Abstract
Antibiotics have been administered to agricultural animals for disease treatment, disease prevention, and growth promotion for over 50 y. The impact of such antibiotic use on the treatment of human diseases is hotly debated. We raised pigs in a highly controlled environment, with one portion of the littermates receiving a diet containing performance-enhancing antibiotics [chlortetracycline, sulfamethazine, and penicillin (known as ASP250)] and the other portion receiving the same diet but without the antibiotics. We used phylogenetic, metagenomic, and quantitative PCR-based approaches to address the impact of antibiotics on the swine gut microbiota. Bacterial phylotypes shifted after 14 d of antibiotic treatment, with the medicated pigs showing an increase in Proteobacteria (1–11%) compared with nonmedicated pigs at the same time point. This shift was driven by an increase in Escherichia coli populations. Analysis of the metagenomes showed that microbial functional genes relating to energy production and conversion were increased in the antibiotic-fed pigs. The results also indicate that antibiotic resistance genes increased in abundance and diversity in the medicated swine microbiome despite a high background of resistance genes in nonmedicated swine. Some enriched genes, such as aminoglycoside O-phosphotransferases, confer resistance to antibiotics that were not administered in this study, demonstrating the potential for indirect selection of resistance to classes of antibiotics not fed. The collateral effects of feeding subtherapeutic doses of antibiotics to agricultural animals are apparent and must be considered in cost-benefit analyses.
Keywords: intestinal microbiota, microbiome shifts, swine bacteria, BioTrove microarray, metagenomics
Antibiotics are the most cost-effective way to maintain or improve the health and feed efficiency of animals raised with conventional agricultural techniques (1, 2). In addition to improving feed efficiency, antibiotics are commonly given to livestock, poultry, and fish for disease treatment and prevention. The sum of agricultural antibiotic use reportedly accounts for as much as half of all antibiotics produced in the United States (3). Despite the clear benefits of antibiotics to agriculture, liberal antibiotic use combined with rapid and widespread emergence of both animal and human pathogens resistant to multiple antibiotics has led some to question the prudence of current antibiotic use (4, 5). Studies of environmental and intestinal microbial communities reveal enormous diversity of antibiotic resistance genes (6–8). The addition of antibiotics to feed introduces a selective pressure that may lead to lasting changes in livestock commensal microorganisms. Furthermore, reservoirs of antibiotic resistance genes have been shown to be stable in bacterial communities, even in the absence of antibiotics (9–12). A central concern of increased abundance of antibiotic resistance is the transfer of resistance to pathogens (13). As a result, the Food and Drug Administration recently released a draft guidance recommending restrictions on the use of antibiotics in animal agriculture (14). The Infectious Diseases Society of America testified before a Congressional subcommittee in support of such limitations (15).
Bacteria that inhabit the gastrointestinal tract of animals are important for the maintenance of host health. The intestinal microbiota assists the host in nutrient extraction, immune system and epithelium development, and are a natural defense against pathogens (16). Contrary to these benefits, the gut microbiota may antagonize future disease treatment by facilitating the dissemination of resistance alleles across distantly related organisms. For example, commensal bacteria of the human colon harbor antibiotic resistance genes and can transfer these genes to pathogens (17, 18). In fact, horizontal gene transfer is largely the cause of multidrug resistance in Gram-negative bacteria (19). With the identification of antibiotic resistance genes in commensal bacteria in the human food-chain (20–22), the role of the gut microbiota as a reservoir of resistance genes for animal and food-borne pathogens needs to be explored.
Valuable insights have been gained by culture- and PCR-based approaches to study narrow groups of bacteria or genes, such as erythromycin resistance in swine isolates (23); however, the comprehensive effects of daily feeding of subtherapeutic doses of antibiotics on livestock microbiotas have not been studied. We therefore sought to extensively evaluate the effects of in-feed antibiotics on the entire gut microbiota. Phylotyping, metagenomic, and parallel quantitative PCR (qPCR) approaches were used to track changes in microbial membership and encoded functions, enabling the detection of so-called “collateral” effects of antibiotics (i.e., effects outside of the intended growth promotion and disease prevention). These collateral effects included increases in Escherichia coli populations and in the abundance of certain antibiotic resistance genes.
