Abstract
Almost nothing is known about the mechanisms of dissimilatory metal reduction by Gram-positive bacteria, although they may be the dominant species in some environments. Thermincola potens strain JR was isolated from the anode of a microbial fuel cell inoculated with anaerobic digester sludge and operated at 55 °C. Preliminary characterization revealed that T. potens coupled acetate oxidation to the reduction of hydrous ferric oxides (HFO) or anthraquinone-2,6-disulfonate (AQDS), an analog of the redox active components of humic substances. The genome of T. potens was recently sequenced, and the abundance of multiheme c-type cytochromes (MHCs) is unusual for a Gram-positive bacterium. We present evidence from trypsin-shaving LC-MS/MS experiments and surface-enhanced Raman spectroscopy (SERS) that indicates the expression of a number of MHCs during T. potens growth on either HFO or AQDS, and that several MHCs are localized to the cell wall or cell surface. Furthermore, one of the MHCs can be extracted from cells with low pH or denaturants, suggesting a loose association with the cell wall or cell surface. Electron microscopy does not reveal an S-layer, and the precipitation of silver metal on the cell surface is inhibited by cyanide, supporting the involvement of surface-localized redox-active heme proteins in dissimilatory metal reduction. These results provide unique direct evidence for cell wall-associated cytochromes and support MHC involvement in conducting electrons across the cell envelope of a Gram-positive bacterium.
Keywords: electricigen, Fe(III) reduction
The importance of bacteria in mediating iron redox cycles is well-established (1), and the mechanisms used by some Gram-negative dissimilatory metal-reducing bacteria (DMRB) have been extensively characterized. Recently, Gram-positive DMRB have been recognized (2–4) and shown to be the dominant DMRB in some environments (2, 3). In contrast to Gram-negative DMRB, however, the mechanisms of metal reduction by Gram-positive DMRB are unknown. Mechanisms used by Gram-negative DMRB include indirect reduction through the use of electron shuttles (5, 6) or iron chelators (7), or direct reduction through surface mediated electron transfer involving cytochromes (1, 8) or conductive pili (9, 10). Thermincola potens strain JR was isolated from a thermophilic microbial fuel cell (MFC) and physiological characterization revealed that it coupled acetate oxidation to hydrous ferric oxide (HFO) or anthraquinone-2,6-disulfonate (AQDS) reduction (11). Remarkably, T. potens failed to use soluble iron, such as ferric citrate or ferric nitrilotriacetic acid, suggesting an obligate utilization of insoluble iron as an electron acceptor (11). A similar preference for insoluble iron was observed with hyperthermophilic Geobacteraceae (2). Supernatant swapping experiments in MFCs and live/dead staining of anode biofilms support a direct mechanism of electron transfer, and cyclic voltammetry of MFC supernatants indicated the absence of redox active shuttles (11).
The abundance of multiheme c-type cytochromes (MHCs) in the T. potens genome is unusual among sequenced Gram-positive bacteria (12). T. potens has 32 genes coding for MHCs, with one predicted to have as many as 58 heme motifs (12) (Table S1). Although genetic, biochemical, and spectroscopic evidence have identified a role for c-type cytochromes in iron reduction by Gram-negative species (13), until recently, no clear correlation between the presence of c-type cytochromes and iron reduction has been reported outside the Proteobacteria. Within the Firmicutes, c-type cytochromes were expressed during metal reduction in one Gram-positive bacterium, Carboxydothermus ferrireducens (14, 15). Furthermore, the genomes of several Firmicutes and Actinobacteria have abundant MHC genes, suggesting that these proteins may be important to some Gram-positive species (16).
In this article we present biochemical and biophysical evidence that T. potens has cell wall or surface-localized MHCs. Gene deletions to demonstrate the involvement of these proteins in metal reduction are not currently possible. However, physiological characterization indicates that T. potens is an obligate DMRB, and therefore, the MHCs expressed under respiratory conditions (HFO or AQDS) are likely necessary for this metabolism (11). Cells for electron microscopy, low pH extraction, resonance Raman, surface-enhanced Raman spectroscopy (SERS), and trypsin shaving were grown with AQDS because of concerns about the stability of cell proteins during iron extraction (17) and possible interferences of HFO with the analytical procedures. Preliminary studies showed that the AQDS and HFO cell proteomes were similar for T. potens, indicating that reduction of these alternate compounds is mediated by a similar mechanism, as previously found for Geobacter and Shewanella (18). The identification of cell wall/surface cytochromes is unprecedented in Gram-positive bacteria, but suggests a general mechanism of electron transfer across cell walls.
Results
Electron Microscopy Supports Cell-Surface Localization of Cytochrome-Mediated Metal Reductase Activity and Lack of Crystalline S-Layer.
