Abstract
Filamin A (FLNA) is an actin-binding protein with a well-established role in the cytoskeleton, where it determines cell shape and locomotion by cross-linking actin filaments. Mutations in FLNA are associated with a wide range of genetic disorders. Here we demonstrate a unique role for FLNA as a nucleolar protein that associates with the RNA polymerase I (Pol I) transcription machinery to suppress rRNA gene transcription. We show that depletion of FLNA by siRNAs increased rRNA expression, rDNA promoter activity and cell proliferation. Immunodepletion of FLNA from nuclear extracts resulted in a decrease in rDNA promoter-driven transcription in vitro. FLNA coimmunoprecipitated with the Pol I components actin, TIF-IA, and RPA40, and their occupancy of the rDNA promoter was increased in the absence of FLNA in vivo. The FLNA actin-binding domain is essential for the suppression of rRNA expression and for inhibiting recruitment of the Pol I machinery to the rDNA promoter. These findings reveal an additional role for FLNA as a regulator of rRNA gene expression and have important implications for our understanding of the role of FLNA in human disease.
Filamin A (FLNA) is a well-characterized cytoskeletal protein that regulates cell shape and migration (1–3). FLNA cross-links actin and acts as an intracellular signaling scaffold by binding to a variety of signaling network components (4). Mutations in FLNA are associated with a wide range of human genetic disorders, including skeletal, craniofacial, and cardiovascular defects (4). FLNA consists of an N-terminal actin-binding domain and 24 Ig domains that form a rod-like structure (3). Dimerization occurs via the C-terminal Ig domain 24 to form a Y-shaped structure with two N-terminal actin-binding domains that serve to cross-link actin filaments. In addition to its well-characterized conventional role in the cytoplasm, FLNA also has a role in the nucleus (4). Both full-length FLNA and proteolytic fragments of FLNA have been shown to interact with nuclear proteins. A proteolytically derived C-terminal fragment of FLNA interacts with a small number of transcription factors (5, 6). A small amount of FLNA has been reported in the nucleus, where it participates in the DNA damage response by interacting with BRCA2 (7, 8).
The nucleolus is the site of ribosome biogenesis (9, 10). Transcription of the major rRNAs by RNA polymerase I (Pol I) occurs in the nucleolus (11, 12). Regulation of rRNA gene transcription is central to ribosome production and, therefore, to cell growth and proliferation. Transcription of rRNA genes is initiated at the rDNA promoter by a Pol I multiprotein complex. Regulation of transcription initiation is primarily determined by regulatory mechanisms that affect the assembly of this complex on the rDNA promoter. Moreover, signaling pathways that affect cell growth and proliferation are known to directly target components of the Pol I machinery. For example, both ERK and JNK signaling pathways phosphorylate the Pol I initiation factor TIF-IA to activate and inhibit transcription, respectively (13, 14).
While exploring the colocalization of FLNA with potential transcription factor partners, we noticed that FLNA was present in nucleoli. Here we show that FLNA is an abundant nucleolar protein that associates with the Pol I transcription machinery to suppress rRNA transcription. The FLNA actin-binding domain is essential for suppression of rRNA expression and to prevent recruitment of the Pol I machinery to the rDNA promoter. We propose a model in which FLNA inhibits rRNA transcription by inhibiting recruitment of the Pol I machinery to the rDNA promoter.
Results and Discussion
FLNA Is a Nucleolar Protein.
