Abstract
In the dorsal spinal cord, distinct interneuron classes relay specific somatosensory information, such as touch, heat, and pain, from the periphery to higher brain centers via ipsilateral and contralateral axonal pathways. The transcriptional mechanisms by which dorsal interneurons choose between ipsilateral and contralateral projection fates are unknown. Here, we show that a single transcription factor (TF), BARHL2, regulates this choice in proprioceptive dI1 interneurons by selectively suppressing cardinal dI1contra features in dI1ipsi neurons, despite expression by both subtypes. Strikingly, dI1ipsi neurons in Barhl2-null mice exhibit a dI1contra cell settling pattern in the medial deep dorsal horn, and, most importantly, they project axons contralaterally. These aberrations are preceded by ectopic dI1ipsi expression of the defining dI1contra TF, LHX2, and down-regulation of the dI1ipsi-enriched TF, BARHL1. Taken together, these results elucidate BARHL2 as a critical postmitotic regulator of dI1 subtype diversification, as well as its intermediate position in the dI1 genetic hierarchy.
Keywords: neurogenesis, neuronal differentiation, Atoh1/Math1, Robo3/Rig1, BarH/BarH-like
Spinal neurons that process sensory input and motor output are broadly distributed in the dorsal and ventral spinal cord, respectively (1, 2). Relay interneurons of the dorsal spinal cord process and transmit specific somatosensory modalities, such as tactition, proprioception, and nociception, from the periphery to intraspinal and supraspinal brain targets, such as thalamus and cerebellum, via ipsilateral and contralateral axonal pathways (3). The dorsal interneurons are vastly heterogeneous, but they can be classified into six early-born (dI1–dI6) and two late-born (dILA and dILB) groups, each derived from a specific progenitor domain in the dorsal spinal neural tube and exhibiting stereotypic function, cell settling pattern, molecular profile, and axonal targeting (4).
Of these, the proprioceptive dI1 interneurons relay positional information about the trunk and limbs to the cerebellum via spinocerebellar and cuneocerebellar tracts (3). The dI1 neurons are derived from the ATOH1+, dorsal-most progenitor domain of the spinal cord that flanks the roof plate (3, 5). As dI1 neurons exit cell cycle, they migrate ventrally to settle in the deep dorsal horn and segregate into the ipsilaterally projecting dI1i subtype in the lateral deep dorsal horn and contralaterally projecting dI1c subtype in the medial deep dorsal horn (3, 5, 6). The two subtypes are further differentiated by their LIM homeodomain transcription factor (TF) profiles: dI1i neurons express LHX9 but not LHX2, and dI1c neurons express LHX2 and very low levels of LHX9 (5, 6).
Several regulatory genes expressed by the roof plate, such as the signaling molecule GDF7 and the LIM homeodomain TF LMX1A, or by dI1 progenitors, such as ATOH1, regulate the specification and generation of dI1 interneurons (3, 5, 7–11). Others execute dI1 subtype-specific properties, such as midline crossing of dI1c axons by LHX2 and LHX9 via regulation of the commissural axonal receptor ROBO3 (6). However, the TFs that regulate the binary diversification of dI1 neurons into dI1i and dI1c subtypes remain unknown. The mammalian Bar class TFs, BARHL1 and BARHL2, both ATOH1 downstream targets, are potential candidates because their ectopic expression in the dorsal spinal cord specifically induces commissural axonal targeting (12–14). However, the expression of Barhl1 and Barhl2 by both dI1i and dI1c neurons provides a significant argument against their role in subtype divergence (6, 13, 14). Although targeted deletion of Barhl1 has no discernable effect on dI1 identity, or subtype divergence (15), BARHL2's function in dI1 neurons itself has not been previously identified.
We thus used an in vivo loss-of-function strategy and generated mice with targeted deletion of Barhl2. Barhl2-nulls exhibit a dramatic increase and corresponding decrease in dI1 neurons exhibiting dI1c and dI1i properties, respectively. The supernumerary dI1c neurons are attributable to a dI1i-to-dI1c fate switch, because dI1i neurons settle in the medial deep dorsal horn and project axons across the midline into the contralateral ventral funiculus (VF). Intriguingly, this failed dI1 subtype divergence is characterized by ectopic dI1i expression of the dI1c markers LHX2 and ROBO3, and down-regulation of the dI1i-enriched TF, Barhl1. Taken together with the preserved expression of upstream regulators like Gdf7 and Atoh1 in Barhl2-nulls, our results reveal the central function of BARHL2 in the postmitotic divergence of dI1 neurons into distinct dI1i and dI1c subtypes, and establish its intermediate position in the dI1 genetic hierarchy.
Results
Barhl2 Is Expressed in Postmitotic dI1 Neurons of the Developing Spinal Cord.