Piglets were birthed at the National Animal Disease Center in Ames, IA, and housed together in highly-controlled, decontaminated rooms to avoid cross contamination among the medicated animals, nonmedicated animals, and other resident barn animals. Neither the piglets nor the sow were exposed to antibiotics before the study. This design was to ensure that the inoculum for the piglets would come horizontally from their mother, minimizing variability so that effects of antibiotic treatment could be detected. At 18 wk of age, one group of littermates received ASP250 feed (medicated) and the other received the same but unamended feed (nonmedicated) for 3 wk. ASP250 is an antibiotic feed additive containing chlortetracycline, sulfamethazine, and penicillin that is commonly given to swine for the treatment of bacterial enteritis and for increased feed efficiency. Fecal samples were collected just before treatment (day 0), and after 3, 14, and 21 d of continued treatment. Day 0 samples were used to describe the swine intestinal microbiome before antibiotic treatment period.
Results
Shifts in Community Membership with ASP250.
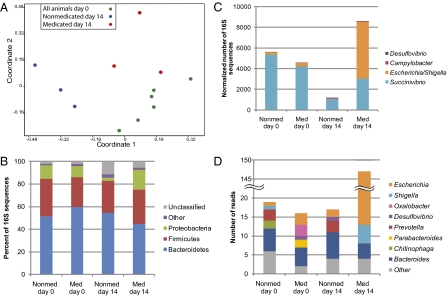
We collected 133,294 sequences of the V3 region of the 16S rRNA gene from a total of 12 fecal samples. Data from pigs of the same treatment and sampling date were grouped to appraise an antibiotic effect on community membership. As reported for a mammalian intestinal environment (24), and recently in a swine metagenome (25), the majority of classifiable sequences (75–86%) belonged to the Bacteroidetes, Firmicutes, and Proteobacteria phyla (Table S1). Of the Bacteroidetes, the Prevotella genus was consistently abundant, as was shown to be a feature of the swine microbiome (25). The Bray-Curtis index was calculated for all sample combinations and an analysis of similarities (ANOSIM) was performed. A nonmetric multidimensional scaling (NMDS) plot of these data indicated divergence of the day 14 samples from the day 0 samples (P < 0.01), and the medicated microbiome diverged from the nonmedicated (P < 0.05) (Fig. 1A), demonstrating changes in microbial community membership over time and with treatment.
Fig. 1.
Shifts in fecal bacterial community membership with antibiotic treatment. (A) NMDS analysis of Bray-Curtis similarity coefficients calculated from 16S rRNA gene sequence data from individual animals at days 0 and 14 shows the similarity among replicate pig fecal samples. (B) Phylum-level composition of fecal microbial communities. Data were pooled for a given treatment and time point and are shown as percentage of abundance. (C) Genus-level composition of Proteobacteria, shown as the total number of sequences (normalized to 50,000 total reads). (D) Predicted genera of COG3188 homologs found in the swine metagenomes based on BLASTx analysis. COG3188 was overrepresented in the medicated metagenome vs. the nonmedicated metagenomes.
Specific changes in the microbial community associated with ASP250 treatment included a decrease in the abundance of Bacteroidetes, along with members of Anaerobacter, Barnesiella, Papillibacter, Sporacetigenium, and Sarcina genera. Members of the Deinococcus-Thermus and Proteobacteria phyla increased with ASP250 treatment as well as Succinivibrio and Ruminococcus genera (Table S1). The increase in Proteobacteria abundance with in-feed ASP250 was particularly striking: from 1% of the population in nonmedicated animals to 11% of the population with antibiotic treatment (Fig. 1B). Specifically, E. coli populations were the major difference between medicated and nonmedicated animals, comprising 62% of the Proteobacteria in medicated animals (Fig. 1C). The increase in E. coli was confirmed in the metagenomic data (Fig. 1D) and by qPCR targeting the uidA gene of E. coli (P < 0.05). A separate study using 12 pigs similarly treated but with analysis by culture-based techniques further established that swine fed ASP250 have an increased E. coli population at 14 d posttreatment, showing a 20- to 100-fold greater E. coli abundance in medicated than nonmedicated swine (Fig. S1).
Shifts in Functional Gene Abundance with ASP250.
DNA samples from the feces of nonmedicated and medicated pigs at days 0 and 14 were isolated, and samples of like treatment and sampling date were pooled for pyrosequencing. Metagenome sequences (1,202,058 total) were analyzed in MG-RAST for SEED subsystems (26), and in-house for clusters of orthologous groups (COGs). All metagenomes showed functional stability over time by both COG and subsystem analyses (Fig. S2). The most abundant SEED subsystem of known function was carbohydrate metabolism, mirroring what was previously reported for the swine metagenome (25). A statistical analysis of COGs revealed shifts in microbial community functions with ASP250: the medicated metagenome contained 169 COGs that were significantly more abundant than in the nonmedicated metagenomes (Table S2). Three COGs (0477, permeases of the major facilitator superfamily; 1289, predicted membrane protein; 3570, streptomycin 6-kinase) contain swine metagenomic genes that are annotated as resistance genes in the antibiotic resistance gene database (ARDB). Three of the COGs with the lowest P value (3188, 3539, and 3121) contained genes related to P pilus assembly, and additionally among the statistically significant COGs are transposases (0675, 1662, and 4644).