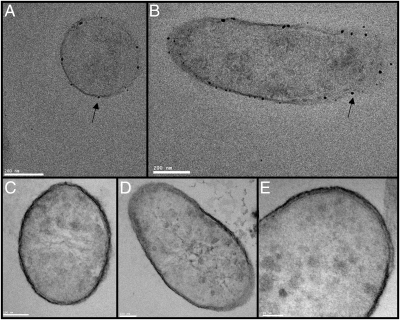
Incubation of T. potens with Ag+ before thin sectioning and staining resulted in the precipitation of Ag(0) on the cell surface (Fig. 1 A and B). Similar observations were made previously with Geobacter sulfurreducens (19). Ag precipitation was inhibited by potassium cyanide added before incubation with Ag+, implicating involvement of heme proteins. Cyanide binds to ferric hemes, inhibiting redox cycling by cytochromes and thereby blocking electron flow through electron transport chains. Previous electron microscopic analysis revealed the ultrastructure of the T. potens cell envelope (11). A 16-nm thick low-density inner wall zone is surrounded by a 17-nm thick high-density outer wall zone. No S-layer was observed. In support of this finding, a crystalline protein S-layer was also not apparent in transmission electron microscopy (TEM) images (Fig. 1 C–E). This finding is similar to the observations with Thermincola ferriacetica, another metal-reducing member of the Thermincola genus (20). In contrast, Thermincola carboxydophila is not a DMRB and does have a crystalline S-layer (21).
Fig. 1.
TEM images of thin sectioned T. potens. Intact cells were treated with Ag+ (A and B) or KCN before Ag+ (C–E) before thin-sectioning. (A) Ag precipitation (indicated by arrows) visible on the cell surface (A and B) is inhibited by the addition of KCN (C–E). A crystalline S-layer is not visible on the cell surface. Cells were stained with three stains: 0.5% Ruthenium red (polysaccharides), 2% Osmium tetroxide (lipids), and 2% Uranyl acetate (proteins and nucleic acids).
Low pH or Denaturant Extraction of a Multiheme Cytochrome from Intact Cells.
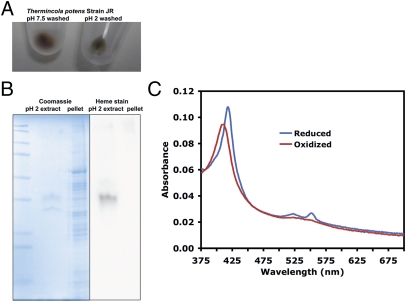
Treatment of T. potens with denaturants such as 8 M Urea, 5 M guanidine HCl, or 0.2 M glycine pH 2 buffer led to a loss of red color in cell pellets (Fig. 2A), and extraction of one major and one minor protein band on Coomassie-stained SDS-Page gels (Fig. 2B). Chelators, salts, and detergents did not extract proteins from intact T. potens cells (Table S2). Microscopic inspection of cells showed that less than 5–10% lysis occurred during the extractions. The major protein band displayed heme peroxidase activity (Fig. 2B), and UV-visible spectra of the neutralized low pH extract revealed characteristic cytochrome absorbance peaks (Fig. 2C). In-gel tryptic digests and LC-MS/MS identified the major protein band as TherJR_1122, a hexaheme cytochrome, and the minor protein band as TherJR_2793, a flagellin homolog. Based on Bradford assays, the dialyzed low pH extract was estimated to contain ∼5% of the total cellular protein, and both TherJR_1122 and TherJR_2793 were sensitive to trypsin when extracted from cells (Fig. S1).
Fig. 2.
Low pH treatment of T. potens cells solubilizes a multiheme cytochrome, TherJR_1122. (A) Cells suspended in neutral pH buffer retain a red color after pelleting, but cells suspended in acidic buffer lost the red color after pelleting. (B) Low pH extract consists of one major band (TherJR_1122, MHC) displaying peroxidase activity, and one minor band (TherJR_2793, flagellin). (C) UV-visible electronic absorption spectra of dithionite reduced and air oxidized TherJR_1122 extracted from T. potens cells.
It seems likely that TherJR_1122 is noncovalently associated with cell-wall polymers or cell-wall proteins. Denaturants, low pH, chelators, and high salt have all been used to extract crystalline S-layers from Gram-positive and some Gram-negative cells (22–24). These treatments disrupt interactions between S-layer homology domains and the underlying cell-wall polymers (24). T. potens lacks a crystalline S-layer, but has a number of proteins with S-layer homology domains. These domains bind to cell-wall polymers to anchor proteins or multiprotein complexes to the cell (24, 25). It is noteworthy that proteins with S-layer homology domains identified in the trypsin-shaving experiments (described below) are not readily extracted with denaturants or low pH. It may be that these proteins are covalently attached to the peptidoglycan surface through an unidentified mechanism or that their large size traps them in the cell-wall matrix after treatment with chaotropic buffers. In contrast, TherJR_1122 is of a size and conformation and is anchored to the cells in a way that enables its extraction.
SERS Supports Surface/Cell-Wall Localization of Multiheme Cytochromes.
Heme proteins have distinct Raman bands from porphyrin bending and stretching modes. These peaks can be resonantly enhanced with an excitation wavelength that falls within the heme absorbance spectrum. The Raman signals can also be enhanced when the protein lies close to nano-roughened gold or silver surfaces (Fig. 3 and Fig. S2) (26). This process is termed SERS, and has served to visualize cell-surface Raman active species in several bacteria (19, 27–29). The SERS effect decays exponentially within a few nanometers from the metal surface, but the enhancements can be as great as 104- to 1014-fold (26). Therefore, SERS is a powerful, direct in situ method to probe the bacterial surface.
