Abstract
Myosin Va (myoV) is a processive molecular motor that transports intracellular cargo along actin tracks with each head taking multiple 72-nm hand-over-hand steps. This stepping behavior was observed with a constitutively active, truncated myoV, in which the autoinhibitory interactions between the globular tail and motor domains (i.e., heads) that regulate the full-length molecule no longer exist. Without cargo at near physiologic ionic strength (100 mM KCl), full-length myoV adopts a folded (approximately 15 S), enzymatically-inhibited state that unfolds to an extended (approximately 11 S), active conformation at higher salt (250 mM). Under conditions favoring the folded, inhibited state, we show that Quantum-dot-labeled myoV exhibits two types of interaction with actin in the presence of MgATP. Most motors bind to actin and remain stationary, but surprisingly, approximately 20% are processive. The moving motors transition between a strictly gated and hand-over-hand stepping pattern typical of a constitutively active motor, and a new mode with a highly variable stepping pattern suggestive of altered gating. Each head of this partially inhibited motor takes longer-lived, short forward (35 nm) and backward (28 nm) steps, presumably due to globular tail-head interactions that modify the gating of the individual heads. This unique mechanical state may be an intermediate in the pathway between the inhibited and active states of the motor.
Keywords: molecular motors, regulation, single molecules
Myosin Va (myoV) is a double-headed processive molecular motor that transports intracellular cargo (e.g., vesicles, secretory granules) along actin tracks. As it moves, myoV takes multiple hand-over-hand steps powered by its hydrolysis of ATP (1, 2). The motor is formed by the dimerization of two heavy chains, with each heavy chain having four distinct structural domains: (i) an N-terminal head which has both an actin-binding site and active site; (ii) a long neck, containing six IQ domains, that is stabilized by binding of calmodulin, and serves as a rigid lever arm; (iii) an α-helical coiled-coil dimerization domain; and (iv) a C-terminal globular tail domain (GTD) responsible for cargo binding and regulation/inhibition of the motor’s activity [for review see (3)]. What effect the GTD has upon myoV stepping dynamics is still a matter of debate.
Hydrodynamic (4–6) and structural studies (7, 8) provide a mechanistic view of GTD-specific motor regulation. MyoV is both enzymatically and mechanically “off” when folded into a compact structure (approximately 15 S), and “on” when extended (approximately 11 S). Inhibition is due to the GTD simultaneously binding to both heads, which may prevent the motor from completing its enzymatic cycle by interfering with the release of hydrolysis products from the catalytic site (9, 10). To activate the motor, the electrostatic GTD-head interactions must be disrupted. In vitro this has been shown to occur either by increasing the ionic strength or by binding calcium to calmodulin, which leads to a major rearrangement of calmodulin bound to the myoV lever arm (4–6, 11). In vivo, there is evidence that calcium binding activates myosin Vb (12), and it is generally assumed that cargo binding to the GTD will activate the motor. In support of cargo-mediated activation, a processive tripartite complex consisting of full-length myoV (FL-myoV), melanophilin, and Rab27a-GTP, the adapter proteins for melanosome transport, was observed at 50 mM KCl (13). This suggests that even at low ionic strength, high affinity interactions between adapter proteins and the GTD can compete with and disrupt the GTD-head interactions, allowing unfolding and activation of the motor. Therefore, myoV regulation may be viewed as an equilibrium between two structural/functional states: folded/inhibited and extended/active. Interestingly, processive FL-myoV motors have been observed in vitro under conditions (i.e., 25 mM KCl) where the motor should have been folded and inhibited (5, 7, 8, 14, 15). These FL-myoV may be: (i) unregulated “rogue” motors; (ii) motors that land on actin while transiently unfolded and thus are mechanically active; or (iii) a new structural state that is functionally distinct and may be physiologically important for efficient cargo transport and delivery. To address this question, we used high spatial and temporal resolution total internal reflectance fluorescence (TIRF) microscopy to characterize the processive behavior of single Quantum-dot (Qdot)- or YFP-labeled Baculo/Sf9-expressed FL-myoV molecules that are capable of autoinhibition, compared to that of a constitutively active, truncated heavy meromyosin construct (HMM-myoV) that lacks the GTD (Fig. S1). Two FL-myoV splice variants that are expressed in mammalian tissue were used to ensure the generality of our results (16–22).
Results and Discussion
Both FL-myoV Isoforms Undergo the Folded-to-Extended Transition.
The two FL-myoV splice variants that were characterized in this study originate from six alternatively spliced exons, named A-F, that lie between the PEST site in the coiled-coil (which marks the end of the HMM-myoV) and the GTD (Fig. S1) (16–22). Some of these exons are predicted to form an α-helical coiled-coil, while others are unstructured (3). The isoform prevalent in brain contains exons ABCE (FLB-myoV), while that found in melanocytes contains exons ACDEF (FLM-myoV) (Fig. S1) (16–22). When structurally characterized by analytical ultracentrifugation, both isoforms showed a similar sedimentation velocity profile, with an approximately 15 S-folded species formed at 100 mM NaCl, and an approximately 10 S extended state favored at 300 mM NaCl (Fig. S2A). Thus, even though these exons are in close proximity to the head domain when myoV is in the folded conformation, alternative splicing does not affect the folding ability of the motor. To better define the range of salt concentrations over which the transition between the folded-to-extended conformational state occurs, we determined the sedimentation velocity profile for the FLB-myoV at 100, 200, 250, and 300 mM KCl (Fig. S2B). At 250 mM KCl or higher, the FLB-myoV is fully extended. At 200 mM KCl, the FLB-myoV sediments at 11.5 S, consistent with a population of predominantly extended molecules in equilibrium with a small amount of folded monomers. The shift between the folded and the extended states thus occurs at physiological ionic strengths (i.e., between 100 and 250 mM KCl).