In situ hybridization reveals the onset of Barhl2 expression in dI1 neurons at embryonic day (E) 10.5 at the dorsal margin of the spinal cord, followed by expression in ventrally migrating dI1 neurons at E11.5. Barhl2 continues to be expressed postmigrationally by both dI1i and dI1c subtypes in the deep dorsal horn at E12.5 but weakens from E15.5 onward (Fig. S1A). To characterize spinal Barhl2 expression more precisely, we analyzed cell type-specific expression of the lacZ reporter in previously generated Barhl2-lacZ knock-in mice (16). Double immunolabeling on transverse E11.5 Barhl2lacZ/+ (heterozygote) cervical spinal cord sections reveals Barhl2-lacZ expression by Tuj1+ postmitotic neurons but not by cycling progenitors (Fig. S1 B–E). Barhl2's dI1-specific expression is confirmed by Barhl2-lacZ colabeling with the dI1 markers LHX2 and LHX9, and absence of colabeling with dI2–dI6 markers, including ISL1 and PAX2 (Fig. S1 F–I). Taken together, Barhl2 is specifically expressed by all postmitotic dI1 neurons, from the time of cell cycle exit to postmigrational settling in the deep dorsal horn. These results are in conformity with previously published Barhl2 expression analysis (13, 14).
Absence of dI1i Neurons in the Lateral Deep Dorsal Horn of Barhl2-Nulls.
BARHL2's aforementioned expression motivated us to investigate its role in postmitotic specification and differentiation of dI1 neurons. We used an in vivo loss-of-function approach, and crossed Barhl2lacZ/+ mice to obtain Barhl2lacZ/lacz (Barhl2-null) mice. Barhl2-nulls were born at Mendelian frequencies and survived up to 3 wk postnatally. In situ hybridization revealed complete absence of Barhl2 transcripts in Barhl2-nulls (Fig. S2).
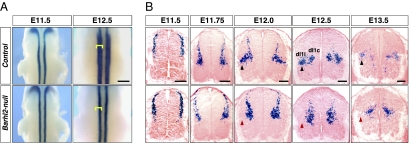
We first characterized the migration and distribution of dI1 neurons in the developing Barhl2-null spinal cord via X-Gal histochemistry on whole-mount and transverse cervical spinal cord sections (Fig. 1). At E11.5, Barhl2-null lacZ+ neurons were normally distributed at the dorsal margin of the spinal cord and in the ventrally migrating stream. Between E11.75 and E13.5, control Barhl2-lacZ+ neurons complete migration to the deep dorsal horn and resolve into dI1i and dI1c neurons in the lateral and medial deep dorsal horn, respectively. Strikingly, Barhl2-null lacZ+ neurons overwhelmingly settle in the medial deep dorsal horn. The lateral dI1i subset is absent. The absence of dI1i is not attributable to cell death or failure in generation; Barhl2-nulls do not exhibit a change in the total number of Barhl2-lacZ+ neurons at E11.5 (per side: control = 50.00 ± 3.56; Barhl2-nulls = 47.75 ± 5.38; P = 0.5114; n = 4 sections from 2 mice) or E12.5 (per side: control = 90.82 ± 7.51, Barhl2-nulls = 84.20 ± 10.09; P = 0.495; n = 6 sections from 3 mice). These results collectively suggest that Barhl2-null dI1i neurons either migrate erroneously to the medial deep dorsal horn or are transfated to dI1c identity.
Fig. 1.
dI1i neurons are absent in the lateral deep dorsal horn of the Barhl2-null spinal cord. (A) X-Gal histochemistry on whole-mount embryos. At E11.5, Barhl2-lacZ+ dI1 neurons in Barhl2lacZ/+ controls and Barhl2lacZ/lacZ-null spinal cords are similarly distributed in one column. At E12.5, there are two lacZ+ columns in the controls; in contrast, Barhl2-nulls only have one column medially (yellow brackets). (B) X-Gal histochemistry on transverse cervical spinal cord sections. Between E11.5 and E13.5, control dI1 neurons migrate to the deep dorsal horn and resolve into the lateral dI1i (black arrowheads) and medial dI1c subtypes. In contrast, Barhl2-null dI1 neurons overwhelmingly migrate to and settle in the medial deep dorsal horn starting from E11.75. The lateral dI1i subtype is absent (red arrowheads). (Scale bars: A, 1 mm; B, 100 μm.)
Ectopic LHX2 Expression in dI1i Neurons of Barhl2-Nulls.