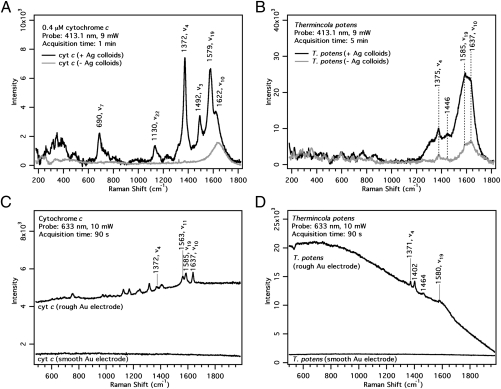
Fig. 3.
Resonance Raman, SERS and SERRS spectra of cytochrome c and T. potens with 413.1 nm laser excitation (A and B) and 633-nm laser excitation (C and D) (A) 0.4 μM cytochrome c and (B) T. potens in the presence (black traces) and absence (gray traces) of Ag colloids. (C) Cytochrome c and (D) T. potens on smooth and electrochemically roughened Au electrodes.
The resonance Raman spectra of cytochrome c with 413.1-nm excitation show the ν4 band at 1,372 cm−1 from the Cα-N stretching mode along with other heme skeletal markers (Fig. S2). In the presence of Ag colloids, the cytochrome c peaks are further enhanced (Fig. 3A and Fig. S2). We used the 1,372-cm−1 (ν4) band as a marker for surface cytochromes. The Raman spectra of T. potens with and without added Ag colloids are shown in Fig. 3B. Here the marker peak at 1,375 cm−1 is clearly enhanced, suggesting that the cytochromes are close to the cell envelope surface.
To further confirm that cytochromes are indeed localized on the cell wall of T. potens, we performed SERS on electrochemically roughened Au electrodes with 633-nm laser excitation. This wavelength was chosen because it is off resonance with the cytochrome absorption spectrum, and hence, any signal enhancement can be attributed solely to the SERS effect (from the Au). A smooth Au surface was used as a control.
On a polished Au electrode with negligible SERS effect, no peaks were observed from both cytochrome c and T. potens (Fig. 3 C and D). However, on a roughened Au surface, the ν4 and other bands belonging to the cytochromes were clearly visible. Because there is no resonance enhancement, these signals must have been enhanced by the roughened Au substrate. This finding is confirmed by the elevated background of the SERS spectra, a phenomenon associated with the SERS effect. The observation of SERS cytochrome signals in T. potens indicates that these proteins are localized on the outer cell wall.
We cannot completely rule out cytochrome peaks from lysis, but all cells were washed and fixed with glutaraldehyde before SERS, and T. potens can clearly interact with metals without lysing, as indicated by its ability to grow by HFO reduction and to precipitate Ag on its cell surface. The 1,402- and 1,464-cm−1 peaks observed in the 633-nm T. potens SERS spectrum cannot be assigned to porphyrin modes or glutaraldehyde, and may be because of other cell-surface molecules.
Trypsin Shaving Supports Surface/Cell-Wall Localization of MHCs.
We used a trypsin shaving approach to identify T. potens surface proteins. This approach was effective for identifying surface proteins in Gram-positive organisms, such as Staphylococcus aureus or Bacillus subtilis (30–32). Other studies relied on spectral counting to identify proteins as surface-exposed based on the prevalence of peptides from these proteins after the cells have been incubated with trypsin (30, 31). A false-positive control, in which cells were incubated without trypsin, was used to identify cytoplasmic proteins leaking or “shed” from lysing cells (30). In our T. potens experiments, rarely were more peptides observed for a protein in shed controls compared with trypsin-shaved cells (Dataset S1). It may be that less lysis occurs in T. potens cells or that the shed proteins are more tightly associated with the cell surface than for mesophilic bacteria previously used. T. potens does not readily grow on agar plates, and therefore, we could not easily determine the effect of trypsin on viability, but microscopy and cell counting revealed no fewer intact cells after trypsin treatment. However, trypsin-shaved cells showed a much slower rate of HFO reduction (Fig. S3) in washed cell experiments, suggesting a functional role for cell-surface proteins in metal reduction.
The low number of peptides observed in the false-positive (shed) condition led us to seek an alternative control to identify surface proteins. Toward this end, we sonicated an equivalent number of cells before incubation with trypsin. Any proteins that are more abundant in the periplasm or cytoplasm relative to the surface should yield more peptides in this lysed cell condition compared with the trypsin condition. The comparison of trypsin-shaved to lysed cells is a stringent filter for the identification of surface proteins as the relative abundances of the surface proteins do not change between the trypsin and lysed cell conditions, but trypsin will have more access to buried and cytoplasmic proteins in the lysed cell condition.