Most FL-myoV Molecules Bind Actin but Remain Stationary at Low Ionic Strength.
At low ionic strength the GTD-head interactions result in a highly inhibited motor in solution that hydrolyzes ATP 100-fold slower than the fully active molecule (4, 5, 9). Whether or not inhibited motors bind to actin filaments in the presence of MgATP is still uncertain (6–10). One-headed binding of the folded conformation to actin was shown by negative staining in the presence of MgADP and phosphate (7). While Thirumurugan et al. confirmed binding of the inhibited motor to actin under similar conditions in electron microscopic images, actin decoration was not observed in the presence of high (≥1 mM) MgATP (8). It is worth noting that even constitutively active HMM-myoV, with its high duty ratio, poorly decorated actin at 1 mM MgATP (23), suggesting that the conditions used to image actin-bound FL-myoV in the electron microscope prevent the full extent of such binding to be realized. Under our experimental conditions (25 mM KCl, 1 mM MgATP), actin filaments were decorated with stationary, Qdot-labeled FL-myoV, unlike HMM-myoV (Movies S1 and S2). Even if inhibition traps both heads in a weakly-bound-ADP/Pi state (10), the folded conformation could still decorate actin, knowing that myoV binds actin 10–20-fold more tightly in the weakly bound state compared to myosin II (24). Therefore, the stationary motors, which remained so for the entire recording period (≤ 2 min), reflect a fully inhibited mechanical state that binds actin.
FL-myoV Switches Between Fast and Slow Processive Velocities in an Ionic Strength-Dependent Manner.
The majority (84%) of FL-myoV that interact with actin are stationary at 25 mM KCl, in contrast to HMM-myoV where only 7% are stationary. However, the remaining 16% of FL-myoV are processive, as we reported previously using the same FLM-myoV construct but with YFP at the C-terminus (5). This result was surprising since the motor was expected to be folded and completely inhibited under these conditions. Normalized to HMM-myoV, the number of such processive FL-myoV motors was 10% at 25 mM KCl, increasing to 25% at 100 mM KCl. This observation is consistent with the FL-myoV shifting its equilibrium from the inhibited to the active state as ionic strength is increased. When displacement versus time traces of both FL-myoV splice variants were compared to HMM-myoV at 25 mM KCl (Fig. 1 A and B and Fig. S3A), the movement of FL-myoV was significantly different. The FL-myoV processive runs are broken up into periods of fast and slow (< 50 nm/s) velocity, whereas the HMM-myoV displacement traces are linear with a constant velocity (Movies S3 and S4). This difference was not as apparent at the highest ionic strength (200 mM KCl) where both the FL-myoV and HMM-myoV are characterized by constant velocity processive runs (Fig. 1 A and B and Fig. S3A). These periods of fast and slow velocity were not an artifact of the Qdot labeling strategy, because a C-terminal YFP-tagged FL-myoV (YFP-FL-myoV) (Fig. S1), previously shown to be completely regulated (5), also demonstrated a mixture of fast and slow velocities during a processive run (Fig. S3D).
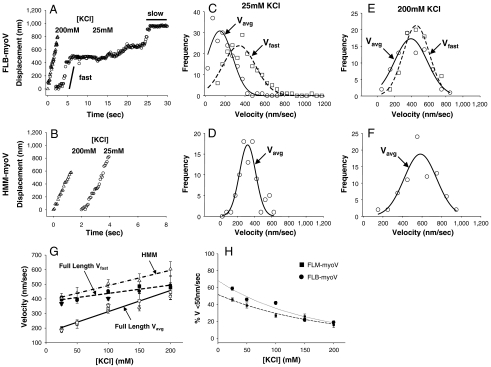
Fig. 1.
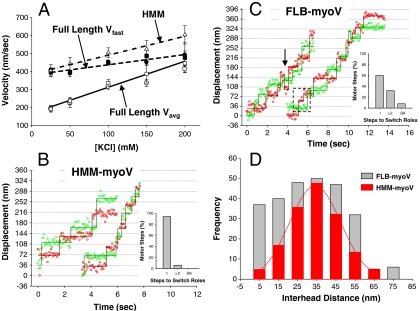
Ionic strength dependence of myoV displacements and velocities. Typical displacement versus time traces for FLB-myoV (A) and HMM-myoV (B) at 200 mM (triangles) and 25 mM (circles) KCl at 1 mM ATP. In A, solid lines highlight the periods of fast and slow processive movement (C–F) Velocity histograms for FLB-myoV and HMM-myoV at 25 mM and 200 mM KCl. Circles represent Vavg and squares represent Vfast. At 25 mM KCL Vavg and Vfast for FLB-myoV are 147 ± 10 nm/s and 343 ± 12 nm/s (n = 175), respectively, while Vavg for HMM-myoV is 320 ± 18 nm/s (n = 145). At 200 mM KCl, Vavg and Vfast for FLB-myoV are 394 ± 22 nm/s and 456 ± 17 nm/s (n = 80), respectively, while Vavg for HMM-myoV is 583 ± 19 nm/s (n = 115). (G) Vavg and Vfast as a function of ionic strength for the FL-myoV. FLB-myoV (open and closed circles), FLM-myoV (open and closed squares), YFP-FL-myoV (open and closed down triangles) and Vavg for HMM-myoV (open up triangles). Vavg of the FL-myoV is significantly slower than Vavg of HMM-myoV at all [KCl] (ANOVA, p ≤ 0.05). (H) The percentage of time that the FL-myoV spend traveling at slow processive speeds (%Vslow) vs. KCl concentration. Values are reported as mean ± S.E.M.
