 fl allele, 5′-TAGCCCTGTAGAGTTTTACTGG-3′ and 5′-GATAGACTATATACTAGGTACATGG-3′; Trpm7 null allele, 5′-AGACCAGGAGAAAATGCTCTGG-3′ and 5′-ATAGACTATATACTAGGTACATGG-3′; and Cre, 5′-GTATAGCCGAAATTGCCAGGATCAG-3′ and 5′-ACCCGGCAAAACAGGTAGTTATTCG-3′.
Genomic DNA quantitative PCR (qPCR) was carried out on 10-μL volumes using SYBR GreenER qPCR Supermix (Qiagen) in a C1000 cycler-converted CFX96 Real-Time PCR Detection System (Bio-Rad). Each PCR assay was done in triplicate, side by side with a β-actin control. Relative expression level was determined using the ΔΔCt method of Bio-Rad CFX Manager software. The primer sequences were as follows: Trpm7 exon 17, 5′-GGCAGTTGAATTACTGGAACAG-3′ and 5′-GGTCTAAGTCTTGAAGAAACTG-3′; Trpm7 exon 12, 5′-GGATCCTTGGAACAGGCTATG-3′ and 5′-GTTATAAAGTTCTTCCAGTCTGG-3′; andβ-actin exon 2: 5′-GCCATGGATGACGATATCGCTG-3′ and 5′-GATGGAGGGGAATACAGCCCG-3′.
Histology and Immunostaining.
Tissues were fixed overnight with 4% (vol/vol) paraformaldehyde (PFA) in PBS at 4 °C for paraffin-embedded sections. Paraffin-embedded sections (∼5 μm) were stained with H&E, periodic acid–Schiff reagent, or von Kossa reagent (for calcium deposits) at the Children's Hospital Boston Pathology Core and Dana Farber Histopathology Core using standard protocols. Immunofluorescence was performed on paraffin-embedded sections. Tissues were fixed with 4% (vol/vol) PFA in 20% sucrose before optimal cutting temperature-embedded cryosections. Antibodies against AQP1, Calbindin d28K, NCC, and AQP2 were purchased from Millipore and used at 1:200 in staining. Nuclei were counterstained with DAPI.
TUNEL Staining.
Kidney cryosections were stained using the ApopTag plus in situ apoptosis detection kit (catalog no. S7111; Millipore) according to the manufacturer's instructions. Briefly, sections were incubated with digoxigenin-labeled nucleotides and terminal deoxynucleotidyl transferase and then stained with fluorescein-conjugated antidigoxigenin antibody. Apoptotic cells with DNA fragmentation were positively stained (green), and nuclei were counterstained with DAPI (blue).
Western Blot Analysis.
Mouse tissue was homogenized and lysed in PBS lysis buffer containing 1% Nonidet P-40, 2 mM EDTA, and protease inhibitor mixture (1 tablet per 10 mL; Roche). Protein concentration of the lysate was quantified using Coomassie Protein Assay Reagent (Thomas Scientific). Two hundred micrograms of total protein was loaded onto SDS/PAGE, transferred to a nitrocellulose membrane, and stained with a rabbit anti-TRPM7 antibody. Positively stained signals were detected by Amersham ECL reagents (GE Healthcare).
Genotyping and Genomic DNA Real-Time PCR.
Mouse-tail DNA or tissue DNA was prepared using the mouse-tail kit for PCR genotyping. Genomic real-time PCR was done in a 10-μL volume using SYBR GreenER qPCR Supermix in the C1000 cycler-converted CFX96 Real-Time PCR Detection System. Each PCR assay was done in triplicate with β-actin, side-by-side. Relative expression levels were analyzed using CFX Manager software and normalized to the control.
Real-Time RT-PCR.
Total RNA was purified using an RNeasy Mini Kit (Qiagen). First-strand cDNA was synthesized using Retroscript (Ambion). Real-time PCR in 10-μL volumes used SYBR GreenER qPCR Supermix in a C1000 cycler-converted CFX96 Real-Time PCR Detection System. Each RT-PCR assay was done in triplicate with β-actin, side-by-side. Relative expression levels were analyzed using CFX Manager software and normalized to the designated control. The primer sequences are as follows: for Trpm7 exon 16–17, 5′-GACCTGGTAGATGATACTTCAG-3′ and 5′-GTGAGTAATTTCATAGCCATCG-3′, and for β-actin exon 3–4, 5′-GAACCCTAAGGCCAACCGTG-3′ and 5′-GTACGACCAGAGGCATACAG-3′.
Cell Culture.
