Abstract
More than 120 human papillomaviruses (HPVs) have now been identified and have been associated with a variety of clinical lesions. To understand the molecular differences among these viruses that result in lesions with distinct pathologies, we have begun a MS-based proteomic analysis of HPV–host cellular protein interactions and have created the plasmid and cell line libraries required for these studies. To validate our system, we have characterized the host cellular proteins that bind to the E7 proteins expressed from 17 different HPV types. These studies reveal a number of interactions, some of which are conserved across HPV types and others that are unique to a single HPV species or HPV genus. Binding of E7 to UBR4/p600 is conserved across all virus types, whereas the cellular protein ENC1 binds specifically to the E7s from HPV18 and HPV45, both members of genus alpha, species 7. We identify a specific interaction of HPV16 E7 with ZER1, a substrate specificity factor for a cullin 2 (CUL2)-RING ubiquitin ligase, and show that ZER1 is required for the binding of HPV16 E7 to CUL2. We further show that ZER1 is required for the destabilization of the retinoblastoma tumor suppressor RB1 in HPV16 E7-expressing cells and propose that a CUL2–ZER1 complex functions to target RB1 for degradation in HPV16 E7-expressing cells. These studies refine the current understanding of HPV E7 functions and establish a platform for the rapid identification of virus–host interactions.
Keywords: ubiquitylation, proteomics, retinoblastoma protein, keratinocyte, cervical cancer
The many types of human papillomaviruses (HPVs) that have been described exhibit considerable diversity. The HPVs are DNA viruses with a tropism specific for squamous epithelial cells. More than 120 HPVs have been identified and cloned to date, and these share a conserved genomic structure with eight to 10 ORFs encoded on one strand of a small double-stranded circular DNA genome (1). The ORFs involved in fundamental processes such as DNA replication or capsid formation are well conserved. Other ORFs, such as E6 and E7, have some conserved features but are more divergent at the nucleotide and protein level. Consistent with these differences, the lesions that are caused by infection with different HPVs and the propensity for these lesions to progress to cancer vary as well (2). A subset of the HPVs are the primary etiological agent in the development of cervical cancer, and other HPVs cause genital or cutaneous warts or other skin lesions. Relatively little is known about how these sequence differences translate into different biological outcomes in infected human cells. Thus, there exists an opportunity to systematically define features of diverse HPVs and to understand at the molecular level how their varied genetic compositions result in different disease states.
The standard phylogeny of the HPVs is based on the sequence of the L1 gene, and a virus with an L1 DNA sequence that differs by 10% or more from other HPV L1s is designated as a separate type (1). Similar HPV types are grouped into species and further into genera. The majority of the HPVs identified and sequenced to date sort into genus alpha and genus beta, with most alpha HPVs exhibiting a mucosal tropism and most beta HPVs able to infect cutaneous cells. Mucosal HPVs in species 7 and 9 include virus types 16, 18, 31, 33, 45, and 52, and infection with one of these can cause lesions that have a high risk of progression to cervical cancer. These high-risk HPVs cause the majority of all cervical cancers worldwide, accounting for 250,000 cancer deaths annually (3). HPV16 in particular is found in 50% of cervical cancers (4). The species 10 mucosal HPVs are also found in cervical cancers, although these low-risk virus types (including HPVs 6b, 44, and 74) are responsible for fewer cancers and for the majority of genital warts (4). A subset of the alpha-type HPVs, including those in species 4, are associated with cutaneous lesions that typically do not progress to cancer.
Much of the current understanding of HPV biology has been gained from the study of protein–protein interactions (PPIs), and the majority of this work has focused on the alpha HPVs. With the exception of the DNA helicase function of the E1 protein, no enzymatic activities have been associated with the HPV-encoded proteins (2), suggesting that HPV infection must alter conditions in the host cell largely by changing the PPI landscape in that cell. Many examples of such virus–host PPIs have been documented. One example is the binding and inactivation of a hypophosphorylated form of the retinoblastoma tumor suppressor (RB1) by HPV E7 proteins (5–7). This allows RB1–E2F complexes to dissociate and the G1-S phase checkpoint to be bypassed, promoting a cellular environment conducive to the replication of the viral DNA in differentiated host cells. To counter the apoptotic effects of such dysregulated cell growth, a second key interaction is the binding of high-risk HPV E6 proteins to the cellular ubiquitin ligase E6AP, resulting in the targeted degradation of the tumor suppressor p53 (8, 9). Many other HPV–host PPIs have been documented, but certainly many others remain to be discovered. These will provide insight into the molecular mechanisms by which different HPV types exhibit different host cell tropisms and pathologies.
To date, fewer studies have focused on the biology of the genus beta HPVs compared with alpha HPVs. These virus types sort into at least five distinct species (1) and were initially identified in the lesions of patients with the genetic disease epidermodysplasia verruciformis (EV) (10, 11). EV is characterized by the early development of multiple hyperproliferative lesions on sun-exposed regions of patients’ skin, and the cells in these lesions typically contain beta-PV DNA. By the time an patient with EV has reached middle age, the likelihood that some of these lesions will have progressed to squamous cell carcinoma is quite high. This observation provided the first connection between HPVs and cancer. HPV DNA is also frequently detected in nonmelanoma skin cancer (NMSC) samples, and HPV DNA is detected even more often in the actinic keratoses that can precede and progress to squamous cell carcinoma (12). This has led to the hypothesis that, even if beta-HPVs are not the direct cause of NMSCs, they may be important in promoting skin cancer. It is of note that beta-HPVs are found quite frequently in healthy skin samples, and that few cells in an HPV-positive NMSC indeed harbor HPV DNA. This suggests that, if beta-PVs do contribute to skin cancers, it is likely at an early stage, and that the presence of the virus in every cell is not required for later expansion and progression of the tumor. One attractive idea is that beta PVs function as a cofactor with UV irradiation in the development of NMSCs, and that viral products allow for the evasion of apoptosis in UV-damaged cells. This allows a population of damaged cells that would otherwise be eliminated to persist, perhaps acquiring additional mutations and progressing to cancer (12, 13).
The analysis of immunoprecipitation-MS (IP-MS) data with CompPASS software has allowed the robust and high-throughput identification of important PPIs in several biological systems (14–18). By using this technology, we have designed and implemented a system for the detection of interactions between HPV and host cellular proteins. Our system is based on the expression of epitope-tagged HPV ORFs in N/Tert-1 cells, human keratinocytes immortalized by hTERT (19). The tools we have created include a library of full-length, sequence-verified ORFs cloned from 20 different HPV genomes and a collection of N/Tert-1 stable cell lines, each expressing an HA-tagged version of a viral or cellular protein. These reagents, in conjunction with the existing CompPASS algorithm, allow us to define with high confidence the binding partners of the HPV proteins in their natural host cell type.