To identify themes among differentially represented COGs between the medicated and nonmedicated metagenomes, COGs of Table S2 were clustered by their respective COG category. Only one COG functional category, energy production and conversion (C), was found more frequently (P < 0.05) in the medicated metagenome than in the nonmedicated metagenomes (Table S3).
Pervasive Antibiotic Resistance in the Absence of Antibiotic Exposure.
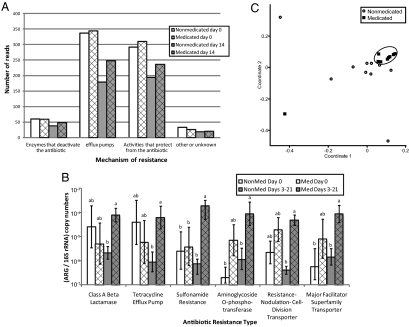
The discovery that resistance-related COGs fluctuated with antibiotic treatment led to further scrutiny of the metagenomes by BLAST against the ARDB (27). All metagenomes, regardless of antibiotic treatment, harbored sequences similar to diverse antibiotic resistance genes representing most mechanisms of antibiotic resistance: efflux pumps, antibiotic-modifying enzymes, and modified or protected targets of the antibiotic (Fig. 2A). This analysis detected 149 different resistance genes in the day 0 metagenomes.
Fig. 2.
Changes in diversity and abundance of antibiotic resistance genes (ARG) in swine feces with antibiotic treatment. (A) Metagenomes were analyzed by BLASTx against the ARDB, and the number of reads were normalized to 100,000 total reads per metagenome. (B) Differences in the abundance of resistance genes were assessed by calculating the ratio of resistance gene copy number (ARG) to 16S rRNA gene copy number per sample as detected by qPCR. Columns denoted by the same letter are not statistically significant (P > 0.05) within each resistance type. Error bars represent the SEM. (C) Bray-Curtis similarity coefficients were calculated from qPCR-derived resistance gene abundance data and plotted in a multidimensional scaling graph. The distance between points indicates the degree of difference in the diversity of resistance genes between samples. The medicated sample outlier (square) is from one medicated pig on day 21. Measures for day 0 samples are not shown.
The finding of diverse fecal antibiotic resistance genes in the nonmedicated metagenomes was supported by parallel qPCR analysis. A rich array of 57 resistance genes was detected at least once in the swine fecal samples by qPCR. Samples from nonmedicated animals showed a total of 50 different resistance genes, but few were shared between animals: only five [ermA, ermB, mefA, tet(32), and aadA] were detected in 66% of the samples and none were found in more than 80% of the samples. No enrichment of these genes was observed in the medicated animals, even though tet(32), a ribosomal protection protein, is known to confer resistance to an administered antibiotic (tetracycline). Samples from medicated animals yielded more homogenous resistance gene diversity: 38 genes were detected in at least one medicated sample, 19 were detected in 66% of samples, and 10 [mefA, ermA, ermB, tet(32), tet(O), aadA, aph(3′)-ib, bcr, acrA, and bacA] were detected in at least eight of nine of the samples.
qPCR and Metagenomic Analyses Reveal Shifts in Resistance Gene Richness and Abundance in Medicated Pigs.
Statistical analysis of the ARDB results showed 23 genes to be differentially represented in the medicated and nonmedicated metagenomes (Table 1). The 20 genes that were more abundant in the medicated metagenome were associated with efflux, sulfonamide resistance, and aminoglycoside resistance, the latter of which represents resistance to a class of antibiotics not present in ASP250 (Table 1).
Table 1.
