Abstract
Floral organs are specified by the combinatorial action of MADS-domain transcription factors, yet the mechanisms by which MADS-domain proteins activate or repress the expression of their target genes and the nature of their cofactors are still largely unknown. Here, we show using affinity purification and mass spectrometry that five major floral homeotic MADS-domain proteins (AP1, AP3, PI, AG, and SEP3) interact in floral tissues as proposed in the “floral quartet” model. In vitro studies confirmed a flexible composition of MADS-domain protein complexes depending on relative protein concentrations and DNA sequence. In situ bimolecular fluorescent complementation assays demonstrate that MADS-domain proteins interact during meristematic stages of flower development. By applying a targeted proteomics approach we were able to establish a MADS-domain protein interactome that strongly supports a mechanistic link between MADS-domain proteins and chromatin remodeling factors. Furthermore, members of other transcription factor families were identified as interaction partners of floral MADS-domain proteins suggesting various specific combinatorial modes of action.
Keywords: protein complex isolation, transcriptional regulation, chromatin activation, histone marks
Flower development is one of the best understood developmental processes in plants. According to the classic ABC model (1), floral organs in the model plant species Arabidopsis are specified by the combinatorial activity of three functional gene classes. The A class genes represented by APETALA1 (AP1) and APETALA2 (AP2) specify sepal identity, and together with B class genes APETALA3 (AP3) and PISTILLATA (PI), they determine the identity of petals. The C class gene AGAMOUS (AG) alone determines carpel identity and, together with B class genes, it specifies stamen identity. The ABC model was extended to the ABCE model, in which E class genes [SEPALLATA1-4 (SEP1-4)] are required for the specification of all four types of floral organs (2, 3). Based on genetic and yeast n-hybrid protein interaction data, it was later proposed in the “floral quartet model” that floral organs are specified by combinatorial protein interactions of ABCE-class MADS-domain transcription factors, which are thought to assemble into organ-specific quaternary protein complexes that bind to two CArG boxes, DNA consensus sequence CC[A/T]6GG, in regulatory regions of target genes (4, 5). E-class proteins have a special role in this model as major mediators of higher-order complex formation. Although interactions that were predicted in this model were further supported by additional in vitro DNA-binding assays and protoplast FRET-FLIM experiments (6–8), formation and composition of these complexes in endogenous tissues remained unknown.
Heterologous interaction studies in yeast and genetic data suggest recruitment of transcriptional coregulators such as SEUSS (SEU) and LEUNIG (LUG) by floral MADS-domain proteins (9). Ovule-specific MADS-domain protein complexes were found to form higher-order interactions with BELL1 (BEL1), a member of the homeobox family of transcription factors in a yeast-based screen (10). Also, interactions between other plant MADS-domain proteins and proteins that are functionally analogous to polycomb group (PcG) proteins, as well as putative components of histone-deacetylase complexes, have been reported, suggesting these types of interactions play a role in the activity of the transcriptional regulatory complexes (11–13). Unraveling the in planta interactome of floral homeotic MADS-domain proteins could, therefore, advance our understanding of the mechanism and specificity underlying target gene regulation by these proteins.
In this study, we identified MADS-domain protein complexes by immunoprecipitation, followed by mass spectrometry (MS) and label-free quantification. Our results indicate that MADS-domain proteins interact not only with each other but also with non-MADS transcriptional regulators. Chromatin remodeling and modifying factors represent the most prominent group among these interactors.
Results
Floral Homeotic MADS-Domain Proteins Interact in Floral Tissues.