Although it is a reasonable assumption that the trypsin/lysed ratios will recapitulate the cellular localization of proteins, additional factors may affect the spectral counts observed for a given protein. The abundance of peptides from a protein is influenced by the accessibility and number of trypsin cleavage sites. For example, burial of protease sites in a protein-protein complex or membrane will slow the proteolysis rate. Coelution on the liquid chromatography (LC) and spectral interference of peptides from other proteins can also mask peptides in the LC-MS/MS analysis. For MHCs, peptides that are covalently linked to heme confound identification by LC-MS/MS.
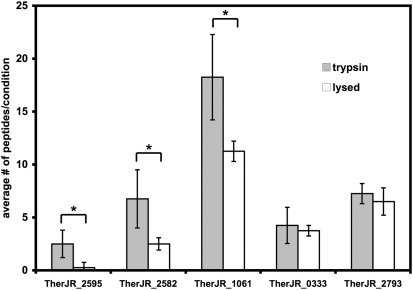
To analyze the effect of trypsin shaving, we combined the results of four independent experiments. A total of 2,576 peptides were observed. The complete dataset is given in Dataset S1. For further analysis, we focused on the set of proteins for which greater than 10 peptides were observed in the combined results of the four independent experiments (Table S3 and Dataset S1). We calculated the ratio of the number of peptides observed in the trypsin shaved to lysed cells conditions. A trypsin/lysed ratio greater than 1 indicated surface localization for a protein. In confirmation of this approach, all proteins observed with trypsin/lysed ratios of 0.9 or greater had N-terminal signal peptides, twin-arginine translocation signals, or in the case of flagellin, are known to be secreted. Three proteins had trypsin/lysed ratios greater than 1.5: TherJR_2595, a nonaheme MHC, and two proteins with S-layer homology domains, TherJR_2582, and TherJR_1061 (Fig. 4, Table S3, and Dataset S1). These three proteins consistently yielded more peptides in trypsin shaving compared with lysed cell controls (Fig. 4). Flagellin (TherJR_2793), a decaheme MHC (TherJR_0333), two periplasmic hydrogenase subunits (TherJR_0764-765), and an extracellular solvent binding protein (TherJR_0861) had trypsin/lysed ratios close to 1. The next highest trypsin/lysed ratio was 0.5 (Table S3) for TherJR_2994, a cobalamin binding protein with likely cytoplasmic localization. The S-layer homology protein, TherJR_2871, had a lower trypsin/lysed ratio of 0.38. This ratio is lower than other S-layer homology proteins, but TherJR_2871 is homologous to a middle cell wall protein from Brevibacillus brevis (33), and may be buried on the cytoplasmic face of the cell wall in trypsin treatments, but released in sonicated, lysed cells.
Fig. 4.
Average number of peptides observed in trypsin (shaded bars) and lysed cell (open bars) conditions for proteins with the highest trypsin/lysed ratios. Across four biological replicates, TherJR_2595, TherJR_2582, and TherJR_1061 were found to have significantly more peptides in trypsin-shaved samples compared with lysed cell samples. *P < 0.05, Student's two-tailed T-test.
A number of core metabolic proteins likely associated with the cytoplasmic membrane or cytoplasm were identified in the trypsin experiments. Consistent with inner localization, these proteins had trypsin/lysed ratios of less than 0.5. ATP synthase (TherJR_2887), and Wood-Ljungdahl pathway proteins (TherJR_1157-1160) were abundant in proteomic datasets and had trypsin/lysed ratios of 0.19, 0.26, 0.31, 0.37, and 0.13. Similarly, EF-Tu (TherJR_0276), adenylate kinase (TherJR_1985), and GroEL (TherJR_0775) were also abundant (trypsin/lysed ratios of 0.43, 0.31, and 0.16, respectively).
The hexaheme MHC, TherJR_1122, and the decaheme MHC, TherJR_1117, were only observed in the trypsin fraction, but fewer peptides were observed for these two proteins (eleven and two peptides observed, respectively, across the four biological replicates) (Table S4 and Dataset S1). TherJR_1122 is readily extracted from T. potens with denaturants (Fig. 2), but may be resistant to protease in intact cells because of a close association with other cellular components or because of a protease-resistant conformation when associated with cells. Consistent with this hypothesis, TherJR_1122 can be extracted at similar yields from trypsin shaved cells, but is sensitive to trypsin when denatured in SDS/PAGE gels for in-gel digests or when trypsin is added to neutralized low pH extracts (Fig. S1). TherJR_1117 has an additional N-terminal transmembrane region after the hydrophobic signal sequence, and may serve as a quinol dehydrogenase that initiates the extracellular electron transport chain, but could be shielded from trypsin by its inner membrane localization.
We also analyzed tryptic peptides from lysed T. potens cells grown on HFO and found that a similar set of proteins were present (Dataset S1). Of particular interest, the identical set of S-layer homology domain proteins and MHCs were found in HFO and AQDS grown cells (Table S4 and Dataset S1).