To characterize how the two phases of FL-myoV processive motion affect the overall velocity of all myoV motors, we calculated the average velocity (Vavg) for a processive run (Fig. 1 and Fig. S3). In addition, a velocity for only the fast processive periods (Vfast) (Fig. 1 and Fig. S3) within FL-myoV and YFP-FL-myoV runs was calculated (see Materials and Methods and Fig. S4). Vavg for FL-myoV increased by 200% with increasing KCl concentration (Fig. 1G), but never attained the velocities observed for HMM-myoV at the comparable KCl concentration. The slower Vavg for the FL-myoV is due to the contribution of the intermittent slow (< 50 nm/s) periods of movement, which are as much as 60% of the motor’s travel time at 25 mM KCl (Fig. 1H). The kinetics of switching between slow and fast processive movement; i.e., kfast→slow and kslow→fast, was characterized from the fits to the exponential distribution of times spent in slow and fast processive periods within a run (Fig. S5), with kfast→slow = 1.8 ± 0.3 s-1 and kslow→fast = 0.8 ± 0.1 s-1 at 25 mM KCl. With increasing ionic strength (i.e., 100 mM KCl), the motor spends more time in the fast processive mode as a result of a 50% reduction in kfast→slow = 0.9 ± 0.2 s-1 without any change in kslow→fast = 0.9 ± 0.2 s-1. Thus, the contribution of the slow period to Vavg is reduced with increasing ionic strength and decreases to as little as approximately 17% at 200 mM KCl (Fig. 1H). When the slow processive periods are not included in Vavg, the FL-myoV and YFP-FL-myoV Vfast approach the velocities of HMM-myoV over the range of salt concentrations (Fig. 1G). In solution at 200 mM KCl, the motor spends the majority of time in the extended conformation (Fig. S2B), which may be related to why the slow processive periods are detected less frequently at this KCl concentration (Fig. 1H). Rapid switching between fast and slow processive periods could still exist that are below our temporal resolution and thus explain why Vfast for FL-myoV does not attain the faster HMM-myoV velocities.
Our observation that the number of processive FL-myoV runs increases with ionic strength suggests that these motors exist in an ionic strength-dependent equilibrium between mechanically inhibited and active states that is determined by the folded-to-extended equilibrium in solution. However, once bound to actin, the active state itself can be characterized by an ionic strength-dependent equilibrium between fast and slow processive movement that determines Vavg. The kinetics of this mechanical equilibrium are related to but not necessarily identical to the kinetics governing the equilibrium between folded and extended conformations in solution in the absence of actin.
FL-myoV Run Lengths Are Ionic Strength-Independent and Shorter than HMM-myoV.
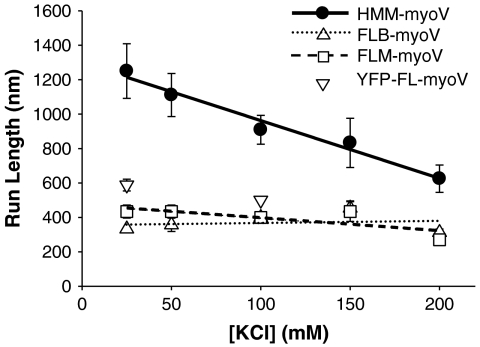
Another characteristic of a processive motor is run length, the distance it travels during an encounter with its cytoskeletal track. For the FL-myoV, characteristic run lengths were significantly shorter than HMM-myoV between 25 and 200 mM KCl (Fig. 2 and Fig. S6). While characteristic run lengths for HMM-myoV decreased by a factor of two over the range of ionic strengths, as previously reported (25), characteristic run lengths for FL-myoV were constant (approximately 400 nm).
Fig. 2.
Ionic strength dependence of run lengths and termination rates at 1 mM ATP. Characteristic run length constants of FL- and HMM-myoV as a function of ionic strength. FLB-myoV (up triangles, dotted line), FLM-myoV (squares, dashed line), and YFP-FL-myoV (down triangles at 25 and 100 mM KCl only) have significantly shorter characteristic run lengths than the HMM-myoV (filled circles, solid line) at all KCl concentrations tested (T test; p ≤ 0.002).
Run lengths are limited by the motor’s termination rate, so the shorter FL-myoV run lengths may result from the motor terminating its run at a faster rate than HMM-myoV during either a fast and/or a slow period of processive motion. Therefore, the period of time that the full-length motor traveled at either the fast or slow velocity prior to termination was determined, and the distribution of such lifetimes fit to exponential decays to estimate the termination rates (kterm) from these two distinct modes of processive movement (Fig. S7). During Vfast , FLB-myoV terminates at a rate of 1.3 s-1 and 0.9 s-1 at 25 and 100 mM KCl, respectively, with rates of termination during Vslow of 0.7 s-1 and 1.4 s-1 at 25 and 100 mM KCl, respectively. These rates are 1.5–2.2-fold faster than for HMM-myoV, which has termination rates of 0.5 s-1 to 0.6 s-1 at 25 and 100 mM KCl, respectively. Thus, in addition to the FL-myoV having slower velocities, the shorter run lengths can be attributed to higher termination rates from these processive states as well. This result implies that the gating between the two heads is altered compared to HMM-myoV, leading to a greater probability of detachment. Alternatively, the GTD may interact with one or both heads of the FL-myoV during its processive motion and adopt a conformation that is susceptible to run termination (see model below).
Stepping Dynamics of FL-myoV Suggests Altered Gating Between Heads.