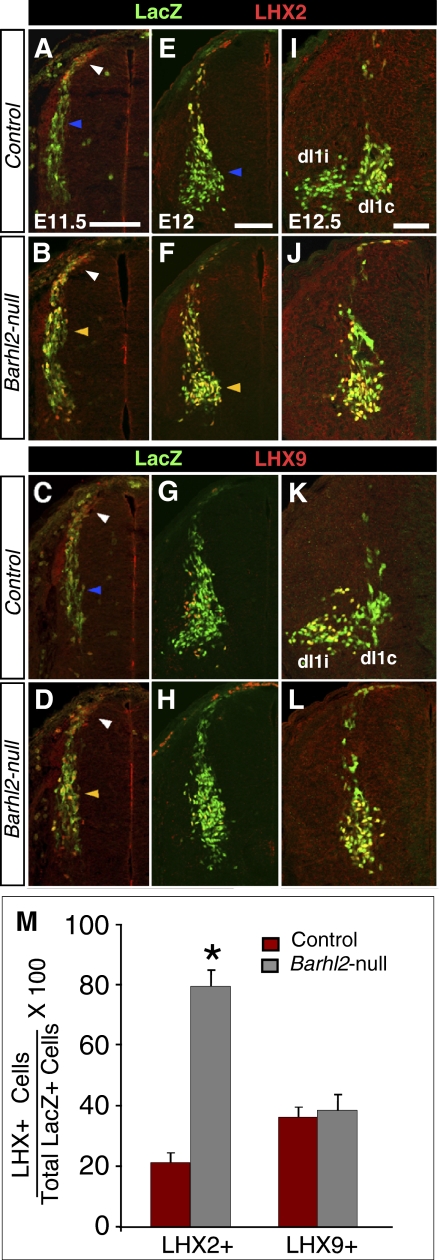
To investigate these possibilities, we first correlated the medial deep dorsal horn cell settling of all dI1 neurons with dI1 subtype-specific gene expression changes in Barhl2-nulls. LHX2 and LHX9 are differentially expressed by dI1i and dI1c subtypes on segregation, with LHX2 exclusively expressed by dI1c neurons (5, 6, 8, 12). Barhl2-nulls exhibit a robust increase in dI1 neurons expressing LHX2. In E10.5 controls, Lhx2 and Lhx9 are coexpressed by newly postmitotic dI1 neurons at the dorsal margin. This early expression of LHX2 and LHX9 is unaltered in Barhl2-nulls (Fig. S3). At E11.5, the presumptive dI1i neurons, which express LHX9 but not LHX2, migrate ventrally toward the deep dorsal horn (Fig. 2 A and C). These neurons likely arise from the more dorsal newly postmitotic neurons after they switch off LHX2 (8). Strikingly, the presumptive Barhl2-null dI1i neurons ectopically express LHX2, and thus continue to express both LHX2 and LHX9 (Fig. 2 B and D).
Fig. 2.
Ectopic LHX2 expression in dI1i neurons of Barhl2-nulls suggests dI1i-to-dI1c respecification. (A–D) In E11.5 Barhl2lacZ/+ controls, newly postmitotic neurons at the dorsal margin of the spinal cord (white arrowhead) express LHX2 and LHX9, but the ventrally migrating dI1i neurons express only LHX9 (blue arrowhead). In Barhl2lacZ/lacZ-nulls, dI1i neurons ectopically express LHX2 (yellow arrowhead). LHX2 at the dorsal margin and LHX9 are unchanged. (E–H) At E12.0, LHX2 continues to be ectopically expressed by dI1i neurons that have migrated to the medial deep dorsal horn (yellow arrowheads) in Barhl2-nulls. LHX2 expression in dI1c neurons and LHX9 is unperturbed. (I–L) At E12.5, Barhl2-expressing dI1 neurons segregate into lateral LHX2−/LHX9+ dI1i neurons and medial LHX2high/LHX9low dI1c neurons. In Barhl2-nulls, dI1 neurons fail to segregate into dI1i and dI1c groups. (M) Quantitation in I–L reveals an approximately fourfold increase (*P = 0.0001) in Barhl2-LacZ+ neurons expressing LHX2 in Barhl2-nulls. There is no change in Barhl2-LacZ+ neurons expressing LHX9 (P = 0.21). (Scale bars: 100 μm.)
At E12.0, there are two populations of dI1 neurons: (i) the LHX2−/LHX9+ presumptive dI1i neurons that reach the medial deep dorsal horn and (ii) the LHX2+/LHX9− presumptive dI1c neurons that emerge from the ATOH1+ progenitor domain and migrate ventrally toward the deep dorsal horn (Fig. 2 E and G). In Barhl2-nulls, the dI1i neurons in the medial deep dorsal horn continue to express LHX2 ectopically, whereas dI1c neurons exhibit a LHX2+/LHX9− profile similar to controls (Fig. 2 F and H). Finally, at E12.5, dI1 neurons resolve into the lateral LHX2−/LHX9+ dI1i neurons and the medial LHX2high/LHX9low dI1c neurons (Fig. 2 I and K). In contrast, dI1 neurons in Barhl2-nulls fail to resolve into these two groups and accumulate in the medial deep dorsal horn (Fig. 2 J and L). Quantitation at E12.5 reveals a dramatic, approximately fourfold increase in the percentage of Barhl2-lacZ+ dI1 neurons expressing LHX2 in Barhl2-nulls compared with controls (Fig. 2M; controls = 21.4%, n = 7 sections from 3 mice; Barhl2-nulls = 79.4%, n = 10 sections from 3 mice; P = 0.0001). There is no change in the percentage of dI1 neurons expressing LHX9 in Barhl2-nulls (control = 36.6%; Barhl2-nulls = 39.5%; P = 0.21; per group; n = 10 sections from 3 mice). To summarize, absence of BARHL2 results in ectopic LHX2 expression, specifically in dI1i neurons. Of note, Barhl2-null dI1 neurons do not ectopically express dI2–dI6 markers (Fig. S4).