Here we describe the validation of this system and an analysis of cellular interacting partners of the E7 proteins from 17 different HPV types. We detect previously reported interactions and identify a number of previously unknown interactions between E7 and cellular proteins. Some of the cellular factors bind to E7 proteins from all HPV types tested, whereas others are restricted to specific E7s from a single HPV species or type. One such type-specific interaction is the binding of HPV16 E7 to ZER1, a substrate specificity factor for a CUL2 ubiquitin ligase complex that we demonstrate to be involved in the HPV16 E7-mediated degradation of RB1. This work establishes and validates a tool for the study of HPV–host cell interactions and further defines the pathway by which RB1 is degraded in HPV16-positive cells, a critical component of both the normal HPV life cycle and the progression to cervical cancer.
Results
Identification of Interactions Between HPV and Host Cellular Proteins.
The diversity among the papillomaviruses and the hypothesis that the cellular protein interactions in which their encoded proteins engage will define some of their biological differences led us to develop and implement a systematic, increased-throughput method for defining interactions between HPV and host cellular proteins. This builds on technologies for the rapid and robust detection of PPIs in human cells, specifically the use of CompPASS software coupled with IP-MS/MS detection of proteins in whole-IP eluates (14–18, 20–24). CompPASS analysis is based on a statistics table, a library of IP-MS/MS data generated in a specific cell line from a large set of IP experiments. The stats table is then used for comparisons that separate bona fide protein interactors termed high-confidence interacting proteins (HCIPs) of a given tagged bait from potential background interactors.
We established our PPI identification system (Fig. 1A) in N/Tert-1 cells, a human keratinocyte cell line that was generated from primary foreskin keratinocytes and immortalized by hTERT (19), as squamous epithelial cells are the natural host of HPV infection. The majority of studies that have used CompPASS technology thus far have used data generated in 293 and 293T cells. Although robust and facile, these cells are not optimal for the study of cellular interactions of HPV proteins because they are kidney epithelial cells and they harbor other viral proteins, including adenovirus E1A in 293 and additionally SV40 large T antigen in 293T cells.
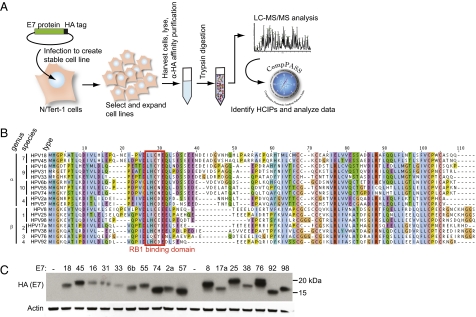
Fig. 1.
Systematic identification of interactions between HPV E7s and cellular proteins. (A) Schematic of IP-MS-CompPASS identification of HPV E7 HCIPs. (B) Protein sequence alignment of 17 E7 ORFs used in this study. Sequences were aligned by ClustalW and amino acids colored according to the ClustalX color scheme. (C) Western blot of stable E7 expression in N/Tert-E7-FlagHA stable cell lines, detected with anti-HA antibody.
To establish this system in a cell line not yet used in CompPASS studies, we generated 37 N/Tert-1 cell lines each expressing a single human ORF tagged with HA (Dataset S1). These HA-tagged baits were immunopurified, and the resulting protein complexes were analyzed by LC-MS/MS, resulting in the data set used to populate a CompPASS statistics table for the analysis of interactions in N/Tert-1 cells. To validate the combination of a new stats table and a new cell line, we analyzed the data from the N/Tert-HA-BAP1 cells in two ways: first, analyzed against an existing statistics table containing data from 293 cell IP-MS/MS experiments, then compared with the new N/Tert-1 data set (Fig. S1). Both analyses identify known interactors of BAP1 (15) as HCIPs, and only the comparison with N/Tert-1 cell data removes the likely nonspecific interactors such as myosins, laminins, and other proteins including MVP and AHNAK that were consistently identified in all N/Tert-1 data sets regardless of the HA-tagged bait.
For the expression of HPV proteins in N/Tert-1 cells, we obtained 20 cloned HPV genomes from the Reference Center for Human Papillomaviruses and additional laboratories (Dataset S2). These types were chosen to encompass the diversity of the HPVs from two genera and include 11 HPVs from genus alpha [HPV18 and 45 from species 7; HPV16, 31, 33, and 52 from species 9; HPV6b, 55 (a subtype of HPV44), and 74 from species 10; and HPV2a and 57 from species 4] and nine HPVs from genus beta (HPV5, 8, 20, 25, and 98 from species 1; HPV17a and 38 from species 2; HPV76 from species 3; and HPV92 from species 4).
The initial studies described here focus on the identification and characterization of interactions between HPV E7 proteins and the proteome expressed in N/Tert-1 cells. Seventeen HPV E7 ORFs were subcloned from the viral genomes into Gateway entry vectors (Dataset S3), then recombined into retroviral expression vectors (Dataset S4) and used to generate stable N/Tert-1 cell lines. E7 ORFs were used for these initial experiments because they have been characterized in many previous studies, resulting in known cellular interacting partners that can serve as positive binding controls. Also, as one of the key HPV oncoproteins, E7 is a likely candidate to participate in virus genus-, species-, and type-specific PPIs that could determine differences in the oncogenic potential of the various HPVs. The E7s encoded by the 17 virus types in the study share several conserved regions and diverge elsewhere. Each E7 contains the LXCXE motif responsible for binding to RB1 (5) (Fig. 1B). We therefore generated a library of 17 N/Tert-E7 cell lines, each stably expressing a single E7 ORF (Fig. 1C) with C-terminal Flag and HA epitope tags. It should be noted that no two E7 ORFs included in the study are identical in protein sequence; as predicted from the established HPV phylogeny, many similar sequence elements are found in E7s from closely related types.
HPV E7 Proteins Exhibit Conserved and Virus Type- and Species-Specific Interactions with Cellular Proteins.