We next characterized the FL-myoV stepping dynamics during the fast and slow periods of processive movement to determine if they are distinct from that of HMM-myoV (Fig. 3). Twenty-five mM KCl was chosen because movement is equally distributed between Vfast and Vslow (Fig. 1H). Since velocities and run lengths of the FL-myoV were identical for the two splice variants over all ionic strengths (Fig. 1G, 2), we chose only to characterize the stepping dynamics of FLB-myoV.
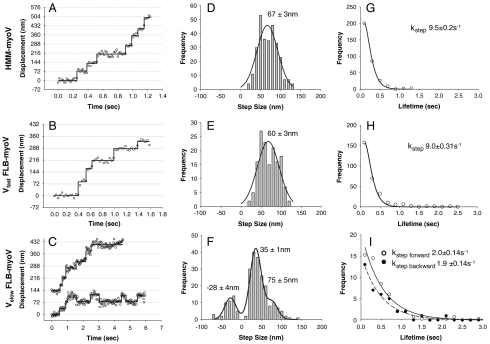
Fig. 3.
Stepping dynamics of HMM-myoV and FLB-myoV. Displacement vs. time traces for HMM-myoV captured at 60 fps (A), during a Vfast period captured at 30 fps (B) and two different traces one of which shows a period of dynamic stall during a Vslow period captured at 30 fps (C) for FLB-myoV. Step size distributions and their mean ± S.E. of HMM-myoV (n = 290) (D), during Vfast periods for FLB-myoV (n = 266) (E), and Vslow periods for FLB-myoV (n = 235) where the distribution was fitted (R2 = 0.94) by the sum of three Gaussians (F). Step lifetime frequency histograms for HMM-myoV (G), for steps during Vfast (H) and Vslow (I) of FLB-myoV with forward steps (open circles) and backward steps (closed circles) as indicated. The stepping rate (kstep) is the exponential decay constant ± S.E.M. Conditions: 25 mM KCl, 1 mM ATP.
When only one motor domain was labeled with a Qdot, displacement versus time traces for the FLB-myoV during fast processive periods show that individual heads take 60 ± 3 nm steps at a rate of 9.0 ± 0.3 s-1 (Fig. 3 B, E, and H). This was not significantly different than HMM-myoV, which takes 67 ± 3 nm steps at a rate of 9.5 ± 0.2 s-1 (Fig. 3 A, D, and G). The approximate 10 s-1 rate is consistent with ADP release being rate-limiting (5, 6, 26). Although we and others have previously measured steps of 72–74 nm for HMM-myoV (1, 2, 27) through the use of fluorescent probes attached to or near the head at low MgATP (≤ 4 μM), the lower values and more broadly distributed steps reported here are the result of the lower signal to noise that accompanies the fast stepping rate of the motor at 1 mM MgATP.
The stepping dynamics of FLB-myoV during the slow processive periods were dramatically different due to the appearance of backward steps interspersed between shorter forward steps (Fig. 3C). For the traces shown, the motors’ average velocities were at or below the 50 nm/s threshold used to identify these slow processive periods. During such periods the distribution of step sizes for FLB-myoV is described by the sum of three Gaussians (Fig. 3F), with peaks at -28 ± 4 nm for backward steps, a major peak at 35 ± 1 nm for short forward steps, and a minor peak at 75 ± 5 nm, which could be “normal” steps or two 35-nm steps in rapid succession. To determine if this stepping pattern is unique to the FL-myoV during the slow processive period, which at low temporal resolution gives the appearance of being stationary (Fig. 1A), we analyzed the displacement characteristics of the occasional stationary HMM-myoV (Fig. S8A). These nonmotile HMM-myoV are truly stationary with no discernable steps, as was the case for the majority of nonmotile FL-myoV that decorated actin filaments (Fig. S8B and Movie S1). Therefore, the FL-myoV’s slow period of processive movement is the result of a forward bias in the number of positive step displacements (Fig. 3F) and not merely a dynamic stall between forward and backward steps, although such periods are observed occasionally (Fig. 3C).
The lifetime distributions for the short forward and backward steps, when fit to single exponentials (Fig. 3I), gave similar rates of 2.0 ± 0.1 s-1 and 1.9 ± 0.1 s-1, respectively. These rates are five times slower than the forward stepping rate of either HMM-myoV or the full-length motor during the fast processive periods. Based on our previous studies, HMM-myoV rarely takes a backward step under unloaded conditions, but does so in response to resistive loads (28). With approximately 20% of the FLB-myoV steps being backwards during Vslow, the altered gating/coordination of the two heads may reflect an effective resistive load due to the GTD interacting with one or both of the heads, or potentially with the actin filament, although no evidence of GTD-actin interactions have been reported.