While investigating the nuclear localization of the C-terminal fragment of FLNA fused to GFP, we observed that FLNA was present in discrete bodies within the nucleus (Fig. 1A). Subsequent immunofluorescence staining for endogenous FLNA and the nucleolar marker protein fibrillarin demonstrated that FLNA is present in the nucleoli of ∼75% of SaOS-2 cells (Fig. 1B and Fig. S1E). FLNA is also present in the nucleoli of HeLa cells (Fig. S1A). Nucleolar staining was also observed in nucleoli of SaOS-2 cells with two different FLNA antibodies (Fig. S1B). Immunofluorescence confocal microscopy of primary bone marrow stromal cells also demonstrated that FLNA is a nucleolar protein (Fig. S1D). To confirm the immunofluorescence data, protein extracts were prepared from purified intact nucleoli and analyzed by Western blotting (Fig. 1C). This analysis confirmed that the full-length 280-kDa FLNA protein is present in the nucleolus (Fig. 1C). Immunofluorescence staining and Western blotting analysis were subsequently performed with purified nucleoli derived from an FLNA− cell line (M2) and its clonal derivative (A7), which stably express FLNA (15). FLNA was observed in nucleoli from FLNA+ cells (A7) but not from FLNA− cells (M2) (Fig. 1 D and E). We also tested the specificity of FLNA nucleolar localization by disrupting the integrity of the nucleoli with actinomycin D (AMD) (Fig. 1F). At the concentration used, AMD specifically inhibits Pol I transcription, and treatment of cells with AMD results in the segregation of nucleolar components to the nucleoplasm (16, 17). As expected, immunofluorescence analysis of SaOS-2 cells treated with AMD resulted in segregation of the nucleolar protein nucleophosmin (NPM) throughout the nucleoplasm (Fig. 1F) (18, 19). A similar analysis demonstrated that FLNA nucleolar retention was also lost upon treatment with AMD, whereas cytoplasmic FLNA appeared unaffected (Fig. 1F). This effect was observed in all cells containing nucleolar FLNA (Fig. S1E). Altogether, these data demonstrate that FLNA is a bona fide component of actively transcribing nucleoli. These findings are also supported by a large-scale mass spectrometry analysis of the human nucleolus in which peptides corresponding to FLNA were identified (Nucleolar Online Proteomics Database, http://www.lamondlab.com/NOPdb3.0).
Fig. 1.
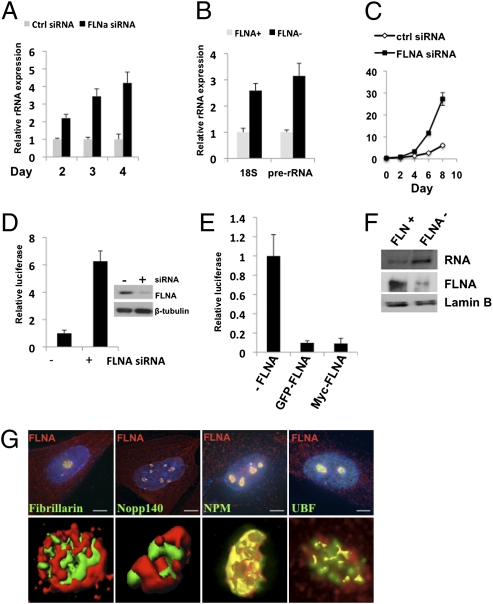
FLNA is a nucleolar protein. (A) Immunofluorescence microscopy of HeLa cells transfected with the C-terminal 100-kDa fragment of FLNA fused to GFP (FLNA-C-GFP). (Scale bars, 10 μm.) (B) Immunofluorescence microscopy showing colocalization of endogenous FLNA A (red) and the nucleolar marker fibrillarin (green) in SaOS-2 cells. (Scale bars, 10 μm.) (C Upper) Western blot analysis of total (T), nuclear (N) and nucleolar (No) protein fractions from SaOS-2 cells showing full-length FLNA in the nucleolus. (Lower) Immunofluorescence staining of the isolated nucleoli. (D) Immunofluorescence microscopy showing specific staining of FLNA (using mouse monoclonal antibody, MAB1678, Millipore) and fibrillarin in nucleoli isolated from FLNA+ (A7) cells. Fibrillarin was present in nucleoli from both cells lines, whereas FLNA was only present in A7 nucleoli. Some background FLNA staining was present in M2 cells. (Scale bars, 2 μm.) (E) Western blot analysis of total (T), nuclear (N) and nucleolar (No) protein fractions from FLNA+ (A7) cells showing full-length FLNA in the nucleolus. FLNA was not detectable in any of the fractions from FLNA− (M2) cells. (F) Nucleolar retention of FLNA depends on ongoing Pol I transcription. Immunofluorescence analysis of SaOS-2 cells treated with 50 ng/mL of the Pol I inhibitor AMD (Lower) or vehicle only (Upper). The nucleolar protein NPM and nucleolar FLNA redistribute throughout the nucleoplasm in the presence of AMD. Nuclei (blue) were stained with DAPI. (Scale bars, 10 μm.)
FLNA Suppresses Transcription of rRNA Genes.
We next investigated the functional role of FLNA in the nucleolus. Because the nucleolus is the center for rRNA production, we examined the expression of rRNA genes in the presence and absence of FLNA. SaOS-2 cells were transfected with siRNAs targeted against FLNA, and the amount of the 47S pre-rRNA was determined by quantitative RT-PCR (qRT-PCR) (Fig. 2A and Fig. S2A). A marked increase in the level of pre-rRNA was observed upon depletion of FLNA compared with control siRNA; indeed, the amount of pre-rRNA correlated inversely with the amount of FLNA in the cells (Fig. 2A). FLNA− (M2) cells also expressed significantly more pre-rRNA and the spliced 18S rRNA than did FLNA+ (A7) cells (Fig. 2B). Similar increases in rRNA expression were also observed in SaOS-2 and 293T cells transduced with lentiviral FLNA shRNAs (Fig. S2 C and D).