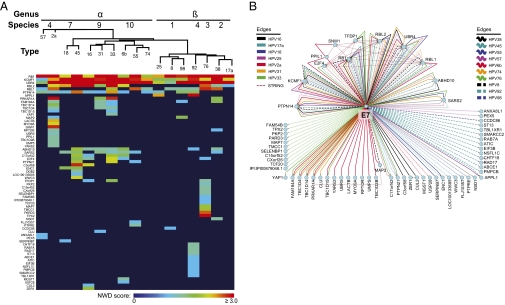
Interaction data were generated from the N/Tert-E7 cell lines and processed in CompPASS by comparison with the statistics table described earlier (Fig. 2 and Datasets S5, S6, and S7). Sixty-eight HCIPs were determined to interact with one or more of the E7 bait proteins, with statistical score cutoffs as described (Materials and Methods). Six of the HCIPs interact with 10 or more of the E7s tested in the studies; these include RB1 and related pocket proteins RBL1 (p107) and RBL2 (p130), UBR4, and KCMF1. As expected, RB1 was detected as a HCIP with each of the E7 baits tested. RBL1 and RBL2 were each detected with 10 of 17 E7 baits, although the 10 E7s positive for the interaction were not the same in each case. Several alternatives are suggested by this observation. It is possible that the limitations of detecting E7-associated proteins in the MS setting prevent us from measuring what may be a conserved interaction (e.g., with RBL1/2) in these cells. This highlights the importance of confirming interactions by IP-Western blotting, as shown later. Alternatively, different E7s may bind preferentially to RBL1 instead of RBL2, or vice versa. The RB1/RBL1/RBL2-associated protein E2F4 was also detected in association with five of the E7 baits, presumably as part of a larger, multiprotein complex. This confirms the IP of multisubunit complexes by this method.
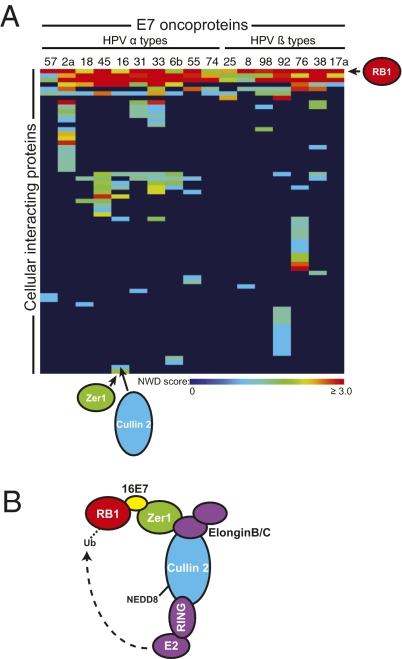
Fig. 2.
HCIPs of HPV E7s. (A) Heat map representing PPIs identified by IP-MS/MS and CompPASS analysis when one of 17 unique HPV E7 proteins was used as a bait. Colors in the heat map represent NWD scores, whereby an NWD score of 1 or greater defines a high-confidence interaction. An interactor is included on the heat map if it was detected at least once in any of several repeat experiments with some of the E7 baits. E7 baits were ordered across the top of the map according to the established HPV phylogeny (1); HCIPs were arranged by using a Manhattan distance hierarchical clustering analysis. (B) Interactome representing HCIPs in complex with various HPV E7 baits.
UBR4 and KCMF1 are also present in the group of HCIPs whose interaction with E7 is highly conserved across virus types. UBR4 was previously reported to bind to E7s from HPV16, 6b, and 11, and to bovine papillomavirus (BPV) E7 (25, 26). These data establish that the interaction of UBR4 with E7 is highly conserved, with each of the E7s tested interacting with UBR4. In preliminary experiments we have determined by IP-MS/MS and CompPASS analysis that UBR4 interacts with KCMF1 in the absence of E7, suggesting the possibility that E7, KCMF1, and UBR4 are present as part of a larger complex in cells. KCMF1 is a protein that contains a RING finger and can function as an ubiquitin-protein ligase (27).
In addition to proteins that are highly conserved in their interactions with E7s, the list of HCIPs includes 16 that interact with two to seven of the E7s tested. The remaining 46 HCIPs were detected in complex with only one of the E7 baits. This distribution is quite similar when the profile of cellular proteins interacting with alpha genus E7s is compared with the beta-genus E7s. Alpha and beta E7 proteins share many of the interactors detected frequently and diverge in the set of HCIPs that interact with a small number of E7s. Some of these HCIPs are likely true type- or species-specific E7 interactors, whereas others are more probably proteins that interact with multiple E7s infrequently, with low abundance, or transiently, limiting our ability to detect them in complex with multiple E7 baits.
Conserved, Species-Specific, and Type-Specific Interactions with HPV E7 Proteins.
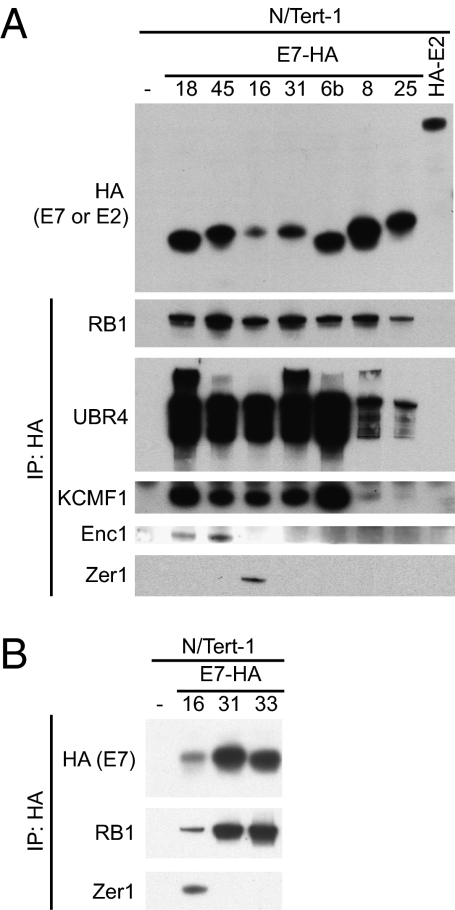
To confirm that we could detect known and novel interactions of cellular proteins with E7s, we selected a subset of the HCIPs introduced earlier for validation and further study. HA-tagged bait and associated proteins were immunopurified from seven of the HPV E7 cell lines, parental N/Tert-1 cells, or cells expressing HA-tagged HPV18 E2. Associated proteins were detected by Western blotting (Fig. 3A). Binding of RB1 to each of the E7 baits, but not controls, served as a positive control for detection of a known interactor. Additionally, we confirm that UBR4 and KCMF1 bind to each of the E7 baits tested, with the amount of KCMF1 recovered in the IP comparable to the amount of UBR4 detected. Of note, the alpha-papillomavirus E7s precipitated more UBR4 and KCMF1 than did the beta-papillomavirus E7s.