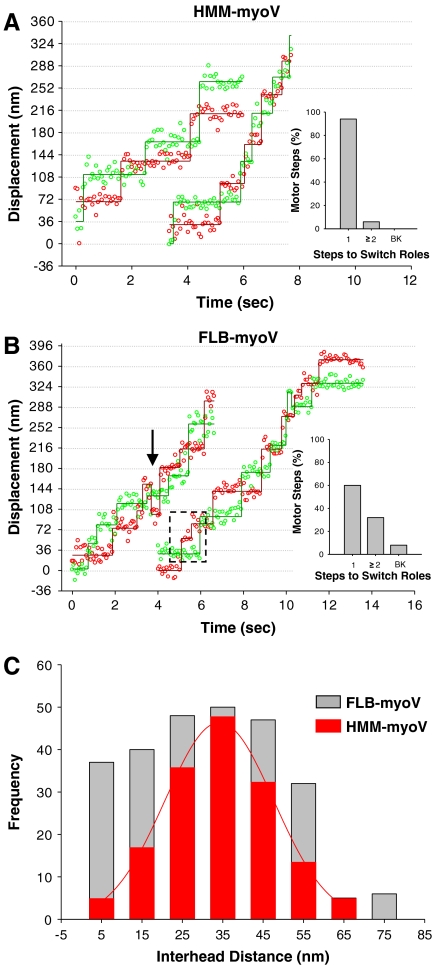
To better understand the gating of the heads during the Vslow processive periods, we labeled both heads of the FLB-myoV and HMM-myoV with different colored Qdots to define the spatial relationships and stepping dynamics of each head relative to its partner (2). The experiments were carried out at 100 mM KCl to increase the probability of observing a processive dual-labeled FLB-myoV motor, and at 10 μM ATP to increase the dwell time of each head, which enhances the certainty in our position detection. However, under these conditions Vavg for FLB-myoV and HMM-myoV are 40–70 nm/s (see Fig. 4A), which makes the delineation between Vslow and Vfast processive periods difficult since Vslow was defined as movement at ≤ 50 nm/s (see above). The merging of Vslow and Vfast at 10 μM ATP suggests that some underlying process governing Vslow may be insensitive to ATP (see model below). Regardless, based on the single Qdot-labeled head stepping data (Fig. 3) we would expect both HMM-myoV-like stepping (Vfast) as well as an emergent stepping pattern associated with altered gating of the motor (Vslow). The displacement versus time trace for HMM-myoV exhibits multiple 72 ± 2 nm, hand-over-hand steps for each head (Fig. 4A), with an interhead distance of 34 ± 2 nm while paused (Fig. 4C, red bars), as previously shown (2). In contrast to this uniform stepping pattern, the stepping dynamics of the full-length motor is quite irregular (Fig. 4B), which is reflected in the interhead distances (Fig. 4C, gray bars) no longer being normally distributed. Although the range of interhead distances spanned by the FLB-myoV molecule matches that of the HMM-myoV, the FLB-myoV pauses more often with either its heads closer together or farther apart.
Fig. 4.
Dual-colored Qdot-labeled FLB- and HMM-myoV stepping dynamics. Representative displacement vs. time traces for dual-labeled HMM-myoV (A) and FLB-myoV (B) at 100 mM KCl and 10 μM ATP, captured at 15 fps. (A) HMM-myoV exhibits almost exclusively (94%) hand-over-hand stepping. The bar chart shows the percentage of motor steps for which it took one (1) or two or more (≥2) steps before the heads switch roles between leading and trailing, and the occurrence of backward (BK) steps. (B) FLB-myoV has a mixture of hand-over-hand (60%) and altered gating (33%) steps where the motor took ≥2 steps before the heads switched roles (highlighted by dashed box). FLB-myoV also took occasional (7%) backward (BK) steps, identified by an arrow in the trace. (C) Interhead distances for the HMM-myoV (red bars) fit to a Gaussian (34 ± 2 nm) and the FLB-myoV (gray bars).
For a more detailed description of the FLB-myoV stepping pattern, we determined how many steps the motor must take before the individual heads swap roles between being a leading versus a trailing head (see Materials and Methods). For hand-over-hand stepping, the number of steps should be one; i.e., each time the motor takes a step the heads exchange roles. Indeed, 94% of HMM-myoV steps (ntotal = 154 steps) are hand-over-hand with the remaining 6% the result of the motor taking two steps before the heads switch roles (Inset, Fig. 4A). This small population is most likely due to one of the heads stochastically taking a rapid and therefore, undetected step. By this criterion, 60% of FLB-myoV steps (ntotal = 264 steps) are hand-over-hand (Fig. 4B, Inset), which corresponds to the percentage of fast movement observed at 100 mM KCl (Fig. 1H). However, for 33% of the steps, the motor takes two or more steps before the heads switch roles. This is fivefold greater than observed for HMM-myoV, suggesting that the gating required for hand-over-hand stepping has been altered, which most likely contributes to the emergence of backward steps but not to the extent that the motor loses its forward stepping bias. The ability of the FLB-myoV heads to maintain their roles as leading and trailing as the motor takes multiple steps can be characterized as a form of inchworm-like stepping. This has been recently reported for myosin VI (29), although the mechanism by which this occurs must differ because the myosin VI was truncated and not a full-length molecule as described here.
A Model of FL-myoV Processivity and Regulation.