Fig. 2.
FLNA suppresses rRNA gene transcription. (A) qRT-PCR showing increased pre-rRNA expression in SaOS-2 cells after transfection with FLNA siRNAs. (B) qRT-PCR showing increased rRNA expression in the M2 cells (FLNA−) compared with A7 cells (FLNA+). (C) Growth curves of SaOS-2 cells after knockdown of FLNA. (D) The activity of a transiently transfected rRNA promoter reporter is stimulated after FLNA depletion by siRNAs in SaOS-2 cells. (Right) Immunoblots show FLNA after siRNA transfection. (E) Transient transfection of M2 cells (FLNA−) with FLNA-expressing plasmids inhibits the activity of the rRNA promoter. The data represent at least three independent experiments performed in triplicate; error bars represent SEM. (F) In vitro transcription assays showing increased transcription from the rRNA promoter reporter in HeLa nuclear extracts after immunodepletion of FLNA (top row). Immunoblots showing FLNA in control and FLNA-depleted HeLa nuclear extracts (middle row). (G) Subnucleolar localization of FLNA. (Upper) Delta Vision deconvolution microscopy of SaOS-2 cells labeled with antibodies against FLNA, shown in red, and the nucleolar proteins: fibrillarin and Nopp140 (DFC proteins), NPM (GC protein), and UBF (FC protein), shown in green. (Scale bars, 2 μm.) (Lower) Nucleolar 3D image reconstructions are shown. Yellow color in UBF and NPM 3D images represents colocalization with FLNA (in which green and red channels are merged). The fibrillarin and Nopp140 signals remained distinct from the FLNA signal.
Cell growth and proliferation depend highly on the availability of rRNAs for ribosome production and subsequent protein synthesis (11). Depletion of FLNA and the concomitant increase in rRNA expression might therefore lead to an increase in cell proliferation. To test this possibility, we compared the proliferation rates of SaOS-2 cells treated with FLNA siRNA and cells treated with control nonspecific siRNA. Cell counts revealed that the proliferation rate of FLNA siRNA-treated cells was significantly greater than that of control cells (Fig. 2C). Increases in cell proliferation were also observed in FLNA siRNA-treated 293T cells and M2 cells (Fig. S2 E and F). Thus, depletion of FLNA leads to an increase in rRNA expression and an increase in the proliferative capacity of the cells.
To determine whether the FLNA-mediated suppression of rRNA expression is caused by a direct effect on Pol I transcription, we initially examined the rDNA promoter activity by using a transfected promoter-reporter plasmid (Fig. 2D). Transcriptional activity was determined by qRT-PCR analysis of the luciferase transcript. Cotransfection of SaOS-2 cells with FLNA siRNAs and the rDNA promoter reporter resulted in a sixfold increase in promoter activity compared with control siRNA transfections (Fig. 2D). When FLNA− M2 cells were cotransfected with the rDNA promoter reporter and an FLNA expression plasmid, the promoter activity was inhibited 10-fold (Fig. 2E). To determine whether the suppressive effect of FLNA was caused by a direct effect on transcription, the activity of the rDNA promoter was analyzed in vitro with HeLa nuclear extracts depleted of FLNA by prior immunoprecipitation (Fig. 2F). When FLNA was depleted from the nuclear extracts, transcription from the rDNA promoter was significantly reduced (Fig. 2F). These findings suggested that the suppressive effect of FLNA is mediated by a direct effect on transcription. We therefore reasoned that FLNA should be present within the subnucleolar compartment where rDNA transcription occurs. The nucleolus is organized into three distinct subcompartments: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC) (10). The FC is enriched with Pol I subunits, and transcription of rDNA occurs primarily at the interface between the FC and DFC (10). Ribosome subunit assembly is completed in the GC, where most other proteins are found (10). SaOS-2 cells were stained with antibodies against FLNA and nucleolar proteins known to be predominantly present in each of the three subnucleolar compartments. Coimmunofluorescence and confocal microscopy revealed that FLNA colocalizes with NPM in the GC and with UBF in the FC but does not colocalize with the DFC proteins fibrillarin and Nopp140 (Fig. 2G). Thus, the presence of FLNA in the FC and GC suggests that it is likely to be involved in the transcription and processing of rRNA. Because a subpopulation of FLNA is present in the FC where transcription occurs, and FLNA can suppress the rDNA promoter as well as expression of the nascent unprocessed 47S rRNA, we conclude that at least one function of FLNA in the nucleolus is the suppression of rRNA transcription.