Fig. 3.
Validation of conserved and specific interactions with HPV E7s. (A) N/Tert-1 cells expressing E7-FlagHA or HA-HPV18 E2 (Top, HA Western blot) were subjected to IP with HA antibody. Immunoprecipitates were separated by SDS/PAGE and Western blotted by using antibodies to RB1, UBR4, KCMF1, ENC1, and ZER1. (B) Additional N/Tert-1 cell lines expressing E7-FlagHA were subjected to IP with HA antibody. Immunoprecipitates were separated by SDS/PAGE and Western blotted by using antibodies to HA, RB1, and ZER1.
Consistent with the initial IP-MS/MS data, other E7-associated HCIPs copurified only with a subset of the E7s. ENC1 was identified in the MS analysis as an interactor of only the species 7 (HPVs 18 and 45) HPV E7 proteins. IP-Western blot experiments confirmed this finding, making ENC1 a validated HPV species 7-interacting protein (Fig. 3A). ENC1, also known as NRP/B, contains BTB and kelch domains (28). Although ENC1 has been reported to bind to RB1 (28), in an IP-MS/MS experiment using HA-tagged ENC1 as the bait and subsequent CompPASS analysis, we did not detect RB1. We also confirmed that ZER1 (Zyg11BL) associated only with HPV16 E7. This was true even when we tested all three of the genus alpha, species 9 E7 proteins included in the study, thus validating that ZER1 is a virus type-specific interactor (Fig. 3B). ZER1 contains a BC-box and a CUL2-box and interacts with elongin B, elongin C, and CUL2 (29), and ELONGIN C is required for the binding of ZER1 to CUL2 (30). ZER1 and HPV16E7 have each been previously shown to interact with CUL2 (29, 31).
HPV16 E7 Associates Specifically with CUL2 Whereas E7–Cullin 3 Interaction Is Conserved Across Virus Types.
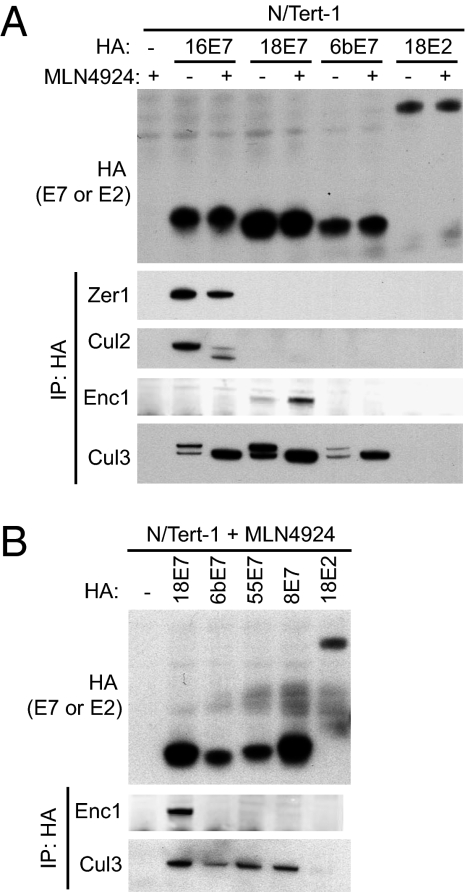
Based on the observation that E7 proteins from two different high-risk species were associated with proteins that are potentially present in cullin-RING E3 ubiquitin ligases, we next examined the possibility that E7s bind to cullins. Cells were treated with MLN4924, an inhibitor of the NEDD8-activating enzyme (32), and protein complexes containing HA were immunopurified and analyzed by Western blotting. MLN4924 was used in these experiments to inhibit cullin neddylation and therefore increase the pool of cullins in complex with their adaptor subunits (20). In cells that were untreated or treated with 1 μM MLN4924 for 4 h before harvest, we detected a virus-specific interaction of HPV16 E7 with CUL2, and consistent with this, we detected ZER1 in complex with E7 only in these samples (Fig. 4A). In the absence of MLN4924, HPV16 E7 binds preferentially to the neddylated, active form of cullin 2 (CUL2; Fig. 4). In contrast, whereas the interaction of ENC1 is specific for species 7 E7s, each E7 that was tested interacted with cullin 3 (CUL3; Fig. 4 A and B). This suggests that ENC1 does not always mediate the interaction of E7 with CUL3. No other BTB proteins were identified as HCIPs by using the criteria presented here, and the only other BTB protein present in the data set is IVNS1ABP. IVNS1ABP (33) was identified in association with HPV45 E7 with statistical scores just below the threshold required for classification as an HCIP in these studies (Dataset S5). We cannot rule out the idea that other proteins not identified in this study may be important mediators of the interaction between CUL3 and other HPV E7s.
Fig. 4.
HPV E7 binding to CUL3 is conserved across virus types, but binding to CUL2 is restricted to HPV 16E7. (A) N/Tert-1 cells expressing E7-FlagHA or HA-HPV18 E2 (Top, HA Western blot) were treated with 1 μM MLN4924 (+) or DMSO control (−) for 4 h and harvested for IP with HA antibody. Immunoprecipitates were separated by SDS/PAGE and Western blotted by using antibodies to ZER1, CUL2, ENC1, and CUL3. (B) N/Tert-1 cells expressing E7-FlagHA or HA-HPV18 E2 (Top, HA Western blot) were treated with 1 μM MLN4924 for 4 h and then harvested for IP with HA antibody. Immunoprecipitates were separated by SDS/PAGE and Western blotted using antibodies to ENC1 and CUL3.
ZER1 Mediates Association of HPV16 E7 with CUL2.