Dramatic Increase in Contralaterally Projecting dI1 Neurons in Barhl2-Null Spinal Cord.
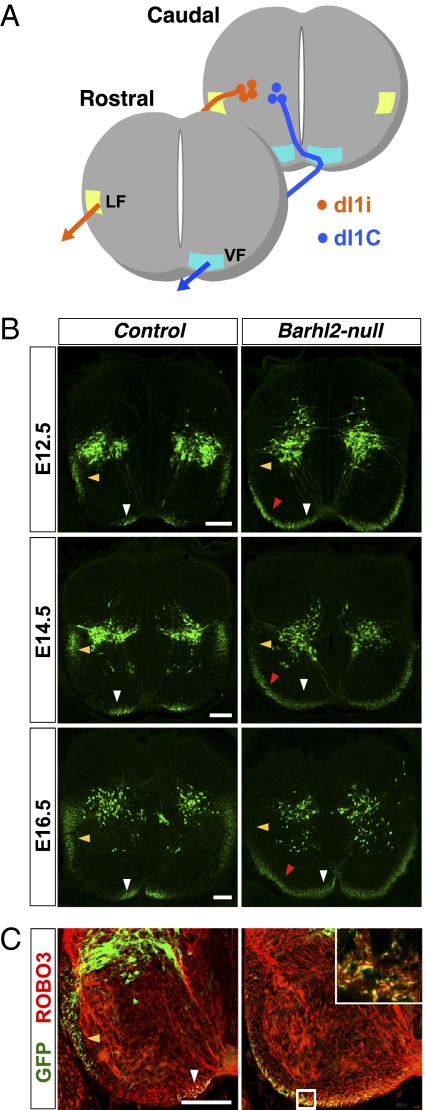
Although ectopic LHX2 expression in dI1i neurons in Barhl2-nulls further bolsters the possibility of a dI1i-to-dI1c fate switch, it does not conclusively prove it; the aberrant settling of all dI1 neurons in the medial deep dorsal horn could also reflect a migration error by dI1i neurons. The dI1i neurons project axons into the ipsilateral lateral funiculus (LF), whereas the dI1c neurons project axons into the contralateral VF (6) (Fig. 3A). A simple migration error is indicated by preserved axonal targeting, whereas a dI1i-to-dI1c fate switch is indicated by reduction of dI1 axons in the LF and a corresponding increase in the VF. We observe the latter in Barhl2-nulls.
Fig. 3.
Genetic lineage tracing reveals a striking expansion of Barhl2+ axons in the VF. (A) Schematic. Medially located dI1c neurons project axons into the contralateral VF close to the midline. Laterally located dI1i neurons project axons into the ipsilateral LF. (B) Barhl2 genetic lineage tracing. GFP (green) immunohistochemistry on E12.5–E16.5 Barhl2cre/+; Z/EG controls reveals dI1i axons in the LF (yellow arrowheads) and dI1c axons in the VF (white arrowheads). In Barhl2cre/lacZ; Z/EG nulls, there is a drastic reduction of GFP+ fibers in the LF and a dramatic expansion of GFP+ fibers in the VF (red arrowheads). (C) GFP (green) and ROBO3 (red) double-immunolabeling indicates the contralateral origin of the ectopic GFP+ fibers spanning the mediolateral extent of the VF in Barhl2cre/lacZ; Z/EG nulls at E12.5. (Inset) Magnified view of a 0.4-μm-thick optical section of the boxed region. (Scale bars: 100 μm.)