Discussion
In the model Gram-negative DMRB, Shewanella oneidensis and G. sulfurreducens, MHCs function as an electron conduit across the periplasm and outer membrane to reduce extracellular electron acceptors, such as humic acids or HFO (13). The Gram-negative cell architecture requires that electron transfers occurs at three distinct locations: (i) from the inner membrane quinone pool to the periplasm (CymA, MacA), (ii) across periplasmic space (MtrA, PpcA), and (iii) across the outer membrane to extracellular acceptors (MtrB/C, OmcA/B, OmcS) (13, 34). The Gram-positive cell architecture imposes different constraints on electron transport. Periplasmic proteins must also transfer electrons from the inner membrane to the cell wall. These proteins may be similar to the periplasmic MHCs of Geobacter and Shewanella. However, transferring electrons across the peptidoglycan/teichoic acid matrix of the cell wall requires a cytochrome that packs into fissures in the cell wall or is anchored to the peptidoglycan. Previous studies suggest that the peptidoglycan matrix permits the diffusion of up to 30-kDa globular proteins (35). Alternatively, an unfolded or extended conformation of a larger protein may also diffuse through the cell wall matrix. Recent structures of MHCs from G. sulfurreducens and S. oneidensis do show extended conformations (36, 37). T. potens lacks sortase homologs, like other Clostridia (38), and none of the cell wall-associated proteins identified in this study have the canonical LPXTG motif recognized by sortases. However, a number of proteins with S-layer homology domains were identified, and these proteins may function like the scaffoldins in cellulosomes (25), binding both cell-wall polymers and other proteins to arrange a multiprotein complex on the cell surface.
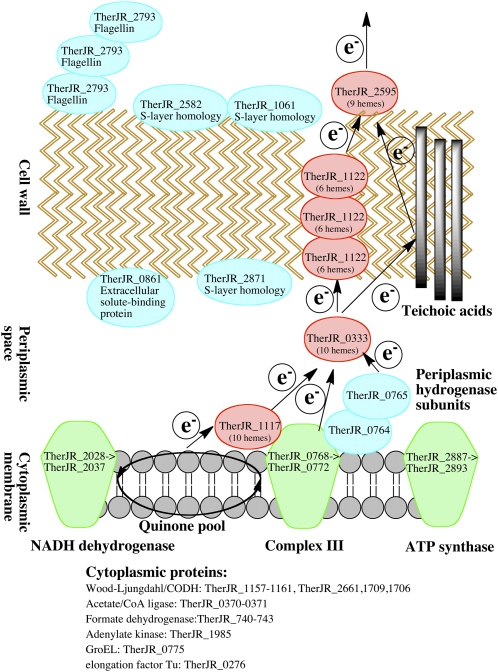
A model for the extracellular electron transfer conduit in T. potens is presented in Fig. 5. Core metabolic proteins in the Wood-Ljungdahl pathway, ATP synthase, and components of putative complex I and III were identified in the trypsin experiments. The extracellular electron transfer may begin at complex I or III, but one decaheme MHC with a N-terminal transmembrane region, TherJR_1117, was observed in the proteomics experiments. Few peptides from TherJR_1117 were observed in the trypsin experiments, but this protein is likely associated with the inner membrane and may function as a quinol dehydrogenase and the first component of the extracellular cytochrome cascade, similar to the role that CymA plays in Shewanella (13). Alternatively, it is possible that electrons may flow through an unidentified protein or the periplasmic hydrogenase subunits, TherJR_0764-0765, to initiate the extracellular electron transfer. In other organisms, the translocation of the large subunit (TherJR_0765) has been shown to be dependent on the Tat sequence of the small subunit (TherJR_0764) (39, 40), and the periplasmic hydrogenase of Desulfovibrio has been suggested to be involved in hydrogen recycling (41). Future work is needed to determine the role of the T. potens periplasmic hydrogenase in dissimilatory metal reduction, and confirm its periplasmic localization. The nonaheme MHC, TherJR_2595, with the highest trypsin/lysed ratio of any protein observed, is likely surface exposed on T. potens, and may be the terminal metal reductase similar to Shewanella MtrC or Geobacter OmcS. The decaheme MHC, TherJR_0333, has a lower trypsin/lysed ratio and may be periplasmic similar to Shewanella MtrA. TherJR_1122 is a hexaheme MHC that can be extracted from T. potens with low pH or denaturants, suggesting a noncovalent association with the cell wall. Few peptides from TherJR_1122 were observed in trypsin experiments, but this may be because the protein is tightly packed in the cell wall with blocked proteolytic sites. The trypsin/lysed ratio for TherJR_1122 was 0.7, and although only 11 peptides were observed for this protein across the four biological replicates, this ratio is consistent with cell wall localization. TherJR_1122 is not homologous to any characterized MHCs, but its loose association with the cell envelope is reminiscent of the Geobacter protein OmcS. OmcS is readily extracted from Geobacter cells with detergents, and similar to TherJR_1122, represents an abundant component of the electron conduit (34).
Fig. 5.
Model for proposed extracellular electron transfer conduit by T. potens. The likely cellular localization of MHCs and other proteins observed in proteomics experiments is indicated.