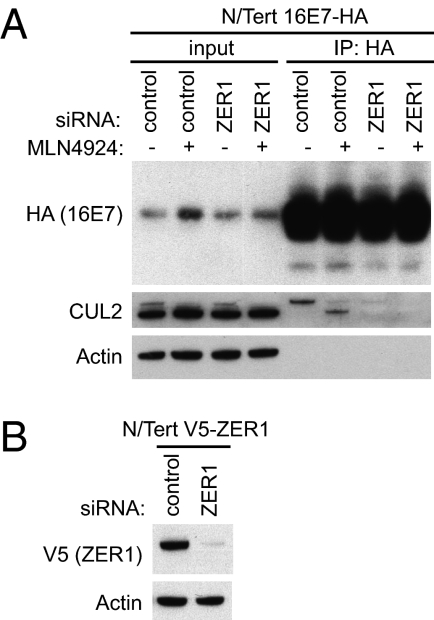
The IP-MS/MS and IP-WB experiments therefore indicate that HPV16E7 interacts with two components of a putative cullin-RING ubiquitin ligase (CRL) complex: CUL2 and ZER1. Based on this observation and previously published results (29, 31), we hypothesized that ZER1, the substrate specificity component of a CUL2-based CRL, mediates the interaction of HPV16 E7 with CUL2 in N/Tert-1 cells. To test this idea, we immunopurified complexes containing HA-tagged HPV16 E7 from N/Tert-16E7 cells that had been transfected with control siRNA or siRNA targeting ZER1. CUL2 was recovered in the HPV16 E7 immunoprecipitates only when ZER1 was present in the cells (Fig. 5), indicating that ZER1 is required for the HPV16 E7 binding of CUL2. This further suggests that ZER1 could function as the adaptor component of a CRL containing CUL2 that is bound by HPV16 E7.
Fig. 5.
ZER1 is required for the interaction of CUL2 with HPV16 E7. (A) N/Tert-1 cells stably expressing HPV16E7-FlagHA were transfected with control siRNA or siRNA targeting ZER1. At 72 h after transfection, cells were treated with 1 μM MLN4924 (+) or DMSO control (−) for 4 h and harvested for IP with HA antibody. Immunoprecipitates were separated by SDS/PAGE and Western blotted by using antibodies to HA, CUL2, and actin. (B) N/Tert-1 cells stably expressing V5-tagged ZER1 were transfected in parallel with control siRNA or siRNA targeting ZER1. At 72 h after siRNA transfection, lysates were harvested and Western blotted by using antibodies to V5 and actin.
HPV16 E7-Mediated Degradation of RB1 Requires ZER1.
HPV E7s are known to bind to the hypophosphorylated form of RB1 (34) that binds E2Fs and blocks cell cycle progression. High-risk HPV types are able to target some of the RB1 present in cells for degradation (35, 36). For HPV16 E7, this degradation has been linked to the interaction of HPV16 E7 with CUL2 (31). This is consistent with the hypothesis that the complex containing CUL2 and ZER1 interacts with HPV16 E7. We tested whether one function of HPV16 E7 in this complex might be to recruit RB1 for ubiquitylation and subsequent degradation by the proteasome. In a cycloheximide chase experiment, we confirmed that RB1 has a shorter half-life in N/Tert-16E7 cells than in parental cells, that depletion of HPV16 E7 by siRNA led to a nearly complete restoration of RB1 levels, and that depletion of ZER1 by siRNA treatment partially restored RB1 levels (Fig. S2A). This confirmed that the differences in RB1 levels observed are caused by protein stability.
Treatment of N/Tert-1 and N/Tert-16E7 cells with the NEDD8-activating enzyme inhibitor MLN4924 led to an increased level of RB1 only in cells that express HPV16 E7 (Fig. 6A), indicating that RB1 is degraded in cells expressing HPV16 E7, but not parental cells, by a CRL-dependent mechanism. HIF1α, a protein that is targeted for degradation via ubiquitylation by a complex containing CUL2 and VHL (37–39), was included as a positive control for MLN4924 activity.
Fig. 6.
ZER1 is required for the HPV16 E7- and CRL-mediated degradation of RB1. (A) Control cells or N/Tert-1 cells stably expressing HPV16 E7-Flag HA were transfected with control siRNA, then, 72 h after transfection, treated with 1 μM MLN4924 (+) or DMSO control (−) for 4 h and harvested. Lysates were separated by SDS/PAGE and Western blotted by using antibodies to RB1, HIF1α, and actin. An asterisk indicates the faster-migrating, hypophosphorylated form of RB1. (B) Control cells or N/Tert-1 cells stably expressing HPV16 E7-Flag HA were transfected with siRNAs as indicated and harvested 72 h after transfection. Lysates were separated by SDS/PAGE and Western blotted by using antibodies to RB1, HA, elongin C (EloC), CUL2, HIF1α, and actin. An asterisk indicates the faster-migrating, hypophosphorylated form of RB1. (C) Proposed model of RB1 interaction with CRL containing CUL2 and ZER1.
To further examine the CRL required for HPV16 E7-dependent RB1 degradation, we measured the level of RB1 in N/Tert-1 and N/Tert-16E7 cells after transfecting cells with siRNAs that target individual components of such a complex. Cells were treated with control siRNA or siRNAs targeting HPV16 E7, ZER1, elongin C, and CUL2. Elongin C is part of CUL2-based CRLs and has been shown to be required for the interaction of ZER1 with CUL2 (30). Depletion of any of these four potential components of the CUL2–ZER1–16E7 ligase complex led to a restoration of the fastest-migrating, hypophosphorylated form of RB1 that is known to be bound by HPV16 E7 (Fig. 6B). This occurs only in HPV16 E7-expressing cells, as RB1 levels do not vary following siRNA treatment in parental N/Tert-1 cells. We therefore propose that HPV16 E7 recruits RB1 to an active CRL complex, as shown in Fig. 6C.
In some experiments, CUL2 knockdown had a more modest effect on the level of hypophosphorlyated RB1, and the siRNA depletion of CUL2 did not affect Rb stability (Fig. S2B). To determine whether a different cullin might complement the loss of CUL2 and mediate the degradation of RB1 when CUL2 is absent, we examined RB1 levels in HPV16E7 cells that had been treated with siRNAs targeting CUL3, CUL5, and both CUL2 and CUL5 (Fig. S3). CUL5 is of particular interest because it shares with CUL2 the ability to bind elongin B and elongin C. None of these knockdowns had an effect on RB1 levels that was significantly different from the effect of transfection with CUL2 siRNAs alone. There are several possible explanations for this observation. First, because CUL2 is a factor central to multiple cellular processes, its knockdown might have indirect effects on RB1 levels or phosphorylation, perhaps leading to cell cycle alterations. Second, an siRNA knockdown experiment does not completely eliminate production of a target protein in cells. Reducing the levels of an adaptor component such as ZER1 in cells may limit the pool of functional ligases to the point at which RB1 degradation is severely compromised, whereas, in contrast, a reduced level of the CUL2 enzymatic component may still allow protein degradation to occur. These effects could explain the variable ability to detect an increased level of RB1 in HPV16 E7 cells even when CUL2 levels are reduced following siRNA treatment.