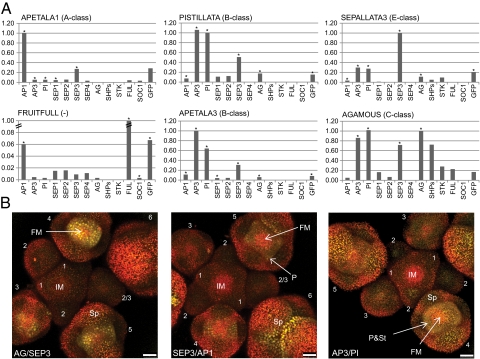
To analyze interactions of floral homeotic MADS-domain proteins in floral tissues, we made use of transgenic plant lines that express the MADS-domain proteins AP1, AG, AP3, and SEP3 from their native promoters linked to GFP as C-terminal fusions (14, 15). Protein complexes were isolated by immunoprecipitation using anti-GFP antibodies and characterized by LC-MS/MS, followed by label-free protein quantification analysis. This approach allowed us to identify proteins that were enriched in immunoprecipitation (IP) samples compared with control samples and provide an approximation of their relative abundance. Our results confirmed all major protein interactions proposed in the floral quartet model (Fig. 1A). We identified the class B floral homeotic proteins AP3 and PI as major interaction partners of each other and found them in similar abundance in the IP samples. Also, putative higher-order complex partners, such as SEP3 (E-class), AP1 (A-class), and AG (C-class) were identified as interaction partners of AP3 and PI. SEP3, which acts as a major mediator of higher-order complex formation (6), appears to be the most abundant interaction partner of class B proteins. SEP3 was also identified as interaction partner of AP1 and AG, whereas its paralog SEP4 was only detected as interaction partner of AP1 and FRUITFULL (FUL), supporting its predominant role in MADS-domain protein complexes that act during floral initiation and sepal development (3).
Fig. 1.
In planta MADS-domain protein interactions. (A) Average MADS-domain protein abundance ratios between the IP samples and the control samples scaled to the ratio of the bait protein. Ratios calculated based on 4–5 most abundant and unique peptides of a particular protein identified by LC-MS/MS are marked with an asterisk. Ratios calculated based on three or fewer identified peptides were not marked. (B) 3D maximum projections of in situ BiFC data using MADS-domain proteins expressed from their own promoters, confirming the interactions between MADS-domain proteins in floral meristems. Left, pAG:AG-eYFP/N + pSEP3:SEP3-eYFP/C. The yellow spots are characteristic of the nuclear localized interaction signal. The signal is positioned in the FM center where stamens and carpels will arise. Center, pSEP3:SEP3-eYFP/N + pAP1:AP1-eYFP/C. Most YFP signal is located in sepal tips and at the edges of the FM from where petals will be formed. Right, pAP3:AP3-eYFP/N + pPI:PI-eYFP/C. Weak YFP signal is found in the meristematic domain giving rise to petal and stamens, which is characteristic for PI and AP3 protein expression patterns (Fig. S1 J and K). 1–6, flower bud stages; FM, flower meristem; IM, inflorescence meristem; P, petal initiation site; Sp, sepal; St, stamen initiation site. (Scale bars, 25 μm.)
Using SEP3-GFP as bait, fruit- and ovule-specific MADS-domain proteins, namely SHATTERPROOF1,2 (SHP1,2) and SEEDSTICK (STK), were identified in addition to the ABC floral homeotic protein classes. This supports the proposed role of higher-order MADS-domain protein complexes in ovule identity specification, referred to as “D-class” function (16). Stamen and carpel complex partners, such as AG and B-class proteins, were more strongly represented than AP1 when using SEP3 as bait. This could reflect the abundance of certain complexes in the inflorescence tissues that were sampled, where the largest relative amount of tissue corresponds to later stages of floral organ differentiation. In the AG-GFP IP, an almost equal amount of AP3/PI and AG proteins were enriched, although one should expect less AP3/PI interacting with AG because of the formation of the carpel identity complexes. This could reflect differences in complex stability, tissue sampling, efficiency of elution from the bait protein in the IP, or estimation of protein levels.