Taken together, the results of this study are unique in providing direct evidence for cytochromes localized to the cell wall or cell surface of a Gram-positive bacterium. Other Gram-positive DMRB species are known (2–4), and the mechanism used by T. potens may be a general strategy for electron transport across cell walls. In future studies, characterization of recombinantly expressed proteins, antibody labeling, and electron microscopy will provide additional information about the function and localization of the MHCs identified in this study. Although it is currently not possible to delete genes in T. potens, a reverse genetic system would confirm the functional roles of the identified MHCs, and the potential contribution of teichoic acids or other cell-wall components to extracellular electron transfer (42–45). The characterization of other Gram-positive DMRB using the techniques of this study will further expand our knowledge of electron transport conduits across bacterial cell walls.
Materials and Methods
Media and Culture Conditions.
T. potens was grown in basal media (11) containing 10 mM sodium acetate and 10 mM AQDS or 80 mM HFO at 55 °C. Cultures were also supplemented with 0.1 g/L yeast extract and an additional 10 μM NiCl2. Cells were harvested in late exponential phase. For experiments with cells grown on HFO, iron was extracted with oxalate, as previously described (17).
Electron Microscopy.
For detecting metallic silver deposition by transmission electron microscopy, T. potens cells from 200 mL of acetate/AQDS cultures were pelleted at 4,000 × g and fixed for 30 min in 0.1 M cacodylate buffer, pH 7.2, containing 2% glutaraldehyde and washed with buffer. Cells were then treated with EnzMet (Nanoprobes) HRP detection kit reagents. Briefly, cells were incubated with 120 μL of Detect A reagent for 2 min followed by 40 μL of Detect B and C respectively. Negative controls were preincubated for 15 min with 0.1 M potassium cyanide (KCN) in 0.1 M cacodylate buffer, pH 6.5, and then incubated with the silver reagents in the presence of KCN. Following this protocol, the cells were treated with 2% osmium and 0.5% ruthenium red for 30 min, processed through a graded ethanol series and embedded in Epon resin. Thin (70–100 nm) sections were collected on Formvar-coated 200-mesh copper grids and post-stained with 2% aqueous uranyl acetate. The sections were imaged at 120 kV using a Tecnai 12 TEM (FEI).
Low pH Protein Extraction.
Bacterial pellets were resuspended in 8 M Urea, 5 M guanidine HCl, or 0.2 M glycine, pH 2 and incubated at 25 °C for 30 min, and cells were pelleted at 10,000 × g for 5 min. The extracts and extracted cell pellets were resuspended in SDS-PAGE loading buffer and loaded onto Bio-Rad Any-Kd Minigels. Low pH extractions were neutralized with 2M Tris HCl, pH 7.5 before SDS-PAGE. SDS-PAGE gels were either stained with Coomassie or transferred to nitrocellulose membranes (Schleicher and Schuell) in a Bio-Rad Mini Trans-Blot cell. Membranes were washed briefly with PBS containing 0.5% Tween 20. Heme peroxidase activity of blotted proteins was assessed by addition of Pierce ECL substrate and luminol and imaging with a Bio-Rad ChemiDoc. In-gel trypsin digests from Coomassie-stained gels were done according to published methods (46) and analyzed by LC-MS/MS.
UV-Visible Spectroscopy.
Low pH extracts from T. potens were dialyzed against 50 mM Tris HCl, pH 7.5 in 10 kDa Slide-A-Lyzer dialysis cassettes (Pierce) overnight at 25 °C before spectral analysis. A Varian Cary 50 Bio spectrophotometer was used for collecting UV-visible electronic absorption spectra. Anaerobic cuvettes were filled with dialyzed extract and sparged with nitrogen. Dithionite was added from a 500 mM anaerobic stock solution to reduce the cytochrome, and the anaerobic cuvette was opened to the air to oxidize the cytochrome.
Resonance Raman and SERS.
T. potens cells were pelleted under N2 at 4,000 × g. For resonance Raman and colloidal surface-enhanced resonance Raman spectoscropy (SERRS), cells were fixed under N2 with 2.5% glutaraldehyde and resuspended to a final concentration of 109 cells/mL and diluted fivefold into spectral buffer (100 mM sodium phosphate buffer, pH 7.5) or Ag colloids prepared according to Lee and Meisel and others (29, 47). Cytochrome c (rabbit heart; Sigma) was prepared from a lyophilized powder and diluted to relevant concentrations in spectral buffer or Ag colloids. 0.1 M NaCl was added to induce aggregation of the Ag colloids. For SERS on Au electrodes, 1–2 μL cytochrome c from a 40 μM stock solution or T. potens cells from a 109 cell/mL cell suspension were spotted onto smooth or electrochemically roughened Au electrodes. A more detailed description of the electrode preparation methods is given in SI Materials and Methods.
Trypsin Shaving.