Discussion
HPV Diversity Is Reflected by Viral Interactions with Host Cellular Proteins.
The large number of papillomaviruses found in nature suggests that a spectrum of differences will be reflected in their molecular biology and host cell interactions. We have established a system (Fig. 1) for the identification of interactions between host cellular proteins and proteins encoded by the HPVs and have used it to begin to define the similarities and differences among these viruses. In the present study, we have used the HPV E7 proteins to validate the function of our system and have identified several known as well as previously unknown interactions between E7s and cellular factors.
The E7 gene has long been understood to have a central role in HPV replication and for the high-risk alpha-virus E7s to drive cancer development when the normal regulation of the virus life cycle is lost. The oncogenic activity of E7 is linked to the binding and inactivation, in some cases via degradation, of RB1. The detection of RB1 in complex with E7 in each of our IP-MS/MS experiments was an early indication that the system functions properly (Figs. 2 and 3). Other functions of E7 have been identified, and several of these also contribute to dysregulation of the host cell cycle (40).
In addition to known interactors of E7s including RB1 and related proteins, the present study revealed interactions with E7. Some of these interactions are conserved and were detected with many of the E7s, suggesting a common role for the interaction in HPV-mediated modulation of host cellular processes. We show here that each E7 tested bound to UBR4 (Figs. 2 and 3), although the effect of this interaction is less clear than that of E7 binding to RB1. For HPV16 E7 and BPV E7, UBR4 binding appears to contribute to cellular transformation and anchorage-independent growth (25, 26). The observation that UBR4 bound to all HPV E7s, including those not associated with cancer, indicates that this interaction may also have a key function in virus replication. Furthermore, our identification of the ubiquitin ligase KCMF1 in complex with UBR4 suggests that the E7–UBR4 interaction may be important in the recruitment of additional cellular factors to complexes containing E7s. We have thus identified interactors of the E7 oncoprotein, although surely our list of interactors is not exhaustive and is limited by features of the system. For example, for some proteins the addition and location of an epitope tag might affect PPIs. We chose to add an epitope tag to the C terminus of E7 because an N-terminal tag has previously been shown to limit binding to UBR4 and to block the ability of BPV E7 and HPV16 E7 to transform cells (25, 26).
HPV Proteins Interact with Cellular CRL Machinery in Multiple Ways.
In contrast to RB1 and UBR4, other cellular proteins are bound by only a subset of the HPV E7s. The idea that different papillomaviruses can have different cellular targets is not new, and a good example is the targeted degradation of the tumor suppressor p53 by E6 proteins from high-risk HPVs only. The identification of such differences has been instrumental in the understanding of HPV biology, thus motivating our search for additional virus type-restricted interactions.
We identified several of these less conserved interactions, including at least one validated species-specific and one validated type-specific interaction with an HPV E7. In each case, an HPV E7 interacted with a potential adaptor subunit of a CRL. In CRL complexes, substrates are recruited to the cullin core via an interaction with an adaptor subunit (41). In the case of CUL2-based CRLs, the core includes CUL2, elongin B, and elongin C, and adaptor proteins contain a BC-box and a CUL2-box (42). In CUL3-based CRLs, adaptor proteins that contain BTB domains recruit substrates. It should be noted that BTB domain-containing proteins have many cellular functions besides recruitment of CUL3 substrates (43). ZER1 (also called Zyg11BL) contains BC- and CUL2-boxes and has at least two human homologues, Zyg11A and Zyg11B (29, 30). ZER1 binds uniquely to HPV16 E7 (Figs. 2 and 3), but interactions with Zyg11A or Zyg11B were never detected in the HPV16 E7 IP-MS/MS experiments. ENC1, a protein that contains BTB and kelch domains, binds to both of the species 7 E7s tested (Figs. 2–4).
These interactions of E7 with CRLs are noteworthy because, in addition to binding and sequestration of RB1, high-risk HPV E7 proteins are able to target a portion of the RB1 protein present in cells for ubiquitin-mediated degradation (35, 44, 45). In uninfected cells, RB1 is quite stable, with a long half-life and a steady-state level that does not change following treatment with proteasome inhibitors. In contrast, HPV16 E7 expression decreases the half-life of RB1 and results in a correspondingly low steady-state level of the protein. Consistent with this observation, it has been shown that HPV16E7 and RB1 are present in a complex with CUL2, a component of the cellular CRL machinery (41), suggesting that this interaction is important for the HPV16 E7-mediated targeting of RB1 for ubiquitylation and subsequent degradation (31).
A number of different viral proteins have been identified in complex with CRLs before, with various functional consequences (46). Here we have shown that HPV16 E7 is unique among the 17 E7 proteins we tested in its ability to bind to CUL2 and ZER1. We further demonstrate that ZER1 mediates the binding of an active, neddylated form of CUL2 to HPV16 E7, and that these three proteins are likely present in a single complex (Fig. 4). We hypothesize that this interaction represents a functional CRL to which HPV16 E7 binds and that this CRL contains ZER1, although we recognize that these data do not prove that the interaction between CUL2 and ZER1 is direct. Consistent with the previous report that HPV16 E7 recruits CUL2 to ubiquitylate and target RB1 for proteasomal degradation (31), we show that the levels of hypophosphorylated RB1 increase with the loss of subunits of this predicted complex (Fig. 6), and we also show that ZER1 is required for the destabilization of RB1 in HPV16 E7-expressing cells (Fig. S2A). Thus, our proposed model refines the previous understanding of HPV16 E7-targeted degradation of RB1, and may in the future define other proteins that are being targeted by this complex. It also suggests that the mechanisms by which other high-risk E7 proteins target RB1 for degradation have yet to be elucidated.
There appear to be many interactions between HPV E7s and the CRL machinery, and we have shown here that CUL3 can interact with each of the E7s we tested (Fig. 4). Although our interest in such an interaction was prompted by the observation that only HPV species-7 E7 proteins bound to the BTB protein ENC1, our further experiments indicated that ENC1 is likely not mediating the interaction of E7 with CUL3 in every case. Additional experiments will be needed to determine what proteins are present in E7–CUL3 complexes and to understand the effects of HPV species-7 E7s binding to ENC1. Our result also suggests that RB1 is not, as previously reported, a binding partner of ENC1 (28). If RB1 did bind to ENC1, we would expect that every E7 in the study would immunopurify ENC, mediated by the E7 interaction with RB1. Although it is possible that the ENC1–RB1 interaction could occur only in the absence of E7, this is not reflected in our data, as HA-ENC1 IP-MS/MS experiments did not in any case recover bound RB1.