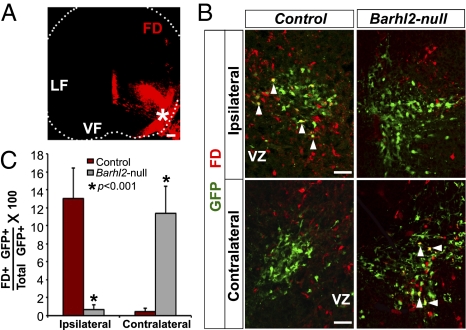
To visualize dI1 axons, we used in vivo lineage tracing and crossed Barhl2cre/+ knock-in mice with conditional GFP reporter lacZ/EGFP (Z/EG) mice (17). As expected, Barhl2-directed GFP signal in E12.5–E16.5 Barhl2cre/+; Z/EG heterozygote controls illuminated dI1i axons in the LF and dI1c axons in the VF close to the midline. In striking contrast, Barhl2cre/lacZ; Z/EG nulls exhibited a drastic reduction of GFP+ axons in the LF and a dramatic expansion of GFP+ axons in the VF that now ectopically span its entire mediolateral extent (Fig. 3B). The contralateral origin of the ectopic GFP+ axons in the VF was further suggested by colabeling with the commissural axonal marker, ROBO3 (18) (Fig. 3C). To test conclusively the hypothesis that the ectopic GFP+ fibers in the lateral extent of the VF of Barhl2-nulls were indeed contralateral in origin, we performed retrograde labeling and injected fluorescein-conjugated dextran (FD) into the E13.5 lateral VF (Fig. 4 A–C). In controls, FD back-labeled GFP+ neurons were ipsilateral to the injection site as expected (control = 13 ± 3.4%; Barhl2-null = 0.66 ± 0.57%; P = 0.0006; per group, n = 6 sections from 3 mice). In striking contrast, FD back-labeled GFP+ neurons in Barhl2-nulls were almost exclusively contralateral to the injection site (control = 0.44 ± 0.54%; Barhl2-null = 11.39 ± 2%; P = 0.0001; per group, n = 6 sections from 3 mice), thereby definitively validating the hypothesis. Of note, the normal expression of the roof plate marker Gdf7 and floor plate markers Ntn1 and Slit2 further bolsters the cell-autonomous nature of the dI1 phenotype in Barhl2-nulls (Fig. S5 B–D).
Fig. 4.
Dramatic increase in contralaterally projecting dI1 interneurons in Barhl2-nulls. (A) E13.5 spinal cord section depicting site of FD crystal placement (*) in the lateral VF and the extent of FD diffusion after 8 h of incubation during retrograde labeling. (B) Retrograde labeling from the lateral VF reveals FD+ back-labeled GFP+ neurons (white arrowheads) ipsilateral to the injection site in controls, as expected, and, strikingly, contralateral to the injection site in Barhl2-nulls. (C) Quantitation. There is a ∼20-fold decrease (P = 0.0001) and ∼26-fold increase (P < 0.0001) in GFP+ neurons back-labeled with FD ipsilateral and contralateral to the FD injection site in Barhl2-nulls, respectively. (Scale bars: 50 μm.)
Taken together, the settling of dI1 neurons in the medial deep dorsal horn, the ectopic LHX2 expression in presumptive dI1i neurons, the near absence of ipsilaterally projecting dI1 neurons, and the supernumerary contralaterally projecting dI1 neurons provide compelling evidence of failed dI1 subtype divergence in Barhl2-nulls attributable to a respecification of dI1i neurons to dI1c fate.
BARHL2 Directly Regulates the Lhx2 Gene.
The ectopic expression of LHX2 in presumptive dI1i neurons motivates the hypothesis that Lhx2 is a direct target of BARHL2. Analysis of the conserved region of the Lhx2 promoter across several species for consensus homeodomain protein binding ACTAATT sequences that contain the core TAAT motif revealed two such sequences in forward and reverse orientations at −3325 and −3248, respectively (15) (Fig. S6A). Next, EMSAs showed that BARHL2 can bind to both these sites and that BARHL2 binding is abolished when these sites are mutated (Fig. S6 B and C). To examine whether BARHL2's binding to these sites can indeed regulate Lhx2 transcription, we cloned the Lhx2 regulation region containing the two binding sites into the luciferase reporter vector and assayed transfected HEK293 cells for luciferase activity (Fig. S6D). Strikingly, cotransfection of Lhx2-luciferase and Barhl2 expression vectors resulted in a robust 68% ± 9.2% decrease in luciferase activity (P < 0.01) compared with baseline controls (Lhx2-luciferase reporter vector only). Intriguingly, mutation (CTTAT to CCGGG) in even one BARHL2 binding site resulted in no change in luciferase activity, thereby suggesting that binding of BARHL2 to both sites is necessary for regulation of the Lhx2 promoter. Taken together, these results are consistent with the idea that BARHL2 might directly repress Lhx2 expression in dI1i neurons.
BARHL2 Is Intermediate in the dI1 Genetic Hierarchy.