The protocol for trypsin shaving was based on previous protocols (30, 31) with modifications. Cell pellets from 200 mL of acetate/AQDS cultures (∼107 cells) were used for each condition. Cells were pelleted at 4,000 × g and washed 3 × 500 μL with 100 mM NH4HCO3 buffer (Sigma) and resuspended in 100 μL 100 mM NH4HCO3 buffer. Cells for the lysed cells control were sonicated aerobically three times for 30 s each and the cell suspension became translucent indicating lysis. Cell lysis was also verified microscopically. Bradford assays (Sigma) were used to determine that 100–200 μg of total protein was used for each condition. Twenty nanograms of trypsin gold porcine protease (Promega) or trypsin agarose beads (Promega) were added to the cell suspensions and the cells were incubated for 90 min at 37 °C. At the end of the trypsin treatment, all cells and cell debris was pelleted at 10,000 × g. Cells from the negative control and trypsin condition were checked by microscopy to confirm that less than 5–10% lysis had occurred. Cysteines were blocked with iodoacetamide and samples were digested overnight before LC-MS/MS (SI Materials and Methods).
LC-MS/MS.
Trypsin-digested proteins were analyzed by LC-MS/MS using a quadrupole time-of-flight mass spectrometer connected in-line with an ultraperformance liquid chromatograph (Waters). The resulting data were searched against the T. potens protein database using ProteinLynx Global Server software (Waters). A more detailed description of the LC-MS/MS methods used in this study is given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the R.A. Mathies, M.A., and J.D.C. laboratories for advice and comments on experimental procedures and the manuscript. This work was funded by independent grants from the Energy Biosciences Institute (to J.D.C. and M.A.); and Ecosystems and Networks Integrated with Genes and Molecular Assemblies, supported by the Office of Science, Office of Biological and Environmental Research of the US Department of Energy under Contract DE-AC02-05CH11231 (to M.A and A.G.). LC-MS instrumentation was acquired with National Institutes of Health Grant 1S10RR022393-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112905109/-/DCSupplemental.
References
- 1.Lloyd JR. Microbial reduction of metals and radionuclides. FEMS Microbiol Rev. 2003;27:411–425. doi: 10.1016/S0168-6445(03)00044-5. [DOI] [PubMed] [Google Scholar]
- 2.Slobodkin AI. Thermophilic microbial metal reduction (in Russian) Mikrobiologiia. 2005;74:581–595. [PubMed] [Google Scholar]
- 3.Wrighton KC, et al. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008;2:1146–1156. doi: 10.1038/ismej.2008.48. [DOI] [PubMed] [Google Scholar]
- 4.Villemur R, Lanthier M, Beaudet R, Lépine F. The Desulfitobacterium genus. FEMS Microbiol Rev. 2006;30:706–733. doi: 10.1111/j.1574-6976.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- 5.Marsili E, et al. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci USA. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bretschger O, et al. Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl Environ Microbiol. 2007;73:7003–7012. doi: 10.1128/AEM.01087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nevin KP, Lovley DR. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl Environ Microbiol. 2002;68:2294–2299. doi: 10.1128/AEM.68.5.2294-2299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lovley DR. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev. 1991;55:259–287. doi: 10.1128/mr.55.2.259-287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Naggar MY, et al. Electrical transport along bacterial nanowires from Shewanella oneidensis MR-1. Proc Natl Acad Sci USA. 2010;107:18127–18131. doi: 10.1073/pnas.1004880107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reguera G, et al. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 11.Wrighton KC, et al. Evidence for direct electron transfer by a Gram-positive bacterium isolated from a microbial fuel cell. Appl Environ Microbiol. 2011;77:7633–7639. doi: 10.1128/AEM.05365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrne-Bailey KG, et al. Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J Bacteriol. 2010;192:4078–4079. doi: 10.1128/JB.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi L, Squier TC, Zachara JM, Fredrickson JK. Respiration of metal (hydr)oxides by Shewanella and Geobacter: A key role for multihaem c-type cytochromes. Mol Microbiol. 2007;65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavrilov SN, Slobodkin AI, Robb FT, de Vries S. Characterization of membrane-bound Fe(III)-EDTA reductase activities of the thermophilic Gram-positive dissimilatory iron-reducing bacterium Thermoterrabacterium ferrireducens (in Russian) Mikrobiologiia. 2007;76:164–171. [PubMed] [Google Scholar]
- 15.Slobodkin AI, Sokolova TG, Lysenko AM, Wiegel J. Reclassification of Thermoterrabacterium ferrireducens as Carboxydothermus ferrireducens comb. nov., and emended description of the genus Carboxydothermus. Int J Syst Evol Microbiol. 2006;56:2349–2351. doi: 10.1099/ijs.0.64503-0. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S, Cavallaro G, Rosato A. A systematic investigation of multiheme c-type cytochromes in prokaryotes. J Biol Inorg Chem. 2010;15:559–571. doi: 10.1007/s00775-010-0623-4. [DOI] [PubMed] [Google Scholar]