Antibiotic resistance genes differentially represented (P < 0.05) in the medicated vs. nonmedicated pig fecal samples as detected by metagenomics [number of sequences in the medicated (n = 1) vs. nonmedicated (n = 3) metagenomes per resistance gene] and qPCR (gene copy number/16S rRNA gene copy number) during the treatment period
| Gene(s) detected by |
|||
| Mechanism of resistance | Metagenomics | qPCR | Confers resistance to |
| More prevalent in the treated metagenome | |||
| ABC transporter system. Macrolide-lincosamide-streptogramin B efflux pump. | lmrA | Lincomycin | |
| Aminoglycoside O-phosphotransferase. Modifies aminoglycosides by phosphorylation. | aph(3′′)-Ib, aph(6′)-Ic, aph(6′)-Id | aph(3′′)-Ib | Streptomycin |
| Class A β-lactamase. Cleaves the β-lactam ring. | blaTEM-1, blaSHV-2 | β-Lactams | |
| Major facilitator superfamily transporter, tetracycline efflux pump. Multidrug resistance efflux pump. | emrD, mdfA, mdtH, mdtL, rosA, tet(B) | tet(B), bcr | Chloramphenicol, tetracycline, deoxycholate, fosfomycin, Florfenicol, sulfathiazole |
| Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. | adeA, amrB, mdtF, mdtN, mdtO, mdtP, oprA, tolC | acrA | Fluoramphenicol, aminoglycoside, macrolide, acriflavine, doxorubicin, erythromycin, puromycin, β-lactams |
| Ribosomal protection protein. Protects ribosome from inhibition by tetracycline. | tet(M) | tet(O) | Tetracycline |
| Sulfonamide-resistant dihydropteroate synthase. Cannot be inhibited by sulfonamide. | sul2 | sul2 | Sulfonamide |
| More prevalent in the control metagenomes | |||
| Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. | mexF | Chloramphenicol, fluoroquinolone | |
| Ribosomal protection protein. Protects ribosome from inhibition by tetracycline. | tetB(P), tet(Q) | Tetracycline | |
The qPCR results mirrored the metagenomic analysis, revealing six resistance-gene types with statistically significantly greater abundance in the medicated animals than in the nonmedicated animals (P < 0.05): tetracycline efflux pumps, class A β-lactamases, sulfonamide resistance genes, aminoglycoside phosphotransferases, and two types of multidrug efflux (Fig. 2B and Table 1). No statistical difference in abundance was found for these six resistance gene types between the medicated and nonmedicated microbiomes on day 0 (Fig. 2B), suggesting that in-feed ASP250 caused the effect. Resistance-gene abundance increased most dramatically in the 3- and 14-d samples (Fig. S3), indicating that antibiotic treatment induced a rapid shift in the abundance of resistance genes.
ASP250 treatment increased the diversity of resistance gene types as detected by qPCR [Shannon indices 1.4 (medicated) and 0.8 (nonmedicated); P = 0.04]. A t test comparing the mean number of resistance genes in the metagenomes at day 14 to the corresponding nonmedicated metagenome confirms this result (P < 0.05). Additionally, the structure of the resistance-gene communities (β-diversity) was altered by antibiotic treatment, as determined by a two-way ANOSIM (P < 0.01) of Bray-Curtis measures; however, the comparison R-value was 0.25, indicating that the degree of separation is limited. Nevertheless, resistance gene diversity converges with ASP250 treatment, presumably because of the selective pressure of the antibiotics (Fig. 2C). Taken together, these results show that feeding antibiotics increases the diversity of resistance genes within an individual sample and homogenizes that diversity between treated samples.
Discussion
We assessed the effect of ASP250 on the swine antibiotic resistome using phylotype, metagenomic, and qPCR approaches. The results show that the swine microbiome harbors diverse resistance genes even in the absence of selective pressure. Five genes in particular were detected at high frequency in both the medicated and nonmedicated microbiomes. These genes could represent a core antibiotic resistome for this cohort of swine. Indeed, it was suggested that tet(32) is abundant in farm animals (28), and our data support that conclusion for swine. The constant selective pressure of 50 y of in-feed antibiotics appears to have established a high background level of resistance in the swine microbiome.
Antibiotic treatment caused a detectable increase in the abundance of resistance genes even above the high background of resistance, and many of these were likely enriched because of direct interaction with the antibiotics in ASP250. For example, sulfamethazine presumably selected for the sulfonamide resistance genes sul2 or sul1, present in eight of the nine medicated samples. Additionally, class A β-lactamases were overrepresented in the medicated animals and confer resistance by cleaving such β-lactam antibiotics as penicillin. Many of the other enriched resistance genes function by exporting chemicals. Such efflux includes but is not limited to antibiotics and may allow bacteria that lack specific resistance genes to survive antibiotic pressure. Multidrug efflux is frequently associated with the medically alarming issue of multiple-drug resistance and can be found on mobile genetic elements (29). In addition to the effects on specific gene families, in-feed antibiotics homogenized the richness of resistance genes among individuals over time. The breadth of the current study enabled the visualization of this intriguing phenomenon despite the tremendous resistance gene heterogeneity across samples.