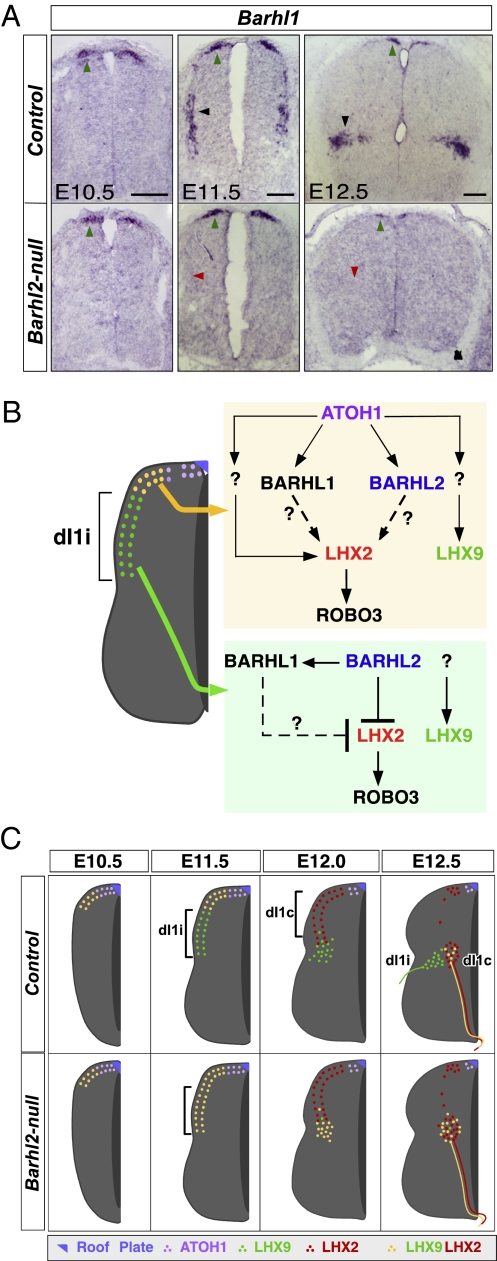
BARHL2's role in dI1 subtype divergence prompts more precise delineation of its position in the dI1 genetic hierarchy. We predicted that BARHL2's postmitotic expression and actions would likely preclude it from regulating the expression of important dI1 genes that are expressed in the roof plate, such as Gdf7, or in dI1 progenitors, such as Atoh1. As expected, the expression of both Gdf7 and Atoh1 is preserved in Barhl2-nulls, confirming their upstream status in the hierarchy (Fig. S5 A and D). To investigate potential BARHL2 downstream targets other than Lhx2, we examined the expression of the other known dI1 marker, Barhl1, in Barhl2-nulls. Barhl1 is expressed by all dI1 neurons at E10.5 and E11.5, but it is largely restricted to dI1i neurons at E12.5 (3, 6, 19) (Fig. 5A). Between E10.5 and E12.5, Barhl1 expression is unaltered at the dorsal margin, but it is absent in all migrating and postmigrational dI1 neurons in Barhl2-nulls, suggesting that BARHL2 is required for maintenance but not initiation of Barhl1 expression in dI1 neurons. Together, these data show that BARHL2 is intermediate in the dI1 genetic hierarchy and that it regulates the expression of select downstream genes, such as Lhx2 and Barhl1, contextually in a subset of dI1 neurons.
Fig. 5.
BARHL2 is intermediate in the dI1 genetic hierarchy. (A) Barhl1 expression is unperturbed at the dorsal margin (green arrowheads) but is lost in dI1i neurons (black and red arrowheads), suggesting that BARHL2 is required for the maintenance but not initiation of Barhl1 expression in dI1 neurons. (Scale bars: 50 μm.) (B) Schematic. Genetic hierarchy and contextual transcriptional regulation by BARHL2 in dI1 neurons. ATOH1 activates Barhl1 and Barhl2 in newly postmitotic neurons at the dorsal margin. Targeted deletion of Barhl1 or Barhl2 does not perturb LHX2, suggesting BARHL-redundant or BARHL-independent activation of LHX2 by ATOH1 at dorsal margin. Other studies show that ATOH1 likely activates LHX9 by non–Barhl-mediated pathways. In dI1i neurons, BARHL2 suppresses LHX2. Also, Barhl1 expression is lost in Barhl2-null dI1i neurons. Thus, BARHL1 cannot compensate for lost BARHL2 function in dI1i neurons. Taken together, BARHL1 and BARHL2 might contextually and redundantly activate LHX2 in newly postmitotic dI1 neurons, and repress LHX2 in dI1i neurons. (C) Summary. The dI1i neurons in Barhl2-nulls ectopically express LHX2, accumulate in the medial deep dorsal horn, and project axons contralaterally. These results indicate respecification of dI1i neurons to dI1c fate, and thus a failure of dI1 subtype divergence.
Discussion
The central finding of this study is that BARHL2 regulates divergence in spinal cord dI1 neurons to generate distinct dI1i and dI1c subtypes. Here, we show that in the absence of Barhl2, the subset of dI1 neurons fated for dI1i identity is respecified to dI1c identity, thereby generating supernumerary dI1c neurons. This failure of dI1i/dI1c divergence is characterized most notably by a striking increase in dI1 neurons that express the dI1c marker LHX2 and project axons contralaterally (Fig. 5C).