FLNA Suppresses Recruitment of the Pol I Machinery to the rDNA Promoter.
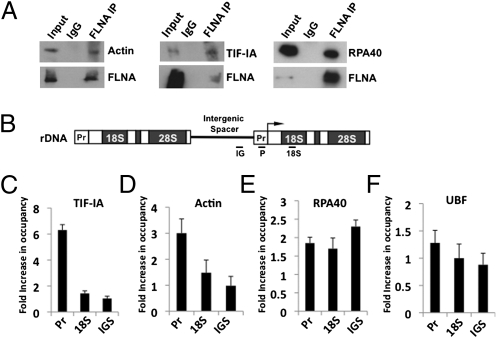
To gain insight into the mechanism by which FLNA suppresses rRNA transcription, we considered FLNA's actin-binding function in the context of transcription. We reasoned that, because FLNA is an actin-binding protein and that actin is an essential component of the Pol I transcription machinery (20, 21), FLNA's ability to suppress rDNA transcription might be mediated via an interaction with the Pol I machinery. Transcriptionally competent Pol I is associated with actin and the initiation factor TIF-IA, a component of the basal transcription machinery required to assemble initiation complexes (20, 21). Recruitment of the Pol I transcription machinery is also stimulated by the transcription factor UBF, which binds the upstream promoter region (11, 12). To determine whether FLNA is physically associated with components of the Pol I machinery, FLNA was immunoprecipitated and the samples were probed for the presence of actin, TIF-IA, RPA40 (a Pol I subunit), and UBF. Actin, TIF-IA, and RPA40 all coprecipitated with FLNA (Fig. 3A). In contrast, FLNA did not coprecipitate with UBF. These data demonstrate that FLNA is physically associated with the Pol I transcription machinery.
Fig. 3.
FLNA is associated with components of the RNA Pol I machinery. (A) Coimmunoprecipitation assays of endogenous FLNA from nuclear extracts showing that FLNA is associated with actin, TIF-IA, and RPA40. (B) rRNA gene organization. ChIP assay primers for IGS (IG), promoter (P), and rRNA coding region (18S) are shown. (C–F) ChIP assays showing differential recruitment of Pol I components to the rDNA in M2 and A7 cells. Chromatin was precipitated with antibodies against TIF-IA (C), actin (D), RPA40 (E), and UBF (F). The relative enrichment was determined by qRT-PCR. Fold occupancy was calculated by dividing fold enrichment in M2 cells by that in A7 cells. The data represent three independent experiments performed in triplicate; error bars represent SEM.
We next examined the role of FLNA in recruiting components of the Pol I machinery to the rDNA promoter. ChIP assays were performed on the rDNA promoter to establish the occupancy of TIF-IA, actin, RPA40, and UBF in FLNA− (M2) and FLNA+ cells (A7). Chromatin was immunoprecipitated from FLNA− and FLNA+ cells with TIF-IA, actin, RPA40, UBF, and nonspecific IgG antibodies; the rDNA was amplified with primers specific for the promoter, the 18S region, and the intergenic spacer (IGS) region (Fig. 3B). This analysis revealed a striking sixfold increase in the occupancy of the rDNA promoter by the initiation factor TIF-IA in the absence of FLNA (Fig. 3C). The increased recruitment of TIF-IA was specific for the promoter because we did not observe changes in its occupancy of the 18S and IGS regions. We also observed an increase in the occupancy of actin at the promoter in the absence of FLNA and an increase in RPA40 on all three regions of the rDNA (Fig. 3 D and E). In contrast, no significant increase in UBF occupancy was observed (Fig. 3F). Thus, in the absence of FLNA, the increased rRNA gene transcription correlates with an increased occupancy of the rDNA promoter by components of the Pol I transcription complex.
Suppression of Pol I Recruitment to the rDNA Promoter Depends on the FLNA Actin-Binding Domain.