One type of resistance, the aminoglycoside O-phosphotransferases, increased in abundance with in-feed ASP250, although they do no confer resistance to the antibiotics therein. This finding suggests an indirect mechanism of selection, perhaps by co-occurrence on mobile elements conferring resistance to ASP250 antibiotics. Ten of the 13 phosphotransferases identified in the medicated swine metagenome are homologous (7 of 10 have 100% amino acid identity) with the streptomycin phosphotransferase on the pO86A1 plasmid in E. coli O86:H- (accession number YP_788126). Resistance genes aggregate on plasmids in response to selective pressure (30), and pO86A1 carries at least two other resistance genes (accession number NC_008460). This congregation of resistance genes on mobile genetic elements could offer a fitness advantage to a bacterium living in the constant presence of antibiotics. However, this would be an undesirable collateral effect of in-feed antibiotics because these resistance gene clusters could be transferred to E. coli or other potential human pathogens in the swine gut or in the agriculture environment. Regardless of the mechanisms of selection, the results show that antibiotic use increased the abundance of resistance genes specific to and beyond the administered antibiotics from a diverse pool of background resistance genes in the swine microbiome, and that this increase was detectable even above a high background of resistance-gene diversity.
The collateral effects of antibiotics extend beyond influencing resistance genes. Statistical analysis of COGs in the swine metagenomes showed that genes encoding virulence, gene-transfer, and energy production and conversion functions are selected by in-feed antibiotics. Specifically overrepresented COGs included some relating to P pilus assembly; the P pilus has been described for attachment and virulence in E. coli (31). Additional COGs of interest in the medicated metagenome included transposases, which are known to participate in the transfer of antibiotic resistance genes (32). These functions could enhance the stability and spread of resistance genes in microbial communities. Additionally, an increase in the abundance of genes encoding energy production and conversion functions could be a factor in growth-promoting properties of at least some antibiotics, but further experiments are required to test this. Antibiotics are thought to improve feed efficiency in agricultural animals primarily by decreasing the bacterial load, which is beneficial to the host by reducing competition for nutrients and decreasing the host's cost of responding to the microbes (2). Analysis of the swine metabolome after antibiotic treatment showed an effect on various biosynthetic pathways, including sugar, fatty acid, bile acid, and steroid hormone synthesis (33). COGs may therefore be useful signposts for identifying microbes and functions important to the performance-enhancing effects of antibiotics like ASP250.
Changes in microbial functions result from changes in microbial membership, and interesting membership shifts were detected. The decrease in Bacteroidetes in the treated animals may relate to the growth-promoting benefits obtained from feeding swine ASP250 as part of their diets. Obese mice have lower levels of Bacteroidetes relative to Firmicutes in their feces compared with lean mice (34). The obese mice have improved energy-harvesting capacity, presumably because of this shift, and perhaps this shift is related to improved feed conversion in swine. In addition, an increase in E. coli prevalence in response to oral antibiotic treatment has been reported for amoxicillin, metronidazole, and bismuth (35), metronidazole (36), and vancomycin and imipenem (37) in the mammalian gut microbiota. However, amoxicillin plus the β-lactamase inhibitor clavulanic acid administered both in the feed and intramuscularly resulted in decreased E. coli in pigs (38), and oral ciprofloxacin yielded decreased Proteobacteria populations in humans during treatment (39). These results are an important reminder of the varying collateral effects of different antibiotics. E. coli are both commensal and pathogenic inhabitants of mammalian gastrointestinal tracts; an increase in E. coli could be beneficial or harmful, either to the host or to the food chain. Additionally, increased E. coli populations associated with excessive weight gain in pregnant women (40) is an unfavorable result in this host but parallels a potential growth-promoting role for this bacterium in livestock. The cost and benefit of a given antibiotic for a desired outcome must therefore be carefully weighed.
Differences among the rarer members of the microbial communities between treatment and control animals are less understood and invite further investigation. Of those that increased with treatment, members of the Deinococcus-Thermus phylum are known for being resistant to environmental stress; these organisms have only recently been identified in the human gut (41). In addition, Ruminococcus spp. are common in ruminants and are frequently found in the hindgut of pigs (42). Adept at degrading cellulose, an increase in Ruminococcus spp. after antibiotic treatment may aid in feed conversion in swine. Taken together, the data suggest numerous possibilities for how the swine gut microbiota might be involved with the improved feed efficiency afforded by certain in-feed antibiotics.