The observed data can be explained by a model in which FL-myoV can adopt multiple structural conformations, with the equilibrium between the various states sensitive to ionic strength (Fig. 5). In the absence of actin, a fully folded, inhibited conformation is in equilibrium with an extended conformation that is capable of processive motion (Fig. 5A). The extended state is favored at higher ionic strength or presumably when bound to cargo (6–8, 30). In the presence of actin and MgATP, both the fully folded and extended conformations can bind actin (Fig. 5 B and C). However, only the extended molecule is processive and must be so from its initial encounter with the actin filament, because stationary actin-bound, inhibited motors never switched to processive motion during the approximately 2-min image acquisition period. Once processive, the full-length motor is in equilibrium between two mechanically distinct conformations. When extended, it is fast-moving (Vfast) and steps hand-over-hand like HMM-myoV (Fig. 5C). At other times, it is slowly processive (Vslow) exhibiting a complex stepping pattern characterized by a significant number of short (approximately 36 nm) forward steps that are inchworm-like (Fig. 4B). During the slow processive period the motor maintains its forward movement despite periods of dynamic stall where the motor alternates between short forward and backward steps (Fig. 3C). What conformation the motor adopts during this period of altered interhead communication and whether or not some of the observed steps are ATP-independent are matters of speculation (Fig. 5D). For example, the GTD may bind transiently to either head and spatially constrain the lever arm swing of the GTD-bound head, limiting the diffusional search of the free head and thus resulting in short forward steps (Fig. 5D1–5D3). Knowing that GTD-head interactions affect ADP release (5, 6, 26), the fivefold slower stepping rate during the slow processive period (Fig. 3I) could arise from the GTD-head interaction acting as a resistive load, which slows ADP release (31) and increases the probability of a backstep (28) as observed here. On the other hand, the motor could alternate between two stable positions on actin (Fig. 5D1 and 5D2), explaining the occasional backward steps and periods of dynamic stall (Fig. 3 C and F). The GTD-head interactions might also result in periods of single-headed binding to actin (Fig. 5D1), as observed in the electron microscope (7), and account for FL-myoV’s higher run termination rates (Fig. 2 and Fig. S7). Finally, an alternate scheme for short forward steps (Fig. 5D2 alternate) that is ATP-independent might involve the leading head being weakly bound to actin due to the tail-head interaction. Upon transient unbinding of the tail, the leading head could then adopt its preferred position on actin, 36 nm forward of the trailing head. At this point, the tail could either fold and interact with the head (Fig. 5D3) or remain extended, allowing the motor to switch to fast processive stepping (Fig. 5C). It is worth noting that kinesin-I undergoes a folded-to-extended transition that modulates the manner in which the kinesin-I motor interacts with the microtubule (32, 33). While extended, the motor is processive, and upon folding presumably interacts with the microtubule in a diffusive manner (34). Interestingly, removal of the cargo-binding domain does not eliminate diffusive behavior of kinesin-I (34). Therefore, transient folding of the FL-myoV may be sufficient to alter the motor’s gating, and stable GTD-head interactions may not be absolutely necessary to observe the unique mechanical state characterized here.
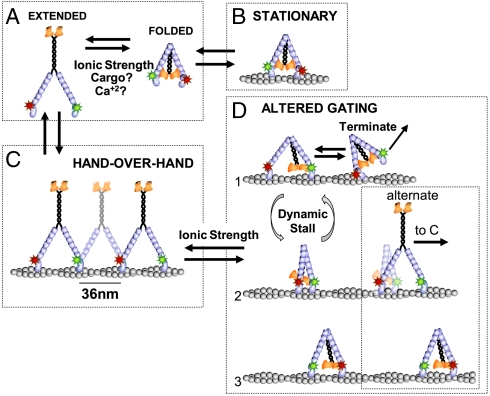
Fig. 5.
Model of FL-myoV folding equilibrium and stepping dynamics. (A) Ionic strength, cargo binding, and changes in Ca+2 concentration affect the folded/ inhibited to extended/active state equilibrium in the absence of actin. (B) The folded motor can bind actin, but remains stationary. Whether one or both heads bind actin is unknown. (C) If the extended motor binds actin, it takes multiple hand-over-hand steps, where the red and green Qdot-labeled heads alternate between being the leading and trailing head. The motor steps along the pseudo actin repeat to the right with a constant interhead distance of approximately 36 nm as observed for the constitutively active HMM-myoV and thus defines the fast FL-myoV processive periods. (D) During the slow processive periods, FL-myoV adopts a conformation with altered gating where the GTD binds to one of the heads (D1) forcing the motor at times to take multiple short steps (D1 to D2) or periods of dynamic stall (D1–D2) before the heads switch roles as leading or trailing head (D2 to D3). An alternate scheme (D2 alternate) for short forward steps is that unfolding of the tail frees the leading head to adopt its preferred position on actin, following which the tail either refolds (D3) or the motor switches to hand-over-hand stepping (C).
These data provide evidence that the globular tail can modulate myoV processivity in the absence of cargo at physiological ionic strength. Although a completely folded and inhibited motor in vivo is a way of conserving ATP when cargo is not bound, the slowly processive motor with altered gating may represent a pathway for facilitating cargo pickup and/or limiting vesicular transport velocity when such modulation is required, providing an additional mode for regulating the motor in vivo. The ability to modulate cargo transport through GTD-head interactions emphasizes the importance of adapter proteins and their association with the GTD as an additional regulatory mechanism of myoV processivity. Finally, the information gained in these studies are necessary entries into the database of full-length myoV movement that will be invaluable as we begin to investigate myoV-mediated vesicle transport inside the cell and in more complex in vitro systems.
Materials and Methods
Expressed Protein and Quantum-Dot Conjugation.
The mouse FLM-myoV, YFP-FL-myoV, and HMM-myoV constructs have been previously described (5, 35). The FLB-myoV splice variant was created by deletion of exons D and F and insertion of exon B between exons A and C using the FLM-myoV as the template (Fig. S1). The FLM-, FLB-, and HMM-myo contained N-terminal biotin tags for conjugation with streptavidin-functionalized Qdots, and C-terminal FLAG tags for purification by affinity chromatography. Heavy chains were coexpressed in the Baculovirus/Sf9 expression system with calcium-insensitive calmodulin light chains (CamΔall) (30).
Purified motors were conjugated to streptavidin-Qdots, with either a 655 nm or 565 nm emission peak (Invitrogen, Carlsbad, CA), in Conjugation Buffer (25 mM imidazole, pH 7.4, 4 mM MgCl2+ , 1 mM EGTA, 200 mM KCl and 10 mM DTT) on ice for 30 min at a 1∶1 molar ratio of motor to Qdot so that on average only one head was labeled. For experiments carried out with dual-labeled heads, 1 μM 655 nm and 1 μM 565 nm Qdots were mixed together, and then conjugated to myoV for a final motor to Qdot ratio of 1∶4 (125 nM myoV to 500 nM Qdots). This fourfold excess of Qdots ensured a small population of motors with differential labeling of the two heads.