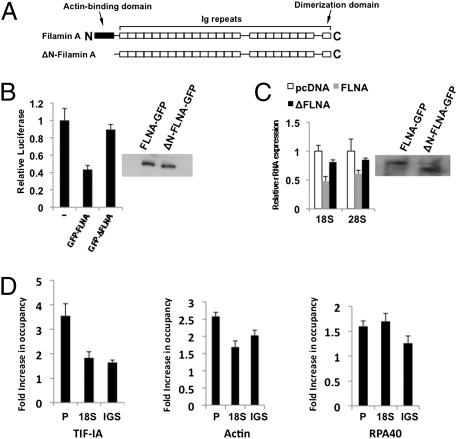
To determine whether the actin-binding function of FLNA is required for suppression of rRNA gene transcription, we deleted the well-characterized N-terminal actin-binding domain of FLNA and tested the resultant protein's ability to suppress rDNA promoter activity and rRNA expression (Fig. 4). When cells were cotransfected with the rDNA promoter reporter and FLNA lacking the actin-binding domain (ΔN-FLNA) (Fig. 4A), suppression of the promoter activity was significantly reduced compared with that of full-length FLNA (Fig. 4B). Moreover, the ability of ΔN-FLNA to suppress endogenous rRNA expression in FLNA− cells was also reduced compared with that of the intact protein (Fig. 4C). The transcriptional suppressive function of FLNA therefore depends on the presence of the actin-binding domain, suggesting that FLNA mediates its effects by interacting with actin.
Fig. 4.
The actin-binding domain of FLNA is required for suppression of Pol I recruitment to the rDNA promoter. (A) Domain structure of FLNA and its deletion derivative lacking the actin-binding domain (ΔN-filamin A). (B) qRT-PCR demonstrating that the FLNA actin-binding domain is required for suppression of the rDNA promoter activity. FLNA− cells (M2) were cotransfected with a promoter reporter and either FLNA-GFP or ΔFLNA-GFP (lacking the actin-binding domain). (C) qRT-PCR demonstrating that the FLNA actin-binding domain is required for suppression of rRNA expression. FLNA− (M2) cells were transfected with FLNA-GFP or ΔFLNA-GFP expression plasmids and isolated by FACS. All graphical data represent three independent experiments performed in duplicate; error bars represent SEM. (D) ChIP assays showing differential recruitment of Pol I components to the rDNA in the presence of FLNA-GFP and ΔFLNA-GFP FLNA. FLNA− (M2) cells were transfected with either FLNA-GFP– or ΔFLNA-GFP–expressing plasmids and isolated by FACS. The relative enrichment was determined by qRT-PCR. Fold occupancy was calculated by dividing fold enrichment in M2 cells transfected with ΔFLNA-GFP by that in M2 cells transfected with FLNA-GFP. The data represent three independent experiments performed in triplicate; error bars represent SEM.
We next determined whether the actin-binding domain of FLNA was required to inhibit recruitment of the Pol I machinery to the rDNA promoter. The occupancy of the rDNA promoter by components of the Pol I machinery was therefore compared in FLNA− cells transfected with either full-length FLNA or the actin-binding domain deletion, ΔN-FLNA (Fig. 4D). This analysis revealed that significantly more TIF-IA, actin, and RPA40 were observed on the rDNA promoter in cells expressing ΔN-FLNA than were observed in cells expressing FLNA (Fig. 4D). These data demonstrate that FLNA-mediated inhibition of the transcriptionally competent Pol I complex to the rDNA promoter depends on FLNA's actin-binding domain.
FLNA Contains a Nucleolar Localization Sequence (NoLS).