Conclusions
The results show that even a low, short-term dose of in-feed antibiotics increases the abundance and diversity of antibiotic resistance genes, including resistance to antibiotics not administered, and increases the abundance of E. coli, a potential human pathogen. Additionally, analysis of the metagenomes implicated functions potentially involved with improved feed efficiency. The study design featured environmental control in a single uniform inoculum source (the mother), control of the host genetics, no exposure of the sow or piglets to antibiotics except for the treatment, and identical diet except for the inclusion of ASP250 in one group. Future studies should include other in-feed antibiotics, multiple litters of swine with robust replication, and the identification of the antibiotic-induced mechanisms that lead to increased feed efficiency. Implications of antibiotic resistance on human and animal health need to be taken into account when discussing agricultural management policies and evaluating alternatives to traditional antibiotics. With the use of antibiotics in animal agriculture at a crossroads, studies like this and others that highlight the collateral effects of antibiotic use are needed.
Materials and Methods
Full protocols are available in SI Materials and Methods.
Swine.
Six pigs (siblings) were used in this study and were split into two groups of three: a group to receive antibiotics and a group to receive no antibiotics. Animals were raised in accordance with National Animal Disease Center Animal Care and Use Committee guidelines. The rooms housing the pigs were decontaminated before the beginning of the study. A pregnant sow was obtained from a hog farm at which she had no prior exposure to antibiotics. The piglets shared a pen with the sow for 3 wk after birth; her feces were therefore the primary bacterial inocula for the piglets. After weaning, all pigs were fed the same diet (TechStart 17–25; Kent Feeds) until the start of the study, at which point the medicated pigs were moved to a new clean room and given the above diet but containing ASP250 (chlortetracycline 100 g/ton, sulfamethazine 100 g/ton, penicillin 50 g/ton). Freshly voided feces was collected from nonmedicated and medicated animals just before treatment (medicated and nonmedicated day 0) and 3, 14 and 21 d after treatment.
DNA Sequencing.
Fecal DNA was isolated by bead-beating, and the V3 region of the 16S rRNA gene was amplified and sequenced. PCR products were sequenced on a 454 Genome Sequencer FLX, using the manufacturer's protocol for FLX chemistry (Roche Diagnostics). For sequencing the metagenome, DNA from the feces was pooled by treatment group (nonmedicated, medicated) for each time point (day 0, day 14). Day 14 samples were sequenced using FLX chemistry and day 0 samples were sequenced using Titanium chemistry (Roche Diagnostics).
Phylotype Analysis.
Only sequences longer than 50 bp were used for phylotype analysis (phylotyping), which totaled 133,294 sequences (70,667 unique sequences) from 12 fecal samples. After binning the samples by barcode, phylogenetic analysis and taxonomic assignments of the V3 portion of the 16S rRNA gene were made using the Ribosomal Database project Web tools (43). Additional phylotype comparisons and hypothesis testing were performed with the software package mothur (44). Bray-Curtis similarity coefficients were calculated from 16S rRNA gene sequence data from individual animals at 0 and 14 d and plotted in an NMDS graph to show the similarity among samples. MDS plots and analysis of similarities statistical tests were done in PAST (45).
Metagenomic Analysis.
Sequences were dereplicated and analyzed by BLAST against the nonredundant database and ARDB (27). The BLAST reports were parsed to extract COG information, and COG frequencies were analyzed in ShotgunFunctionalizeR (46). The ARDB was kindly provided by Liu and Pop (27) so that we could perform BLASTx analyses locally. In both analyses, differences with P < 0.05 were significant, and the significant COGs were labeled with their respective COG category to visualize trends. For ecological analyses, the number of hits was normalized to 100,000 submitted reads and analyzed using NMDS and cluster analyses with the Bray-Curtis similarity measurement in PAST (45).
Quantitative PCR.
Primer sets were grouped into 18 resistance types by subjecting all primer sets to the ARDB BLAST tool (Table S4) or by the BLAST tool in the National Center for Biotechnology Information when no results were obtained by the ARDB BLAST (Table S5). Quantitative PCR primers, reagents, and DNA samples were loaded into six subarrays of OpenArray plates (Applied Biosystems) (47). For each 33 nL qPCR reaction, 1 ng of extracted DNA was added as template. Quantitative PCR reagents and conditions were preformed as previously described (47). Relative gene copy numbers were calculated as follows: gene copy number = 10(26−Ct)/(10/3), where Ct equals the threshold cycle (Table S6). Amplification curves were manually inspected using quality control measures. The abundance of the 16S rRNA gene was determined (48), and E. coli was quantified by using a uidA primer set (49). Copy numbers of the uidA and 16S rRNA genes were calculated in relation to a standard curve, which was generated by using 10-fold dilutions of 108 to 100 copies as template, in triplicate reactions. Those reactions targeting 16S rRNA and uidA were preformed separately from the OpenArray platform.