Analytical Ultracentrifugation.
The sedimentation coefficients of the expressed full-length myoV isoforms were determined using an Optima XL-I analytical ultracentrifuge (Beckman Coulter). Sedimentation velocity runs were performed in the An60Ti rotor at 32,000 rpm and 20°C, in 10 mM Hepes, pH 7.0, 1 mM DTT, 1 mM EGTA, 1 mM NaN3, NaCl (100 or 300 mM) and approximately 2 μM myosin. Sedimentation values were corrected for density and viscosity of the solvent and sedimentation coefficients were determined by curve fitting to one or more species, using the dc/dt program (36).
In Vitro Processivity Assay.
Single molecule in vitro processivity assays, as described previously (25), were carried out using 20 μL flow chambers. Slides were prepared for observing myoV movement by incubation in following solutions for 2 min each, rinsing between each addition with Actin Buffer (25 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 25 mM KCl, 10 mM DTT, 100 ng/mL creatine phosphokinase, 1 mM phosphocreatine and O2 scavengers [final concentrations of 5.8 mg/mL glucose, 45 μg/mL catalase, 66 ug/mL glucose oxidase): (i)1 mg/mL N-ethylmaleimide-inactivated chicken, skeletal muscle myosin in Myosin Buffer (25 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 300 mM KCl and 10 mM DTT); (ii)1 mg/mL BSA in Actin Buffer; (iii) 100 nM TRITC-phalloidin-labeled actin filaments in Actin Buffer. Following flow cell preparation, 20 μl of Qdot-conjugated myoV was then added at a final concentration of 0.125 nM–5 nM in ATP Buffer (Actin Buffer, 500 μg/mL BSA, 10 μM or 1 mM ATP and 25–200 mM KCl).
Data Acquisition and Analysis.
Images were obtained on a Nikon TE2000 inverted microscope outfitted with a 100x, 1.49na PlanApo objective for through-the-objective total internal reflectance fluorescence microscopy (TIRFM) at 25°C. A 488 nm argon laser was used to excite the Qdots. An intensified CCD camera (XR Mega-S30), running Piper Control v2.3.14 software was used to capture image stacks at 15–60 frames/s with 2 × 2 pixel binning (58.5 nm/pixel) (Stanford Photonics, Stanford, CA). A Dual View optical image splitter (Optical Insights, Pleasanton, CA) was used for simultaneous dual-color imaging. On average, five stacks of 1,000 images each were recorded for a single experimental setup. All experiments were carried out three times on three separate days for a given protein derived from two separate protein expressions; 25–100 trajectories were analyzed per experiment.
All image stacks were analyzed using Image J v1.41o (National Institutes of Health, Bethesda, MD). Specifically, single Qdot-labeled motors were analyzed using the MTrackJ plugin. A 9 × 9 pixel area was manually placed over the Qdot of interest and the x,y position of the “bright centroid” determined with 7-nm resolution (Fig. S8B). Run lengths were measured as the distance the myoV traveled from its initial binding to the actin filament until its disappearance—i.e., detachment. Run length histograms were then fit to a single exponential to define a characteristic run length (Fig. S6). The average velocity (Vavg) was calculated by dividing the run length by the total run time (Fig. S4). For each run, periods of fast and slow movement were identified using a script written in R2.9.1. Velocity was determined by sliding a five-frame (335 ms) window over the displacement versus time data one frame at a time with the velocity calculated from the linear regression to the data within the window. A velocity ≤ 50 nm/ sec was defined as slow. A slow period of motion was characterized as one lasting at least five consecutive frames. These periods were tallied in time and the x,y coordinates of the trajectories for these slow periods then removed from the dataset; the remaining coordinates were plotted as a displacement versus time trace and a Vfast reported as the slope of the linear regression (Fig. S4). The percentage of time at the slow velocity (%Vslow) (see Fig. 1H) was calculated by dividing the total time during the run at the slow velocity (i.e., < 50 nm/s) by the time of the entire run. To determine the percentage of active motors for FLB-myoV relative to HMM-myoV control (100%), moving motors in a fixed area of the visual field were tallied over a fixed time frame per length of actin filament at a given concentration of motor-Qdot introduced into the flow chamber.
To characterize the step size and lifetimes of steps generated by a single, Qdot-labeled head, displacement versus time traces were generated by determining the x,y coordinates of the motor of interest using the SpotTracker 2D (37) plug-in for ImageJ (Fig. 3A). Steps were identified in the data using an algorithm published by Kerssemakers et al. (38) so that step size and lifetime histograms could be generated (Fig. 3). To estimate stepping rates from the lifetime data, HMM-myoV lifetime histograms were fit with a gamma distribution with a shape parameter of K = 2, instead of a single exponential (1). This assumes that the stepping rate of both the Qdot-labeled and the visually silent unlabeled head are identical.
For dual-labeled motors, image stacks were first split into the two emission channels (left, 655 nm Qdot; right, 565 nm Qdot) using the ImageJ OI Image Splitter plug-in. Once split, the individual stacks were analyzed using the SpotTracker 2D plug-in to determine the x,y values for each corresponding Qdot. The x,y values for one channel were corrected relative to the other channel for color misalignment (see Supporting Information). These corrected x,y position values were used to generate displacement vs. time traces for each dual-colored pair (Fig. 4).
Supplementary Material
ACKNOWLEDGMENTS.