In summary, our work establishes a robust system for the detection of interactions between HPV and host cellular proteins. We have used this system to further define the mechanism by which HPV16 E7 expression leads to the targeted degradation of RB1, a critical step in the progression from HPV infection to cervical cancer. The identification of unique E7 interactors will also allow for further studies, including determining the downstream effects of CUL3-E7 binding and the specific function of ENC1 binding by a subset of high-risk HPV E7 proteins. Finally, the establishment of this system leaves us poised for the high-throughput identification of HPV–host interactions that will increase our understanding of how molecular differences among the virus types lead to varied consequences of HPV infection.
Materials and Methods
Cloning and Plasmids.
HPV genomes were obtained as plasmid clones (sources are provided in Dataset S2) and propagated in bacteria. DNAs encoding individual HPV ORFs were generated by PCR and recombined into pDONR223 as described (15, 20), and the resulting plasmids are listed in Dataset S3. pDONR-HPV ORF clones were sequence-verified, and 17 HPV E7 ORFs in pDONR223 were transferred by Gateway recombination (Invitrogen) into the MSCV-C-FLAG-HA-PURO vector for retroviral expression (15). Retroviral expression vectors containing HPV E7 ORFs are listed in Dataset S4. Similarly, cellular ORFs in pDONR223 were obtained from the human ORFeome collection (47) and recombined into MSCV-N-HA-IRES-PURO or MSCV-N-V5-IRES-PURO. MSCV-N-HA-IRES-PURO and MSCV-N-V5-IRES-PURO were generated from MSCV-N-FLAG-HA-IRES-PURO (15) by standard site-directed mutagenesis techniques. Additional cellular ORFs in retroviral expression vectors have been previously described (15). A summary of plasmid sources and cellular ORFs used to generate the CompPASS stats table is provided in Dataset S1.
Tissue Culture and Cell Lines.
N/Tert-1 cells (19) were provided by James Rheinwald (Harvard Medical School, Boston, MA) and cultured according to established protocols. Cells were grown in K-SFM (Keratinocyte-SFM) (Invitrogen) prepared according to the manufacturer's instructions with the additions of 100 U/mL penicillin and 100 μg/mL streptomycin, 0.3 mM CaCl2 (final [CaCl2] is ∼0.4 mM), and 4 μg/mL hygromycin B, and two adaptations: EGF was added to a final concentration of 0.2 ng/mL and bovine pituitary extract added to a final concentration of 25 μg/mL Retroviruses were generated by using the viral vectors described earlier according to published protocols (15, 20) and used to infect low-passage N/Tert-1 cells. Following infection, N/Tert-1 cells expressing cellular and HPV ORFs were selected and maintained in K-SFM prepared as described earlier with the addition of 0.5 μg/mL puromycin. Cell lines were maintained at no greater than 35% confluence to limit cellular differentiation.
For growth to high density for use in IP experiments, N/Tert-1 cell lines were grown to 35% confluence as described earlier, and then media were replaced with a 1:1 mixture of K-SFM and DF-K medium that was subsequently replenished every 48 h. DF-K medium is 0.5× calcium-free, glutamine-free DMEM (Invitrogen), 0.5× Ham F-12 (Invitrogen), 0.2 ng/mL EGF, 25 μg/mL bovine pituitary extract, 1.5 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were harvested at approximately 80% confluence and snap-frozen in liquid nitrogen then stored at −80 °C until the time of processing. Four 15-cm tissue culture dishes of N/Tert-1 cells were used for each IP-MS/MS experiment.
To enhance the stability of CRL complexes, cells were treated with 1 μM MLN4924 (Takeda Millennium) or DMSO control for 4 h before harvest. For protein stability experiments, cells were treated with 40 μg/mL cycloheximide and harvested by direct lysis in 1× reducing sample buffer [50 mM Tris (pH 6.8), 0.2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 25 mM sodium fluoride, 1 mM sodium orthovanadate, 5 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 50 μM leupeptin, and 100 μM pepstatin A] at the indicated time points.
siRNA Transfections.
N/Tert-1 cells were transfected with siRNA duplexes (Dharmacon/Thermo Scientific) by using DharmaFECT 2 and siRNAs with targets and catalog numbers as follows: nontargeting siRNA no. 1 (D-001810-01), C9ORF60 (ZER1, J-019424–17, -18, -19, -20), CUL2 (J-007277–05, -06, -07, -08), CUL3 (J-010224–06, -07, -08, -09), CUL5 (J-019553–05, -06, -07, -08), elongin C (J-010541–09, -10, -11, -12), and two custom-designed siRNAs targeting HPV16 E7. In several cases, only one or two of the siRNAs tested resulted in efficient knockdown of the target protein in N/Tert-1 cells [C9ORF60 (ZER1, J-019424-19), CUL2 (J-007277–06, -08), CUL3 (J-010224–06, -07), CUL5 (J-019553–05, -08), Elongin C (J-010541-10), and the custom designed siRNA targeting HPV16 E7 with the sequence GGACAGAGCCCAUUACAAUUU]; these validated siRNA duplexes were then used for the experiments in the study. SiGLO red (D-0011630-02; Dharmacon) was used to visualize efficient transfection in a control well. N/Tert-1 cells were transfected according to a published protocol (48) with siRNAs at a final concentration of 40 nM (siRNA-Western blot experiments) or 50 nM (siRNA-IP-Western blot experiments). The protocol was modified for N/Tert-1 transfection by culturing cells in K-SFM lacking antibiotics for 1 d before transfection, transfecting cells, then replacing media containing the transfection reagent at 6 to 8 h after transfection. At 24 h after transfection, media was replaced with K-SFM and DF-K mixed 1:1, and cells were harvested for Western blot or IP as described earlier at 72 h after transfection.
IP.
HA-tagged proteins were immunoprecipitated as previously described (15, 20). For MS analysis, protein-HA resin complexes were washed three times in PBS solution, eluted with HA peptide, and trichloroacetic acid-precipitated as described (15, 20). For Western blot analysis, proteins were eluted from the HA beads by boiling in 1× RSB.
MS and Data Analysis.