All cells were maintained at 37 °C, 5% CO2. Cell culture media were purchased from Invitrogen. MEFs were cultured in DMEM/F12, with 10% (vol/vol) FBS, 1 mM sodium pyruvate, 1 mM l-glutamine, 100 mM nonessential amino acids, 50 U/mL penicillin, and 50 mg/mL streptomycin. iPS cells were cultured in ES cell medium containing KO DMEM with 15% (15 ml/100 ml) FBS certified for ES cell culture, 1 mM sodium pyruvate, 1 mM l-glutamine, 100 mM nonessential amino acids, 50 U/mL penicillin, 50 mg/mL streptomycin, 100 mM 2-mercaptoethanol, and 1,000 U/mL leukemia inhibitory factor (LIF). Media were changed daily. NS cells were cultured in NS cell medium containing Neuralbasal (Invitrogen) supplemented with N-2 and B-27, 1 mM l-glutamine, 100 mM nonessential amino acids, 50 U/mL penicillin, 50 mg/mL streptomycin, 10 ng/mL bovine fibroblast growth factor (bFGF), and 25 ng/mL EGF.
Isolation of MEF Cells.
Pregnant female mice were killed with CO2 asphyxiation at E13.5. The embryos were dissected from the uterine horn, and the brain and internal organs were removed. The remaining embryos were minced and pooled, resuspended in 2 mL of 0.25% Trypsin-EDTA (Invitrogen) in a 15-mL conical tube, and incubated at 37 °C for 30 min with occasional shaking; 5 mL of DMEM/F12 medium was added, and the cell/tissue mixture was pipetted to disaggregate clumps of tissue. After 2 min, the turbid supernatant was aspirated and passed through a 100-μm cell filter. The MEF cells were spun down and cultured in medium as described above.
Generation of iPS Cells.
Concentrated retroviral stock expressing Oct4, Sox2, and Klf4 was generated, and MEFs were reprogrammed. Briefly, 2 million MEF cells were infected with concentrated retrovirus expressing Oct4, Sox2, and Klf4 via spin inoculation. MEF cells were cultured with medium for ES cell culture, as described above. ES cell-like colonies were picked and transferred to 24-well plates. Cells maintaining ES cell-like morphology were expanded and then frozen.
Differentiation of iPS Cells.
For in vitro differentiation of iPS cells, LIF was removed from the culture media to allow formation of embryoid bodies (EBs). At day 5, EBs were transferred to a fresh plate and allowed to reattach for further differentiation. At day 9, the cells were trypsinized and seeded on chamber slides, and they were analyzed the next day using immunofluorescence.
Generation and Differentiation of NS Cells.
iPS cells were seeded on a gelatinized plate in normal ES cell culture medium and grown to near confluence. The medium was changed to NS medium the following day. Cells were trypsinized and replated in an uncoated plate in NP cell culture medium 4–5 d later. The supernatant containing NS cell aggregates was transferred to a new plate 3–5 d later, where they grew into a monolayer of NS cells. Differentiation of NS cells to neurons was in laminin-coated plates. NS cells were seeded in regular NS medium; the next day, bFGF and EGF were withdrawn from the medium. The medium was changed every 2–3 d, and cells were analyzed at day 7. NS cells were differentiated to astrocytes by addition of 1% FBS to the normal NS medium. The medium was changed every 2–3 d, and cells were analyzed at day 7.
Cell Proliferation Assay.
For the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethonyphenol)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay, the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega) was used following the manufacturer's instructions. Briefly, cells were cultured in a 96-well plate at the density suggested by the manufacturer. Ten microliters of the MTS reagent was added into each well, cells were incubated at 37 °C for 3 h, and absorbance was measured at 490 nm with a microplate reader. All experiments were done in triplicate with the “no-cell” well as the blank control.
Inductively Coupled Plasma-MS Analysis for Mg2+ Content.
Trpm7fl/fl and Trpm7−/− NS cells were snap-frozen in liquid nitrogen until processed for inductively coupled plasma (ICP)-MS analysis, as described previously (7). Cell pellets were lysed in 1% HNO3, diluted in deionized H2O (18 MΩ) to 0.1% HNO3, and analyzed by quadrupole ICP-MS (X-Series ICPMS with CCT; Thermo Electron). The data reduction was carried out using independently calibrated internal standards. The K+ and Mg2+ data (in parts per billion) were used to calculate the Mg2+/K+ ratio and the intracellular molarity of total Mg2+ by normalizing to [K+] assumed as 120 mM.
AxV Apoptosis Assay.
iPS cells were trypsinized for 5 min, dispersed to remove aggregates, and washed twice with Dulbecco PBS. Cells were suspended in binding buffer at 1 million cells/mL. Apoptosis assays used the AxV FITC Apoptosis detection kit (Sigma). Briefly, 500 μL of cells was stained with 5 μL of AxV FITC and 10 μL of PI for 10 min, and then immediately analyzed on a flow cytometer. Data were analyzed using FlowJo (National Institutes of Health).
Electrophysiology.