Using AP1-GFP as bait, we also identified a lowly abundant interaction with SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which is a major regulator of floral transition (17). In addition, SOC1 was identified as a major interaction partner of FUL, supporting the existence of a FUL/SOC1 protein complex acting in floral transition. Using FUL-GFP as bait, we also identified several floral homeotic proteins, in particular AP1 and SEP proteins, as FUL interaction partners. In contrast to endogenous FUL, some expression of the FUL-GFP transgene has been observed in stages 1 and 2 floral buds, and later in whorl 2 and 3 (14), which might explain the observed interactions of FUL and floral MADS-domain proteins. Remarkably, using AP1 or FUL as bait, most tagged protein appeared not to be present in a heteromeric complex, because interaction partners are far less abundant than the bait protein (Fig. 1A). This could reflect the presence of these proteins in a homodimeric or monomeric form or a low stability of heteromeric complexes for these proteins during the biochemical isolation procedure. Although AP1 and FUL fusions to GFP can complement the respective mutant phenotypes (14, 18), it remains possible that the level of transgenic AP1- and FUL-GFP is elevated or stabilized compared with that of endogenous protein. This could potentially result in an overrepresentation of these proteins relative to their interaction partners in these transgenic lines.
To obtain detailed spatial information on in planta interactions of MADS-domain proteins, we applied the bimolecular fluorescence complementation (BiFC) assay (19) using MADS-domain proteins expressed from their endogenous promoter and fused to either the N-terminal or C-terminal half of eYFP. Using this method, we confirmed the interactions of SEP3 and AG, SEP3 and AP1, and AP3 and PI in floral meristems (Fig. 1B and Fig. S1 A–I). The interactions were mainly detected at stages of meristem development when floral organ identities are initially specified. Whereas AG/SEP3 and AP1/SEP3 heterodimers showed preferentially nuclear localization, the AP3/PI heterodimer shows an even distribution throughout the whole cell.
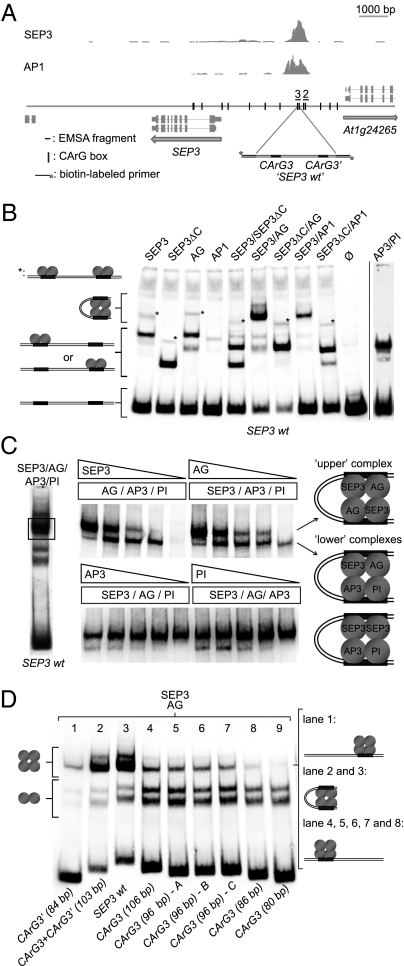
Formation of Quaternary MADS-Domain Protein Complexes on the DNA.
It is still not well understood how heteromeric higher-order MADS-domain protein complexes assemble and associate with their target DNA. To date, only DNA-binding homotetrameric and quartet-like complexes consisting of a SEP3 homodimer and AP3/PI heterodimer have been reconstituted in vitro (7, 8).
We identified a regulatory region in the SEP3 promoter that was bound in planta by several MADS-domain proteins, such as AP1, SEP3, FUL, and AG (Fig. 2A and Fig. S2 A and B) (20, 21), and chose this region to study the DNA-binding of higher-order MADS-domain protein complexes. The distal SEP3 promoter region between −2.6 and −3.1 kb containing these MADS transcription factor binding sites is required for the positive autoregulation of SEP3 in an inducible system and triggers enhancement of expression in floral tissues (Fig. S2 C–G), and is also bound by AP1 during early floral meristematic stages (21).
Fig. 2.