Statistical Analysis of qPCR Results: Abundance and Diversity.
All qPCR data were normalized between samples by dividing the gene copy number by 16S rRNA copy number and subsequently natural log-transformed to achieve normal distribution. A repeated-measures ANOVA model was used to determine if treatment or time was significantly related to the abundance of antibiotic resistance genes and Shannon diversity in different samples. The best covariance structure of the residuals for each response variable was determined and used for repeated measures ANOVA testing (SAS v9.2; SAS Institute). A Bonferroni adjustment was not used in the comparison of resistance genes or resistance gene types because of excessive reduction in power of tests; therefore, the reported P values were not corrected for multiple comparisons.
Shannon diversity was calculated using PAST ver. 1.87 (45) using data normalized between samples (resistance gene copy number/16S rRNA gene copy number). Bray-Curtis coefficients were calculated for each of the samples using the natural log-transformed data (50). A two-way ANOSIM was calculated using these data, considering treatment and time as the two factors. Two-way ANOSIM analysis and NMDS plots were completed using the Bray-Curtis measure for β-diversity.
Supplementary Material
Acknowledgments
The authors thank Sam Humphrey, Uri Levine, and Lea Ann Hobbs for technical support; Vince Young for helpful conversations; the Michigan State University Crop and Soil Science statistical consultation center for statistical advice; and Rich Zuerner and Tom Casey for comments on the manuscript. The Michigan State University research was initiated under a grant from Reservoirs of Antibiotic Resistance and was supported by Michigan State University's Pharmaceuticals in the Environment Initiative.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRP004660).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120238109/-/DCSupplemental.
References
- 1.Cromwell GL. Why and how antibiotics are used in swine production. Anim Biotechnol. 2002;13:7–27. doi: 10.1081/ABIO-120005767. [DOI] [PubMed] [Google Scholar]
- 2.Dibner JJ, Richards JD. Antibiotic growth promoters in agriculture: History and mode of action. Poult Sci. 2005;84:634–643. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- 3.Lipsitch M, Singer RS, Levin BR. Antibiotics in agriculture: When is it time to close the barn door? Proc Natl Acad Sci USA. 2002;99:5752–5754. doi: 10.1073/pnas.092142499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy SB. Emergence of antibiotic-resistant bacteria in the intestinal flora of farm inhabitants. J Infect Dis. 1978;137:689–690. [PubMed] [Google Scholar]
- 5.Aarestrup FM, Wegener HC. The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microbes Infect. 1999;1:639–644. doi: 10.1016/s1286-4579(99)80064-1. [DOI] [PubMed] [Google Scholar]
- 6.Allen HK, et al. Call of the wild: Antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 7.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez JL, et al. A global view of antibiotic resistance. FEMS Microbiol Rev. 2009;33:44–65. doi: 10.1111/j.1574-6976.2008.00142.x. [DOI] [PubMed] [Google Scholar]
- 9.Götz A, et al. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salyers AA, Amábile-Cuevas CF. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanton TB, Humphrey SB. Persistence of antibiotic resistance: Evaluation of a probiotic approach using antibiotic-sensitive Megasphaera elsdenii strains to prevent colonization of swine by antibiotic-resistant strains. Appl Environ Microbiol. 2011;77:7158–7166. doi: 10.1128/AEM.00647-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanton TB, Humphrey SB, Stoffregen WC. Chlortetracycline-resistant intestinal bacteria in organically raised and feral Swine. Appl Environ Microbiol. 2011;77:7167–7170. doi: 10.1128/AEM.00688-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 14.US Department of Health and Human Services, Food and Drug Administration, and Center for Veterinary Medicine Draft guidance #209. 2010. Available at http://www.fda.gov/downloads/animalveterinary/guidancecomplianceenforcement/guidanceforindustry/ucm216936.pdf Accessed October 12, 2010.
- 15.The Infectious Diseases Society of America Antibiotic resistance: Promoting judicious use of medically important antibiotics in animal agriculture. 2010. Presentation before the House Committee on Energy and Commerce Subcommittee on Health. www.idsociety.org/WorkArea/DownloadAsset.aspx?id=16796.