- 17.Lovley DR, Phillips EJP. Availability of ferric iron for microbial reduction in bottom sediments of the freshwater tidal potomac river. Appl Environ Microbiol. 1986;52:751–757. doi: 10.1128/aem.52.4.751-757.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voordeckers JW, Kim BC, Izallalen M, Lovley DR. Role of Geobacter sulfurreducens outer surface c-type cytochromes in reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl Environ Microbiol. 2010;76:2371–2375. doi: 10.1128/AEM.02250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis RM, et al. Surface-enhanced Raman scattering from intracellular and extracellular bacterial locations. Anal Chem. 2008;80:6741–6746. doi: 10.1021/ac800838v. [DOI] [PubMed] [Google Scholar]
- 20.Zavarzina DG, et al. Thermincola ferriacetica sp. nov., a new anaerobic, thermophilic, facultatively chemolithoautotrophic bacterium capable of dissimilatory Fe(III) reduction. Extremophiles. 2007;11:1–7. doi: 10.1007/s00792-006-0004-7. [DOI] [PubMed] [Google Scholar]
- 21.Sokolova TG, et al. Thermincola carboxydiphila gen. nov., sp. nov., a novel anaerobic, carboxydotrophic, hydrogenogenic bacterium from a hot spring of the Lake Baikal area. Int J Syst Evol Microbiol. 2005;55:2069–2073. doi: 10.1099/ijs.0.63299-0. [DOI] [PubMed] [Google Scholar]
- 22.Luckevich MD, Beveridge TJ. Characterization of a dynamic S layer on Bacillus thuringiensis. J Bacteriol. 1989;171:6656–6667. doi: 10.1128/jb.171.12.6656-6667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubreuil JD, et al. Structural and biochemical analyses of a surface array protein of Campylobacter fetus. J Bacteriol. 1988;170:4165–4173. doi: 10.1128/jb.170.9.4165-4173.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleytr UB, Beveridge TJ. Bacterial S-layers. Trends Microbiol. 1999;7:253–260. doi: 10.1016/s0966-842x(99)01513-9. [DOI] [PubMed] [Google Scholar]
- 25.Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 26.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Surface-enhanced Raman spectroscopy. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 27.Biju V, et al. Combined spectroscopic and topographic characterization of nanoscale domains and their distributions of a redox protein on bacterial cell surfaces. Langmuir. 2007;23:1333–1338. doi: 10.1021/la061343z. [DOI] [PubMed] [Google Scholar]
- 28.Premasiri WR, et al. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J Phys Chem B. 2005;109:312–320. doi: 10.1021/jp040442n. [DOI] [PubMed] [Google Scholar]
- 29.Culha M, et al. Characterization of thermophilic bacteria using surface-enhanced Raman scattering. Appl Spectrosc. 2008;62:1226–1232. doi: 10.1366/000370208786401545. [DOI] [PubMed] [Google Scholar]
- 30.Solis N, Larsen MR, Cordwell SJ. Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false-positive control. Proteomics. 2010;10:2037–2049. doi: 10.1002/pmic.200900564. [DOI] [PubMed] [Google Scholar]
- 31.Tjalsma H, Lambooy L, Hermans PW, Swinkels DW. Shedding & shaving: Disclosure of proteomic expressions on a bacterial face. Proteomics. 2008;8:1415–1428. doi: 10.1002/pmic.200700550. [DOI] [PubMed] [Google Scholar]
- 32.Cordwell SJ. Technologies for bacterial surface proteomics. Curr Opin Microbiol. 2006;9:320–329. doi: 10.1016/j.mib.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Tsuboi A, Tsukagoshi N, Udaka S. Reassembly in vitro of hexagonal surface arrays in a protein-producing bacterium, Bacillus brevis 47. J Bacteriol. 1982;151:1485–1497. doi: 10.1128/jb.151.3.1485-1497.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta T, Coppi MV, Childers SE, Lovley DR. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol. 2005;71:8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demchick P, Koch AL. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol. 1996;178:768–773. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke TA, et al. Structure of a bacterial cell surface decaheme electron conduit. Proc Natl Acad Sci USA. 2011;108:9384–9389. doi: 10.1073/pnas.1017200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokkuluri PR, et al. Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12 nm protein with interacting hemes. J Struct Biol. 2011;174:223–233. doi: 10.1016/j.jsb.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 38.Paterson GK, Mitchell TJ. The biology of Gram-positive sortase enzymes. Trends Microbiol. 2004;12:89–95. doi: 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Gross R, Simon J, Kroger A. The role of the twin-arginine motif in the signal peptide encoded by the hydA gene of the hydrogenase from Wolinella succinogenes. Arch Microbiol. 1999;172:227–232. doi: 10.1007/s002030050764. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigue A, Chanal A, Beck K, Müller M, Wu LF. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 41.Odom JM, Peck HD. Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria, Desulfovibrio-Sp. FEMS Microbiol Lett. 1981;12:47–50. [Google Scholar]
- 42.Ehrlich HL. Are Gram-positive bacteria capable of electron transfer across their cell wall without an externally available electron shuttle? Geobiology. 2008;6:220–224. doi: 10.1111/j.1472-4669.2007.00135.x. [DOI] [PubMed] [Google Scholar]
- 43.Matias VR, Beveridge TJ. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol. 2005;56:240–251. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- 44.Matias VR, Beveridge TJ. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J Bacteriol. 2006;188:1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matias VR, Beveridge TJ. Lipoteichoic acid is a major component of the Bacillus subtilis periplasm. J Bacteriol. 2008;190:7414–7418. doi: 10.1128/JB.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 47.Lee PC, Meisel D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J Phys Chem. 1982;86:3391–3395. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.