Assembly of the MADS-domain protein complexes in a distal region of the SEP3 promoter. (A) Graphic representation of the SEP3 locus with a 4.1-kb promoter region and the SEP3 and AP1 ChIP-SEQ profiles. Fragments used in the EMSA experiments were flanked with the biotin primers used for amplification and detection. Vertical lines in the sequence map indicate position of the CArG boxes. (B) EMSA of the different MADS-domain protein complexes with the SEP3 wild-type promoter fragment and possible model representations of formed protein–DNA complexes. (C) Left, EMSA of the SEP3/AG/AP3/PI protein mix with the “SEP3 wt” DNA fragment containing two CArG boxes. Center, EMSAs where the concentration of a single protein component was gradually reduced from approximately equimolar amounts to 0. Only the part of the gel containing the slow migrating complexes (rectangle in the left EMSA) is shown. Right, Model representation of the higher-order protein complexes formed in the presence of SEP3, AG, AP3, and PI binding to the SEP3 promoter fragment in vitro. (D) EMSA of the SEP3/AG protein mix with the truncated versions of the SEP3 wild-type DNA fragments. The SEP3 wt fragment was shortened from both 3′ and 5′ ends and contains either a single or double binding site. CArG3 (96 bp) – A, CArG3 (96 bp) – B, and CArG3 (96 bp) – C are different, randomized versions of the 3′-end flanking region of the CArG3 fragment.
Two pairs of CArG boxes (pairs named “2” and “3”) were identified to be located closest to the site of maximum ChIP enrichment (Fig. 2A), of which CArG box pair “3” showed the strongest binding of MADS-domain proteins in vitro (Fig. 2B and Fig. S3A). We choose fragment “3” containing a CArG box pair (CArG 3 and CArG 3′) for further analysis. AG, SEP3, and AP1 proteins bind as homodimers to this sequence, as does the AP3/PI heterodimer (Fig. 2B). When the SEP3 protein was incubated with either AG or AP1, we observed the predominant formation of DNA-binding heteromeric higher-order complexes, which were abolished when using a truncated SEP3 protein (SEP3ΔC) missing the C terminus and the last α-helix of the K-domain that is involved in higher-order complex formation (Fig. 2B). Weak bands corresponding to higher-order complexes were visible in the presence of either SEP3, SEP3ΔC, or AG protein (marked with asterisks in Fig. 2B), which could arise from two MADS dimers binding separately to two DNA-binding sites on this probe. Next, we analyzed the DNA binding of heteromeric higher-order complexes consisting of SEP3, AP3, and PI together with either AP1 (petal specification) or AG (stamen specification). We noticed that two bands were present in the shift corresponding to tetrameric complexes, indicating that at least two different higher-order complexes can potentially be formed on this DNA sequence in the presence of four different MADS-domain proteins (Fig. 2C). The composition of these complexes was analyzed by protein titration experiments. The results show that SEP3 is required for both complexes to form. AG is present in the upper complex, whereas the lower band may correspond to two complexes, one with and one without the AG protein. AP3 and PI proteins are present in the lower complexes because decrease of their concentrations affects only these complexes. When all proteins are present in similar concentrations, the stronger DNA-binding SEP3/AG/SEP3/AG and the weaker SEP3/AG/AP3/PI and/or SEP3/SEP3/AP3/PI complexes are formed (Fig. 2C). Furthermore, we also observed formation of higher-order complexes consisting of SEP3 and AP1, as well as SEP3, AP1, AP3, and PI, on this SEP3 promoter element (Fig. S3B). These results suggest that MADS-domain protein complexes with different composition can coexist within a cell and may compete for interaction partners and DNA-binding sites.
To evaluate the roles of the two CArG boxes in recruiting higher-order complexes, we generated DNA probes where the sequence was gradually shortened. We found that the presence of only one CArG box was sufficient to recruit heteromeric higher-order MADS-domain protein complexes; however, a minimum length of DNA sequence is required (in this case ∼85 bp) (Fig. 2D). This result indicates that additional non-sequence-specific DNA contacts stabilize binding of higher-order complexes to the DNA in the presence of only one CArG box, which supports and extends a previous finding (8).
MADS-Domain Proteins Act Together with Nucleosome Remodelers and Other Transcriptional Regulators.