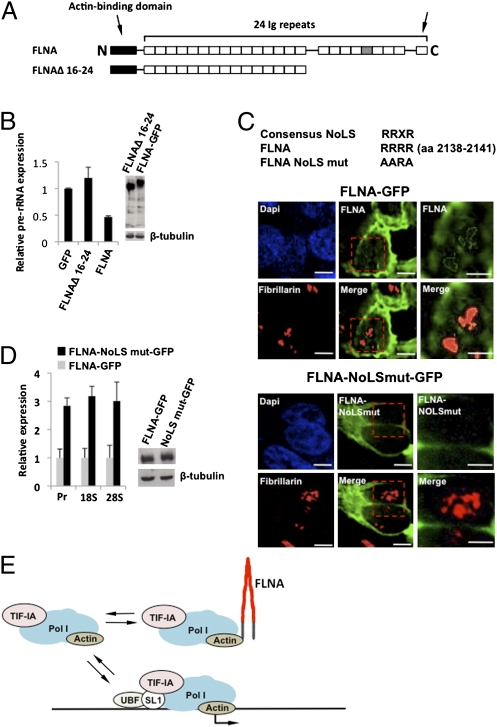
We next examined whether the C-terminal region of FLNA was required for rRNA expression. Deletion of the C-terminal Ig repeats 16–24 resulted in abrogation of FLNA's capacity to inhibit rRNA expression (Fig. 5B). Because we have established that the C-terminal fragment localizes to the nucleolus, we reasoned that this region might determine nucleolar localization of FLNA. Nucleolar retention of proteins is often determined by a NoLS, which is characterized by the consensus sequence RRXR and mediates nucleolar retention of the well-characterized nucleolar proteins such as C/EBPα, Parp2, p14/19Arf, ING1b, Rpp29, and HIV Tat (22). Examination of the primary sequence of FLNA revealed a highly conserved basic amino acid motif (RRRR) located in Ig repeat 20, within the C-terminal region of FLNA (Fig. 5 A and C). This motif corresponds to the consensus NoLS and is identical to that found in Parp2 and C/EBPα (22). To determine whether the RRRR motif in Ig repeat 20 is required for nucleolar retention of FLNA, we mutated the sequence in full-length FLNA, fused it to GFP, and examined its subcellular localization (Fig. 5C). In cells transfected with FLNA-GFP, we observed FLNA in the nucleolus, consistent with our finding that endogenous full-length FLNA resides in the nucleolus. In contrast, mutation of the RRRR motif to AARA abolished FLNA-GFP nucleolar localization (Fig. 5C). Moreover, this mutation abrogated the suppressive effect of FLNA on rRNA expression (Fig. 5D). The RRRR motif is therefore required for nucleolar retention of FLNA and thus its ability to suppress rRNA expression.
Fig. 5.
FLNA contains a nucleolar localization sequence in the C-terminal region. (A) Domain structure of FLNA depicting the C-terminal deletion (FLNAΔ 16–24). Ig repeat 20, containing the NoLS, is colored gray, the arrow on the right indicates the dimerization domain. (B) qRT-PCR demonstrating that the FLNA C-terminal region is required for suppression of rRNA expression. 293T cells stably transduced with FLNA siRNA (Fig. S2D) were transiently transfected with plasmids encoding the indicated proteins and isolated by flow cytometry before qRT-PCR analysis. (C) Microscopic analysis demonstrating the loss of nucleolar retention after mutation of the NoLS in FLNA. The NoLS motif in Ig repeat 20 of the human FLNA (RRRR) was mutated (AARA) in the full-length FLNA fused to GFP as depicted. 293T cells stably transduced with FLNA siRNA were transiently transfected with plasmids encoding the indicated proteins and isolated by flow cytometry before qRT-PCR analysis. For microscopic analysis, GFP and fibrillarin expression were detected at 24 h posttransfection. Images were obtained using a Delta Vision (Applied Precision) microscope. Higher magnifications of the red outlined boxes in Center are shown in Right. (Bars: 10 μm, Left and Center; 5 μm, Right.) (D) qRT-PCR demonstrating that mutation of the NoLS relieves the suppressive effect of FLNA on rRNA expression. 293T cells stably transduced with FLNA siRNA were transiently transfected with plasmids encoding the indicated proteins and isolated by flow cytometry before qRT-PCR analysis. (E) Model depicting the proposed mechanism by which FLNA suppresses rRNA transcription.
Altogether, our findings demonstrate that FLNA is targeted to the nucleolus via the NoLS in the C-terminal region. Within the nucleolus, the actin-binding domain is required to suppress rRNA gene transcription via a mechanism that involves binding to and inhibiting recruitment of the Pol I transcription machinery to the rDNA promoter. Our data support a model in which FLNA exerts its suppressive effect by binding to the Pol I/TIF-IA/actin complex via actin and inhibiting its recruitment to the promoter (Fig. 5E). Because the suppressive effect of FLNA does not completely inhibit rRNA transcription, the data are consistent with a mechanism in which FLNA binds to a population of transcriptionally competent Pol I and prevents its recruitment to the promoter. We suggest that the increase in rRNA gene transcription observed in the absence of FLNA reflects an in vivo mechanism by which relocalization of nucleolar FLNA, or modulation of its function, releases the Pol I complex, thus allowing an increase in rRNA gene transcription. How FLNA is involved in the dynamic regulation of Pol I transcription remains to be elucidated. However, we note that the NoLS we identified in Ig repeat 20 is part of the interaction interface with the adjacent Ig repeat 21 (23), suggesting that modulation of the interaction between Ig repeats 20 and 21, possibly by phosphorylation of nearby serine residues, regulates exposure of the nucleolar localization of FLNA. A similar mechanism has been proposed to expose the integrin-binding region in Ig repeat 21, which is masked by the intradomain interaction between Ig repeats 20 and 21 (23). Intriguingly, the related protein, filamin B (FLNB), lacks the RRRR motif, having RTSR at this position. Although we have not examined the nucleolar localization of FLNB, the absence of a recognizable NoLS suggests that it might be excluded from the nucleolus. However, because FLNB and FLNA can associate in vivo, it is possible that FLNA:FLNB heterodimers exist in the nucleolus (24). Further work is required to establish whether FLNB also has a role in the nucleolus.