We thank S. Previs for her technical assistance, G. Kennedy from the Instrumentation and Model Facility for his microscopy design expertise, and the Warshaw Lab members for their helpful discussions. This work was supported by funds from the National Institutes of Health to J.A., A.M., and S.N. (HL007944), K.T. (GM078097), and D.W. (HL059408, GM094229).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 1363.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109709109/-/DCSupplemental.
References
- 1.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 2.Warshaw DM, et al. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J. 2005;88:L30–32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trybus KM. Myosin V from head to tail. Cell Mol Life Sci. 2008;65:1378–1389. doi: 10.1007/s00018-008-7507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li XD, Mabuchi K, Ikebe R, Ikebe M. Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem Biophys Res Commun. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 5.Lu H, Krementsova EB, Trybus KM. Regulation of myosin V processivity by calcium at the single molecule level. J Biol Chem. 2006;281:31987–31994. doi: 10.1074/jbc.M605181200. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, et al. Regulated conformation of myosin V. J Biol Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 8.Thirumurugan K, Sakamoto T, Hammer JA, 3rd, Sellers JR, Knight PJ. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olivares AO, Chang W, Mooseker MS, Hackney DD, De La Cruz EM. The tail domain of myosin Va modulates actin binding to one head. J Biol Chem. 2006;281:31326–31336. doi: 10.1074/jbc.M603898200. [DOI] [PubMed] [Google Scholar]
- 10.Sato O, Li XD, Ikebe M. Myosin Va becomes a low duty ratio motor in the inhibited form. J Biol Chem. 2007;282:13228–13239. doi: 10.1074/jbc.M610766200. [DOI] [PubMed] [Google Scholar]
- 11.Trybus KM, et al. Effect of calcium on calmodulin bound to the IQ motifs of myosin V. J Biol Chem. 2007;282:23316–23325. doi: 10.1074/jbc.M701636200. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, et al. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Sakamoto T, Zhang F, Sellers JR, Hammer JA., 3rd In vitro reconstitution of a transport complex containing Rab27a, melanophilin and myosin Va. FEBS Lett. 2006;580:5863–5868. doi: 10.1016/j.febslet.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto T, Amitani I, Yokota E, Ando T. Direct observation of processive movement by individual myosin V molecules. Biochem Biophys Res Commun. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 15.Forkey JN, Quinlan ME, Shaw MA, Corrie JE, Goldman YE. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 16.Huang JD, et al. Molecular genetic dissection of mouse unconventional myosin-VA: Tail region mutations. Genetics. 1998;148:1963–1972. doi: 10.1093/genetics/148.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert J, Naeyaert JM, Callens T, De Paepe A, Messiaen L. Human myosin V gene produces different transcripts in a cell type-specific manner. Biochem Biophys Res Commun. 1998;252:329–333. doi: 10.1006/bbrc.1998.9644. [DOI] [PubMed] [Google Scholar]
- 18.Nagashima K, et al. Melanophilin directly links Rab27a and myosin Va through its distinct coiled-coil regions. FEBS Lett. 2002;517:233–238. doi: 10.1016/s0014-5793(02)02634-0. [DOI] [PubMed] [Google Scholar]
- 19.Roland JT, Lapierre LA, Goldenring JR. Alternative splicing in class V myosins determines association with Rab10. J Biol Chem. 2009;284:1213–1223. doi: 10.1074/jbc.M805957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seperack PK, Mercer JA, Strobel MC, Copeland NG, Jenkins NA. Retroviral sequences located within an intron of the dilute gene alter dilute expression in a tissue-specific manner. EMBO J. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner W, Fodor E, Ginsburg A, Hammer JA., 3rd The binding of DYNLL2 to myosin Va requires alternatively spliced exon B and stabilizes a portion of the myosin's coiled-coil domain. Biochemistry. 2006;45:11564–11577. doi: 10.1021/bi061142u. [DOI] [PubMed] [Google Scholar]
- 22.Wu XS, et al. Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 23.Walker ML, et al. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 24.Yengo CM, De la Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of myosin V. Biochemistry. 2002;41:8508–8517. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]
- 25.Baker JE, et al. Myosin V processivity: multiple kinetic pathways for head-to-head coordination. Proc Natl Acad Sci USA. 2004;101:5542–5546. doi: 10.1073/pnas.0307247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheney RE, et al. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 27.Snyder GE, Sakamoto T, Hammer JA, 3rd, Sellers JR, Selvin PR. Nanometer localization of single green fluorescent proteins: Evidence that myosin V walks hand-over-hand via telemark configuration. Biophys J. 2004;87:1776–1783. doi: 10.1529/biophysj.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kad NM, Trybus KM, Warshaw DM. Load and Pi control flux through the branched kinetic cycle of myosin V. J Biol Chem. 2008;283:17477–17484. doi: 10.1074/jbc.M800539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikawa S, et al. Switch between large hand-over-hand and small inchworm-like steps in myosin VI. Cell. 2010;142:879–888. doi: 10.1016/j.cell.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 32.Hackney DD, Stock MF. Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 33.Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 34.Ali MY, Lu H, Bookwalter CS, Warshaw DM, Trybus KM. Myosin V and Kinesin act as tethers to enhance each others’ processivity. Proc Natl Acad Sci USA. 2008;105:4691–4696. doi: 10.1073/pnas.0711531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson SR, Ali MY, Trybus KM, Warshaw DM. Random walk of processive, quantum dot-labeled myosin Va molecules within the actin cortex of COS-7 cells. Biophys J. 2009;97:509–518. doi: 10.1016/j.bpj.2009.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philo JS. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 37.Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- 38.Kerssemakers JW, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]