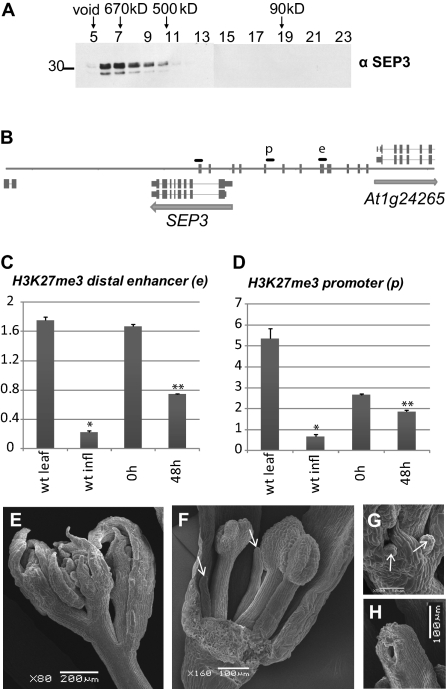
Gel-filtration experiments performed on nuclear protein extract demonstrated that SEP3 is part of a large protein complexes of around 670 kDa, which is far beyond the molecular mass of a MADS heterotetramer (Fig. 3A). Therefore, we analyzed which non-MADS proteins were enriched in the nuclear MADS immunoprecipitates by LC-MS/MS and label-free quantification (Datasets S1 and S2). Among proteins that were consistently enriched in the IP datasets of all MADS-domain proteins, we found several classes of nucleosome-remodeling factors, as well as RELATIVE OF EARLY FLOWERING 6 (REF6), recently characterized as histone H3K27 demethylase (22) (Table 1). This suggests that MADS-domain proteins can recruit or redirect the basic chromatin remodeling machinery to modulate the promoter structure of their target genes. Selected interactions were confirmed by reciprocal complex isolation and coimmunoprecipitation (Fig. S4). The notion that MADS-domain transcription factors recruit the nucleosome remodeling machinery to target gene promoters via more flexible, but in some cases less stable interactions are supported by the finding that interactions of PI with CHROMATIN REMODELING 4 (CHR4) and CHR11/17 are stabilized by the presence of DNA (Fig. S4).
Fig. 3.
Interactions between MADS-domain transcription factors and other transcriptional regulators. (A) Gel filtration reveals that SEP3 is present in large nuclear complexes. (B) SEP3 promoter and genomic locus representation with the quantitative PCR fragments in the distal enhancer site (e) and weaker proximal promoter site (p). Fragments were designed according to ChIP-SEQ profiles of AP1 and SEP3 (see Fig. 2A). Vertical bars indicate CArG box sequences. (C) Enrichment analysis of H3K27me3 at the MADS binding site in the distal SEP3 enhancer (e). ChIP was analyzed by quantitative PCR; material was obtained from inflorescence tissue of 35S:AP1-GR ap1 cal before (0 h) or 48 h after dexamethasone treatment and then subjected to ChIP with antibodies specific to H3K27me3. Results are presented as fold enrichment of input chromatin. Graphs represent average values from triplicates. Error bars represent SE of the mean. Asterisks indicate values that are significantly different from wild-type leaves (*) or from untreated 35S:AP1-GR ap1 cal plants (**) (P < 0.05 using Student t test). (D) Enrichment analysis of H3K27me3 in the proximal SEP3 promoter (p). For both C and D, H3K27me3 signal is reduced 48 h after AP1 induction compared with signal in 35S:AP1-GR ap1 cal uninduced tissues. (E–H) Scanning electron microscopy (SEM) pictures of chr11 chr17 double mutant inflorescences. (E) Overview of an inflorescence showing aberrations in floral organ development. (F) Close-up of a dissected chr11 chr17 flower (sepal in front was removed) with malformed stamens and petals replaced by pin-like structures (see arrow). (G) Close-up of a developing chr11 chr17 flower showing outgrowth of pin-like structures that replace the petals. (H) Incompletely closed carpel.
Table 1.