Mutations in FLNA cause a wide range of developmental disorders in humans, and FLNA-deficient mice display a range of phenotypes, including cardiac, skeletal, and craniofacial defects. It is likely that this wide range of phenotypic defects reflects FLNA's many molecular functions, making it difficult to correlate a particular phenotypic disorder with a specific defect in FLNA function (4). However, we note that mutations in the Pol I regulator, treacle, cause craniofacial defects during development (25). It is therefore possible that the craniofacial defects observed in some FLNA patients may also arise as a consequence of altered Pol I function. Although further studies are required to fully understand the extent of the function of FLNA in the nucleolus, our present findings clearly demonstrate that, in addition to its roles as a cytoskeletal component and a signaling scaffold, it should also be considered as a regulator of Pol I transcription in the nucleolus.
Materials and Methods
Cell Culture and Microscopy.
SaOS-2 cells were grown in McCoy's 5A medium, M2 and A7 cells were gown in RPMI medium 1640, and 293T were gown in DMEM; all complete media were supplemented with 10% FBS and antibiotics. Antibodies for FLNA, TIF-IA, UBF, actin, Lamin B, and β-tubulin were obtained from Abcam unless otherwise stated. The GFP antibody was obtained from Invitrogen, and the RPA40 antibody came from Santa Cruz Biotechnology. IgG was obtained from Sigma-Aldrich. Immunofluorescence staining was performed as described previously (26). Images were processed and analyzed with ImageJ (http://rsb.info.nih.gov/ij). For confocal analysis, images were processed with Zeiss software. For deconvolution analysis, images were deconvolved with 5–10 iterations and prefilter cutoff values (microns) of 0.05. 3D images were obtained with Imaris (Bitplane) software.
Cell Fractionation.
Nuclear and cytoplasmic fractions were purified with the Nuclei EZ Prep Nuclei Isolation Kit (Sigma). Nuclei were subsequently purified by using a dense sucrose cushion (27). Nucleoli were isolated as described in ref. 28. Immunoprecipitation assays were performed as described previously (29).
Plasmids.
The rDNA promoter was amplified from human genomic DNA by PCR and ligated into pGL3 basic (Promega).
RNA Interference and Analysis of Gene Expression.
FLNA siRNA and control siRNAs were obtained from Santa Cruz Biotechnology. SaOS-2 cells were transiently transfected with siRNAs using Oligofectamine Transfection Reagent. Total RNA was prepared from the cells with RNeasy kit (Qiagen). rRNA gene expression was analyzed by qRT-PCR. For reporter gene assays, siRNAs and reporter gene vector were cotransfected with Lipofectamine 2000 transfection reagent. Lentivirus particles of FLNA siRNA and control siRNA (Santa Cruz Biotechnology) were used to generate SaOS-2 and 293T stable cell lines according to the manufacturer's protocol. For analysis of the NoLS mutant, we used a similar approach to that of Wedel et al., whereby overexpression of wild-type proteins via the CMV-driven promoter is sufficient to overcome the silencing effect of the siRNA (30). 293T cells stably transduced with FLNA siRNA (Fig. S2D) were transiently transfected with plasmids encoding FLNA-GFP proteins using Lipofectamine. Cells in which GFP proteins were expressed to sufficiently high levels to overcome the siRNA silencing were isolated by flow cytometry. M2 cells were transiently transfected with expression plasmids using Lipofectamine. After cell sorting by flow cytometry, the expression of FLNA was detected by immunoblotting using an anti-GFP antibody.
In Vitro Transcription Assays.
In vitro transcription assays were performed with HeLa cell nuclear extracts as described previously (31). Reporter vector pGL3-RGP was used as template for transcription. FLNA-depleted nuclear extracts were prepared by using an FLNA antibody-coupled Sepharose column.
ChIP Assays.
ChIP assays were performed as described previously (32). ChIP quantitative PCR was performed with a Bio-Rad Real-Time PCR Detection System.
Cell Proliferation.
SaOS-2 and 293T siRNA-expressing cell lines were used to analyze the effect of FLNA knockdown on cell proliferation. The stable cells were diluted 1/16 and grown in 6-well plates. The cells were harvested and manually counted at different time points.