Trypsin digestion, stage-tip purification, and MS analysis of immunoprecipitated proteins were performed as described previously (15, 20). CompPASS analysis was performed as previously published using an N/Tert-1–derived stats table populated with data generated from 37 stable cell lines, each expressing a single HA-tagged human protein bait. Cell lines used in the stats table are listed in Dataset S1. In these analyses, a protein identified by MS/MS is considered to be a HCIP if its NWD score (normalized, weighted D-score, which is a statistical metric that incorporates the uniqueness, abundance, and reproducibility of a given interactor) is greater than 98% of the NWD scores in the statistics table. For interactome analysis, the HCIPs from all the HPV E7 IP-MS/MS analyses were combined, and interactions among these proteins determined by using the STRING database, with a score cutoff of 700. The resulting interactions were visualized by using Cytoscape (49).
Western Blotting.
Proteins were separated on NuPAGE (Invitrogen) or SDS/PAGE gels and transferred to PVDF. After blocking in 5% nonfat dried milk in Tris-buffered saline solution (pH 7.4) with 0.05% Tween-20, blots were incubated with primary antibodies as follows: RB1 (Calbiochem/EMD), UBR4 (gift from Yoshihiro Nakatani, Dana-Farber Cancer Institute, Boston, MA) (50), KCMF1 (Sigma-Aldrich), ENC1 (BD Transduction Labs), ZER1 (GeneTex), CUL2, CUL3, CUL5 (Bethyl), actin (Millipore), V5 (Invitrogen), HIF1α (Cell Signaling Technology), or elongin C (Novus). Membranes were washed in Tris-buffered saline solution (pH 7.4) with 0.05% Tween-20 and incubated with HRP-coupled anti-mouse or anti-rabbit antibodies or an Alexa 680-coupled anti-mouse antibody and detected by using Western Lightning chemiluminescent substrate or a Li-COR infrared imaging system. HA-tagged proteins were detected by using an HA antibody conjugated to HRP (Roche) and visualized on film.
Supplementary Material
Acknowledgments
We thank members of the P.M.H. and J.W.H. laboratories for helpful discussions and suggestions. We are grateful for the contributions from several laboratories listed in Dataset S2 for the cloned HPV genomes, the UBR4 antibody (Yoshihiro Nakatani, Dana-Farber Cancer Institute), and the N/Tert-1 cells (James Rheinwald, Brigham and Women's Hospital). This work was supported by National Institutes of Health (NIH) National Research Service Award F32AI080075 (to E.A.W.) and a Roche Postdoctoral Fellowship (to E.A.W.); NIH Grants 1RC1 CA145188 (to J.W.H. and P.M.H.) and P01 CA50661 (to P.M.H.); and a National Science Scholarship from the Agency for Science, Technology and Research of Singapore (to M.J.A.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database.
See Author Summary on page 1372.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116776109/-/DCSupplemental.
References
- 1.Bernard HU, et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howley PM, Lowy DR. Papillomaviruses. In: Knipe DM, et al., editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams and Wilkins; 2006. [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Lowy DR, Solomon D, Hildesheim A, Schiller JT, Schiffman M. Human papillomavirus infection and the primary and secondary prevention of cervical cancer. Cancer. 2008;113(7 suppl):1980–1993. doi: 10.1002/cncr.23704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson N, Guida P, Münger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 7.Münger K, et al. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 9.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 10.Jablonska S, Orth G. Epidermodysplasia verruciformis. Clin Dermatol. 1985;3:83–96. doi: 10.1016/0738-081x(85)90052-5. [DOI] [PubMed] [Google Scholar]
- 11.Pfister H. Chapter 8: Human papillomavirus and skin cancer. J Natl Cancer Inst Monogr. 2003;31:52–56. doi: 10.1093/oxfordjournals.jncimonographs.a003483. [DOI] [PubMed] [Google Scholar]
- 12.Akgül B, Cooke JC, Storey A. HPV-associated skin disease. J Pathol. 2006;208:165–175. doi: 10.1002/path.1893. [DOI] [PubMed] [Google Scholar]
- 13.Feltkamp MC, de Koning MN, Bavinck JN, Ter Schegget J. Betapapillomaviruses: Innocent bystanders or causes of skin cancer. J Clin Virol. 2008;43:353–360. doi: 10.1016/j.jcv.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell BC, et al. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol Cell. 2010;40:645–657. doi: 10.1016/j.molcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell ML, et al. NCoR1 mediates papillomavirus E8;E2C transcriptional repression. J Virol. 2010;84:4451–4460. doi: 10.1128/JVI.02390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickson MA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee PC, Sowa ME, Gygi SP, Harper JW. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell. 2011;43:392–405. doi: 10.1016/j.molcel.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litterman N, et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 2011;9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan MK, Lim HJ, Harper JW. SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2011;31:3687–3699. doi: 10.1128/MCB.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeMasi J, Huh KW, Nakatani Y, Münger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc Natl Acad Sci USA. 2005;102:11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huh KW, et al. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci USA. 2005;102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang JH. FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers. FEBS Lett. 2004;578:21–25. doi: 10.1016/j.febslet.2004.10.071. [DOI] [PubMed] [Google Scholar]
- 28.Kim TA, et al. NRP/B, a novel nuclear matrix protein, associates with p110(RB) and is involved in neuronal differentiation. J Cell Biol. 1998;141:553–566. doi: 10.1083/jcb.141.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahrour N, et al. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- 30.Vasudevan S, Starostina NG, Kipreos ET. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 2007;8:279–286. doi: 10.1038/sj.embor.7400895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh K, et al. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 33.Wolff T, O'Neill RE, Palese P. NS1-Binding protein (NS1-BP): A novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imai Y, Matsushima Y, Sugimura T, Terada M. Purification and characterization of human papillomavirus type 16 E7 protein with preferential binding capacity to the underphosphorylated form of retinoblastoma gene product. J Virol. 1991;65:4966–4972. doi: 10.1128/jvi.65.9.4966-4972.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 36.Jones DL, Thompson DA, Münger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 37.Cockman ME, et al. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 38.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 39.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 40.McLaughlin-Drubin ME, Münger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384:335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 42.Kamura T, et al. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: The BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 44.Berezutskaya E, Yu B, Morozov A, Raychaudhuri P, Bagchi S. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 1997;8:1277–1286. [PubMed] [Google Scholar]
- 45.Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barry M, Früh K. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci STKE. 2006;2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- 47.Lamesch P, et al. hORFeome v3.1: A resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith JA, et al. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc Natl Acad Sci USA. 2010;107:3752–3757. doi: 10.1073/pnas.0914818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakatani Y, et al. p600, a unique protein required for membrane morphogenesis and cell survival. Proc Natl Acad Sci USA. 2005;102:15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]