BARHL2's specific function in suppressing commissural fate is notable, and surprising, in light of previous Barhl2 gain-of-function studies that postulated a role in assigning commissural fate in the dorsal spinal cord (12–14). Our loss-of-function analysis reveals that BARHL2's actions occur very specifically in the context of dI1i neurons. Thus, the phenotypic differences observed could perhaps be attributed to novel BARHL2 actions that arise in atypical cellular contexts as a result of ectopic electroporation of the Barhl2 construct, and probably also as a result of its premature expression in cycling progenitors. Our results also further establish the intermediate position of BARHL2 in the transcriptional cascade that controls dI1 identity (Fig. 5B). Postmitotically expressed BARHL2 is downstream of progenitor-expressed TFs, such as ATOH1, that broadly specify dI1 fate (3, 7); as expected, Atoh1 expression is unaltered in Barhl2-nulls, whereas Barhl2 expression is lost in Atoh1-nulls (13). Our results bolster previous studies suggesting that Atoh1 directly recruits Barhl genes to regulate other downstream postmitotic TFs, such as LHX2 and LHX9 (12, 13, 20). Although BARHL2 is dispensable for LHX9 expression in all dI1 neurons, Barhl2 suppresses LHX2 specifically in dI1i neurons. Importantly, this ectopic LHX2 expression precedes the aberrant cell settling of all dI1 neurons in the medial deep dorsal horn, as well as increased contralateral targeting in Barhl2-nulls. Thus, suppressing LHX2 in dI1i neurons, and thereby probably its downstream targets, such as the commissural axon guidance receptor ROBO3, could be crucial for repressing dI1c attributes in dI1i neurons during normal development. Although ectopic LHX2 expression in the dorsal spinal cord results in moderate induction of contralateral targeting (12), Barhl2-directed LHX2 expression, via transgenic or knock-in approaches leading to ectopic expression in dI1i neurons, could provide more accurate insights.
BARHL2's restricted action in dI1i neurons is intriguing but also counterintuitive, because Barhl2 is expressed by all dI1 neurons. For example, at E11.5, BARHL2 regulates LHX2 expression in presumptive dI1i neurons but not in newly postmitotic neurons at the dorsal margin of the spinal cord. One explanation for the dI1i-specific action could be BARHL2's redundant function with BARHL1 (3, 6, 19). Although targeted deletion of Barhl1 does not affect dI1 development, Barhl1 misexpression studies suggest a role in assigning commissural fate and reveal redundant actions with Barhl2, such as ectopic activation of LHX2 in dorsal spinal cord. Both approaches also reveal that BARHL1 does not regulate spinal Barhl2 expression (12, 15). On the contrary, our loss-of-function analysis shows that Barhl2 is required for maintaining but not initiating Barhl1 expression in dI1 neurons; Barhl1 expression is abolished in dI1i neurons, but it is unperturbed at the dorsal margin in Barhl2-nulls. Thus, Barhl1 might compensate for lost Barhl2 function in newly postmitotic neurons at the dorsal margin but cannot do so in dI1i neurons. Analysis of dI1 phenotype in Barhl1; Barhl2-compound nulls could reveal the true extent of redundant BARHL1 and BARHL2 actions, and perhaps an earlier role for Barhl genes at the dorsal margin and in dI1c neurons, such as activation of LHX2 expression.
The maintenance but not initiation of Barhl1 by BARHL2 might be explained by their slightly nonoverlapping spatiotemporal expression in dI1 neurons during spinal cord development. First, Barhl1 onset at E10 precedes Barhl2 onset at E10.5. Second, Barhl1 is expressed in dI1 progenitors, possibly cycling, that are closer to the roof plate, in contrast to Barhl2's strictly postmitotic expression at the dorsal margin (12, 13, 19, 20). This is probably attributable to earlier activation of Barhl1 than Barhl2 by ATOH1 (12). Once Barhl2 expression is triggered, BARHL2, which binds to conserved motifs in Barhl1 promoter and activates Barhl1 expression (15), might then contribute by sustaining Barhl1 expression in more differentiated dI1 neurons, such as dI1i neurons. Of note, BARHL2's activation of Barhl1 and repression of LHX2 in dI1 neurons further corroborate gene-specific and context-dependent transcriptional activation or repression by Barhl genes as seen in other systems, such as the retina or inner ear (21, 22).
BARHL2's key role in dI1 subtype divergence motivates three questions for comprehensive elucidation of dI1 biology. First, is there a converse suppression of dI1i fate in dI1c neurons, or is dI1c fate the default fate for all dI1 neurons? Second, do dI1i and dI1c neurons have different central termination sites, and does BARHL2 inactivation alter central connectivity? Third, what are the behavioral consequences of the dI1i-to-dI1c fate switch observed in Barhl2-nulls vis-à-vis proprioception? Barhl2-nulls survive up to the third postnatal week and exhibit ataxia that is characterized by unstable gait, abrupt limb movements, and bunching of the trunk during tail suspension (Movie S1). However, these anomalies cannot be conclusively attributed to the dI1i/dI1c fate switch, because Barhl2 is also expressed in the other major proprioceptive center, the cerebellum (20). Future analysis of conditional Barhl2 KO mice with spinal cord-specific deletion will more precisely delineate the role of the dI1i-to-dI1c fate switch in manifestation of the aforementioned abnormal proprioceptive behaviors.