Supplementary Material
Acknowledgments
We are grateful to Rakesh Kumar for the pcDNA3.1-FLNa-1795-2647 plasmid and Tom Stossel for cell lines. This work was funded by Association for International Cancer Research Grant 09-0208 (to P.S.). C.L.-C. and D.M.-V. are recipients of Mexican National Council for Science and Technology (CONACyT) doctoral scholarships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.H.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107879109/-/DCSupplemental.
References
- 1.Stossel TP, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 2.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: Promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura F, Stossel TP, Hartwig JH. The filamins: Organizers of cell structure and function. Cell Adh Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou AX, Hartwig JH, Akyürek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113–123. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Berry FB, O'Neill MA, Coca-Prados M, Walter MA. FOXC1 transcriptional regulatory activity is impaired by PBX1 in a filamin A-mediated manner. Mol Cell Biol. 2005;25:1415–1424. doi: 10.1128/MCB.25.4.1415-1424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. A 90 kDa fragment of filamin A promotes Casodex-induced growth inhibition in Casodex-resistant androgen receptor positive C4-2 prostate cancer cells. Oncogene. 2007;26:6061–6070. doi: 10.1038/sj.onc.1210435. [DOI] [PubMed] [Google Scholar]
- 7.Yuan Y, Shen Z. Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J Biol Chem. 2001;276:48318–48324. doi: 10.1074/jbc.M102557200. [DOI] [PubMed] [Google Scholar]
- 8.Yue J, et al. The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 2009;69:7978–7985. doi: 10.1158/0008-5472.CAN-09-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson MO, Hingorani K, Szebeni A. Conventional and nonconventional roles of the nucleolus. Int Rev Cytol. 2002;219:199–266. doi: 10.1016/S0074-7696(02)19014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 11.Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, Yuan X, Frödin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 14.Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19:933–941. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham CC, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 16.Puvion-Dutilleul F, Puvion E, Bachellerie JP. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with a 5′ETS leader probe. Chromosoma. 1997;105:496–505. doi: 10.1007/BF02510486. [DOI] [PubMed] [Google Scholar]
- 17.Grummt I. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 18.Shav-Tal Y, et al. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smetana K, Busch R, Chan PK, Smetana K, Jr, Busch H. Immunocytochemical localization of nucleophosmin and RH-II/Gu protein in nucleoli of HeLa cells after treatment with actinomycin D. Acta Histochem. 2001;103:325–333. doi: 10.1078/0065-1281-00598. [DOI] [PubMed] [Google Scholar]
- 20.Philimonenko VV, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 21.Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008;22:322–330. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller C, Bremer A, Schreiber S, Eichwald S, Calkhoven CF. Nucleolar retention of a translational C/EBPα isoform stimulates rDNA transcription and cell size. EMBO J. 2010;29:897–909. doi: 10.1038/emboj.2009.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lad Y, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26:3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheen VL, et al. Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum Mol Genet. 2002;11:2845–2854. doi: 10.1093/hmg/11.23.2845. [DOI] [PubMed] [Google Scholar]
- 25.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin C, et al. Lamin B1 maintains the functional plasticity of nucleoli. J Cell Sci. 2009;122:1551–1562. doi: 10.1242/jcs.046284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg ME, Bender TP. Current Protocols in Molecular Biology. New York: Wiley; 2007. Identification of newly transcribed RNA. Chap 4, Unit 4.10. [DOI] [PubMed] [Google Scholar]
- 28.Lam YW, Lammond AI. Isolation of nucleoli. In: Celis J, et al., editors. Cell Biology, A Laboratory Handbook. 3rd Ed. San Diego: Academic; 2006. pp. 103–107. [Google Scholar]
- 29.Mendoza-Villanueva D, Deng W, Lopez-Camacho C, Shore P. The Runx transcriptional co-activator, CBFβ, is essential for invasion of breast cancer cells. Mol Cancer. 2010;9:171. doi: 10.1186/1476-4598-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wedel B, Boyles RR, Putney JW, Jr, Bird GS. Role of the store-operated calcium entry proteins Stim1 and Orai1 in muscarinic cholinergic receptor-stimulated calcium oscillations in human embryonic kidney cells. J Physiol. 2007;579:679–689. doi: 10.1113/jphysiol.2006.125641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng W, Malecová B, Oelgeschläger T, Roberts SG. TFIIB recognition elements control the TFIIA-NC2 axis in transcriptional regulation. Mol Cell Biol. 2009;29:1389–1400. doi: 10.1128/MCB.01346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.