Whole-cell currents in fibroblasts or neural precursor (NP) cells were elicited by voltage stimuli lasting 100 ms, delivered every 5 s, using voltage ramps from −100 to +100 mV (0-mV holding potential). The internal pipette solution contained 120 mM Cs-methanesulfonate, 8 mM NaCl, 10 mM 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA), 3.3 mM CaCl2, 2 mM ATP-Na2, and 10 mM Hepes (pH adjusted to 7.2 with CsOH). The extracellular solution was designed to minimize chloride currents and consisted of 140 mM Na-methanesulfonate, 5 mM Cs-methanesulfonate, 1 mM CaCl2, and 10 mM Hepes (pH adjusted to 7.4 with NaOH). All currents were recorded using a MultiClamp 700B amplifier (Molecular Devices) and were acquired with Clampex (pClamp9; Molecular Devices). Data were digitized at 10 kHz and digitally filtered off-line at 2 kHz. All experiments were performed at room temperature.
Data Analysis.
Data are presented as the mean ± SEM. Statistical comparisons were made using the Student's unpaired t test. A P value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Andrew McMahon (Harvard University), Jonathan Epstein (University of Pennsylvania), and Carlton Bates (University of Pittsburgh) for providing Cre lines of transgenic mice. We also thank Mark Fleming, Tonora Archibald, Roderick Bronson, and Li Zhang for assistance with pathology and histology; George Daley and his laboratory for advice and help with iPS cells; Yuko Fujiwara for guidance on mouse i.p. TM injections; and colleagues in the D.E.C. and N.C.A. laboratories for helpful discussions.
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 1365.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120033109/-/DCSupplemental.
References
- 1.Runnels LW, Yue L, Clapham DE. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science. 2001;291:1043–1047. doi: 10.1126/science.1058519. [DOI] [PubMed] [Google Scholar]
- 2.Runnels LW, Yue L, Clapham DE. The TRPM7 channel is inactivated by PIP(2) hydrolysis. Nat Cell Biol. 2002;4:329–336. doi: 10.1038/ncb781. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- 4.Monteilh-Zoller MK, et al. TRPM7 provides an ion channel mechanism for cellular entry of trace metal ions. J Gen Physiol. 2003;121(1):49–60. doi: 10.1085/jgp.20028740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadler MJ, et al. LTRPC7 is a Mg.ATP-regulated divalent cation channel required for cell viability. Nature. 2001;411:590–595. doi: 10.1038/35079092. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, Li M, Yue L. Potentiation of TRPM7 inward currents by protons. J Gen Physiol. 2005;126(2):137–150. doi: 10.1085/jgp.200409185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin J, et al. Deletion of Trpm7 disrupts embryonic development and thymopoiesis without altering Mg2+ homeostasis. Science. 2008;322:756–760. doi: 10.1126/science.1163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elizondo MR, et al. Defective skeletogenesis with kidney stone formation in dwarf zebrafish mutant for trpm7. Curr Biol. 2005;15:667–671. doi: 10.1016/j.cub.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, et al. TRPM7 regulates gastrulation during vertebrate embryogenesis. Dev Biol. 2011;350:348–357. doi: 10.1016/j.ydbio.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 11.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 12.Saxen L. Organogenesis of the Kidney. UK: Cambridge Univ Press, Cambridge; 1987. p. 184. [Google Scholar]
- 13.Costantini F, Kopan R. Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, et al. Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol. 2004;276:403–415. doi: 10.1016/j.ydbio.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grieshammer U, et al. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- 17.Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Chen F, Epstein JA. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 19.Schouwey K, et al. Notch1 and Notch2 receptors influence progressive hair graying in a dose-dependent manner. Dev Dyn. 2007;236:282–289. doi: 10.1002/dvdy.21000. [DOI] [PubMed] [Google Scholar]
- 20.Tonks ID, et al. Tyrosinase-Cre mice for tissue-specific gene ablation in neural crest and neuroepithelial-derived tissues. Genesis. 2003;37:131–138. doi: 10.1002/gene.10242. [DOI] [PubMed] [Google Scholar]
- 21.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 22.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz C, et al. Regulation of vertebrate cellular Mg2+ homeostasis by TRPM7. Cell. 2003;114(2):191–200. doi: 10.1016/s0092-8674(03)00556-7. [DOI] [PubMed] [Google Scholar]
- 24.Cheng H, et al. Transient receptor potential melastatin type 7 channel is critical for the survival of bone marrow derived mesenchymal stem cells. Stem Cells Dev. 2010;19:1393–1403. doi: 10.1089/scd.2009.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takebayashi H, Usui N, Ono K, Ikenaka K. Tamoxifen modulates apoptosis in multiple modes of action in CreER mice. Genesis. 2008;46:775–781. doi: 10.1002/dvg.20461. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura EK, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H, Clapham DE. Mammalian MagT1 and TUSC3 are required for cellular magnesium uptake and vertebrate embryonic development. Proc Natl Acad Sci USA. 2009;106:15750–15755. doi: 10.1073/pnas.0908332106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li FY, et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature. 2011;475:471–476. doi: 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]