Materials and Methods
Barhl2-lacZ and Barhl2-cre knock-in mice were generated previously (16). The Z/EG conditional enhanced GFP reporter mice were purchased from the Jackson Laboratory (stock no. 003920), and genotyping was performed according to protocols provided by the Jackson Laboratory. Mice were maintained in C57BL/6J and 129S6 mixed background. Embryos were identified as E0.5 at noon on the day at which vaginal plugs were first observed. The day of birth was designated postnatal day 0. Results from Barhl2lacZ/lacZ nulls were verified in Barhl2lacZ/cre nulls. All animal procedures in this study were approved by the University Committee of Animal Resources at the University of Rochester. Further experimental details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Goulding and E. Geiman for the retrograde tracing protocol, D. Kingsley for the Gdf7 probe, and M. Tessier-Lavigne for the Ntn11 and Slit2 probes. This work was supported by National Institutes of Health Grant EY013426 and a Research to Prevent Blindness Senior Scientific Investigator Award (both to L.G.), as well as by a Research to Prevent Blindness Challenge Grant to the Department of Ophthalmology at the University of Rochester.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112392109/-/DCSupplemental.
References
- 1.Lee KJ, Jessell TM. The specification of dorsal cell fates in the vertebrate central nervous system. Annu Rev Neurosci. 1999;22:261–294. doi: 10.1146/annurev.neuro.22.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Caspary T, Anderson KV. Patterning cell types in the dorsal spinal cord: What the mouse mutants say. Nat Rev Neurosci. 2003;4:289–297. doi: 10.1038/nrn1073. [DOI] [PubMed] [Google Scholar]
- 3.Bermingham NA, et al. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- 4.Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr Opin Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 5.Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–928. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- 6.Wilson SI, Shafer B, Lee KJ, Dodd J. A molecular program for contralateral trajectory: Rig-1 control by LIM homeodomain transcription factors. Neuron. 2008;59:413–424. doi: 10.1016/j.neuron.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Gowan K, et al. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 8.Lee KJ, Mendelsohn M, Jessell TM. Neuronal patterning by BMPs: A requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 1998;12:3394–3407. doi: 10.1101/gad.12.21.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manzanares M, Trainor PA, Ariza-McNaughton L, Nonchev S, Krumlauf R. Dorsal patterning defects in the hindbrain, roof plate and skeleton in the dreher (dr(J)) mouse mutant. Mech Dev. 2000;94:147–156. doi: 10.1016/s0925-4773(00)00288-4. [DOI] [PubMed] [Google Scholar]
- 10.Millen KJ, Millonig JH, Hatten ME. Roof plate and dorsal spinal cord dl1 interneuron development in the dreher mutant mouse. Dev Biol. 2004;270:382–392. doi: 10.1016/j.ydbio.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Millonig JH, Millen KJ, Hatten ME. The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature. 2000;403:764–769. doi: 10.1038/35001573. [DOI] [PubMed] [Google Scholar]
- 12.Kawauchi D, Muroyama Y, Sato T, Saito T. Expression of major guidance receptors is differentially regulated in spinal commissural neurons transfated by mammalian Barh genes. Dev Biol. 2010;344:1026–1034. doi: 10.1016/j.ydbio.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Saba R, Johnson JE, Saito T. Commissural neuron identity is specified by a homeodomain protein, Mbh1, that is directly downstream of Math1. Development. 2005;132:2147–2155. doi: 10.1242/dev.01781. [DOI] [PubMed] [Google Scholar]
- 14.Saba R, Nakatsuji N, Saito T. Mammalian BarH1 confers commissural neuron identity on dorsal cells in the spinal cord. J Neurosci. 2003;23:1987–1991. doi: 10.1523/JNEUROSCI.23-06-01987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chellappa R, et al. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol Cell Biol. 2008;28:1905–1914. doi: 10.1128/MCB.01454-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Q, et al. BARHL2 differentially regulates the development of retinal amacrine and ganglion neurons. J Neurosci. 2009;29:3992–4003. doi: 10.1523/JNEUROSCI.5237-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 18.Sabatier C, et al. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell. 2004;117:157–169. doi: 10.1016/s0092-8674(04)00303-4. [DOI] [PubMed] [Google Scholar]
- 19.Bulfone A, et al. Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum Mol Genet. 2000;9:1443–1452. doi: 10.1093/hmg/9.9.1443. [DOI] [PubMed] [Google Scholar]
- 20.Kawauchi D, Saito T. Transcriptional cascade from Math1 to Mbh1 and Mbh2 is required for cerebellar granule cell differentiation. Dev Biol. 2008;322:345–354. doi: 10.1016/j.ydbio.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Mo Z, Li S, Yang X, Xiang M. Role of the Barhl2 homeobox gene in the specification of glycinergic amacrine cells. Development. 2004;131:1607–1618. doi: 10.1242/dev.01071. [DOI] [PubMed] [Google Scholar]
- 22.Sud R, Jones CM, Banfi S, Dawson SJ. Transcriptional regulation by Barhl1 and Brn-3c in organ-of-Corti-derived cell lines. Mol Brain Res. 2005;141:174–180. doi: 10.1016/j.molbrainres.2005.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.