- 16.Zoetendal EG, Cheng B, Koike S, Mackie RI. Molecular microbial ecology of the gastrointestinal tract: From phylogeny to function. Curr Issues Intest Microbiol. 2004;5:31–47. [PubMed] [Google Scholar]
- 17.Karami N, et al. Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J Antimicrob Chemother. 2007;60:1142–1145. doi: 10.1093/jac/dkm327. [DOI] [PubMed] [Google Scholar]
- 18.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol. 2001;67:561–568. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leverstein-van Hall MA, et al. Evidence of extensive interspecies transfer of integron-mediated antimicrobial resistance genes among multidrug-resistant Enterobacteriaceae in a clinical setting. J Infect Dis. 2002;186:49–56. doi: 10.1086/341078. [DOI] [PubMed] [Google Scholar]
- 20.Barbosa TM, Scott KP, Flint HJ. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ Microbiol. 1999;1:53–64. doi: 10.1046/j.1462-2920.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- 21.Stanton TB, Humphrey SB. Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl Environ Microbiol. 2003;69:3874–3882. doi: 10.1128/AEM.69.7.3874-3882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Wang HH. Tetracycline resistance associated with commensal bacteria from representative ready-to-consume deli and restaurant foods. J Food Prot. 2010;73:1841–1848. doi: 10.4315/0362-028x-73.10.1841. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Wang GR, Shoemaker NB, Whitehead TR, Salyers AA. Distribution of the ermG gene among bacterial isolates from porcine intestinal contents. Appl Environ Microbiol. 2005;71:4930–4934. doi: 10.1128/AEM.71.8.4930-4934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamendella R, Domingo JW, Ghosh S, Martinson J, Oerther DB. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011;11:103. doi: 10.1186/1471-2180-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010. 2010:pdb prot5368 Available at http://cshprotocols.cshlp.org/ [DOI] [PubMed]
- 27.Liu B, Pop M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melville CM, Scott KP, Mercer DK, Flint HJ. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob Agents Chemother. 2001;45:3246–3249. doi: 10.1128/AAC.45.11.3246-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci. 2009;276:2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol Biol. 2009;532:397–411. doi: 10.1007/978-1-60327-853-9_23. [DOI] [PubMed] [Google Scholar]
- 31.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: Molecular mechanisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 32.Lupski JR. Molecular mechanisms for transposition of drug-resistance genes and other movable genetic elements. Rev Infect Dis. 1987;9:357–368. doi: 10.1093/clinids/9.2.357. [DOI] [PubMed] [Google Scholar]
- 33.Antunes LC, et al. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 35.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pélissier MA, et al. Metronidazole effects on microbiota and mucus layer thickness in the rat gut. FEMS Microbiol Ecol. 2010;73:601–610. doi: 10.1111/j.1574-6941.2010.00916.x. [DOI] [PubMed] [Google Scholar]
- 37.Manichanh C, et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20:1411–1419. doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thymann T, et al. Antimicrobial treatment reduces intestinal microflora and improves protein digestive capacity without changes in villous structure in weanling pigs. Br J Nutr. 2007;97:1128–1137. doi: 10.1017/S0007114507691910. [DOI] [PubMed] [Google Scholar]
- 39.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santacruz A, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 41.Bik EM, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rincon MT, et al. A novel cell surface-anchored cellulose-binding protein encoded by the sca gene cluster of Ruminococcus flavefaciens. J Bacteriol. 2007;189:4774–4783. doi: 10.1128/JB.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electronica. 2001;4:9. [Google Scholar]
- 46.Kristiansson E, Hugenholtz P, Dalevi D. ShotgunFunctionalizeR: An R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737–2738. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- 47.Stedtfeld RD, et al. Development and experimental validation of a predictive threshold cycle equation for quantification of virulence and marker genes by high-throughput nanoliter-volume PCR on the OpenArray platform. Appl Environ Microbiol. 2008;74:3831–3838. doi: 10.1128/AEM.02743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leigh MB, et al. Biphenyl-utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs) ISME J. 2007;1:134–148. doi: 10.1038/ismej.2007.26. [DOI] [PubMed] [Google Scholar]
- 49.Srinivasan S, Aslan A, Xagoraraki I, Alocilja E, Rose JB. Escherichia coli, enterococci, and Bacteroides thetaiotaomicron qPCR signals through wastewater and septage treatment. Water Res. 2011;45:2561–2572. doi: 10.1016/j.watres.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 50.Anderson MJ, Ellingsen KE, McArdle BH. Multivariate dispersion as a measure of beta diversity. Ecol Lett. 2006;9:683–693. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.