List of potential interaction partners enriched in the MADS-GFP IP experiments
| AG-GFP IP |
AP3-GFP IP |
PI-GFP IP |
SEP3-GFP IP |
AP1-GFP IP |
||||||
| Protein name | Log2 ratio | Peptide number | Log2 ratio | Peptide number | Log2 ratio | Peptide number | Log2 ratio | Peptide number | Log2 ratio | Peptide number |
| Nucleosome-associated factors | ||||||||||
| PKL | — | — | — | — | — | — | 2.67 | 4 | — | — |
| CHR4 | 3.12 | 8 | 1.78 | 5 | 2.27 | 5 | 3.47 | 7 | 3.82 | 14 |
| SYD | 0.71 | 2 | — | — | — | — | 1.17 | 2 | 3.1 | 5 |
| BRM | 0.65 | 2 | 0.17 | 2 | — | — | 1.05 | 3 | 2.51 | 4 |
| CHR11 | 2.32 | 19 | 2.09 | 17 | 1.79 | 17 | 2.32 | 19 | 3.38 | 25 |
| CHR17 | 2.8 | 17 | 1.38 | 19 | 2.71 | 16 | 2.29 | 19 | 3.67 | 24 |
| INO80 | 1.06 | 2 | — | — | — | — | — | — | 3.43 | 7 |
| REF6 | 2.34 | 4 | 0.87 | 3 | — | — | 3.22 | 2 | 3.55 | 5 |
| General transcriptional coregulators | ||||||||||
| LUH | — | — | — | — | — | — | 2.93 | 2 | 5.62 | 6 |
| SEU | 0.28 | 2 | — | — | — | — | — | — | 1.61 | 2 |
| Transcription factors | ||||||||||
| KNAT3 | — | — | — | — | — | — | — | — | 4.29 | 2 |
| BLH1 | — | — | — | — | — | — | — | — | 2.34 | 3 |
| BLR | — | — | — | — | — | — | — | — | 1.69 | 3 |
| ARF2 | — | — | — | — | — | — | — | — | 2.28 | 3 |
| SPL8 | 3.04 | 3 | — | — | — | — | 0.78 | 2 | 1.69 | 2 |
All protein enrichment values (log2 ratio) that showed significant differences at False Discovery Rate (FDR) 0.01, except for the AP3-GFP IP, where the FDR threshold was 0.05 because of the higher variability within samples and controls, are bolded. For the results of the detailed statistical analysis with the Student's t-test P values, see Dataset S2.
We also identified previously characterized interaction partners of MADS-domain proteins, the transcriptional coregulator SEU, as well as its interaction partner LEUNIG-HOMOLOG (LUH) (9) (Table 1). Next to basic transcriptional regulators, we identified members of several other transcription factor families as potential MADS interaction partners. AUXIN-RESPONSE FACTOR 2 (ARF2) and SQUAMOSA PROMOTER BINDING PROTEIN LIKE 8 (SPL8) were among the proteins that were enriched in AP1 (and AG) IP samples (Table 1). Analysis of the ChIP-SEQ data of AP1 identified the enrichment of the ARF binding motif (Fig. S5), which is also enriched in SEP3 ChIP-SEQ peaks (20). In addition, we found that the DNA-binding motif of SPL8 (23) was enriched in the AP1 and SEP3 ChIP-SEQ peaks, suggesting that they assemble into complexes that bind to nearby sites in the same genomic region (Fig. S5).
Also, the homeodomain transcription factors BELLRINGER (BLR), KNOTTED-LIKE 3 (KNAT3), and BEL1-LIKE HOMEODOMAIN 1 (BLH1) were identified as complex partners of AP1. Because interactions between BELL-like and KNOTTED-like proteins have been found in yeast two-hybrid (Y2H) experiments (24), our data suggest the formation of larger complexes consisting of MADS and homeodomain transcription factors. Targeted yeast three-hybrid (Y3H) experiments with a selected set of MADS-domain protein dimers revealed that mainly KNAT3, and, to a lesser extent, BLR and BLH1, is found as a direct interaction partner of floral MADS dimers AP1/SEP4 and AP1/SEP3 (Fig. S6A).
Based on genetic data, BLR was previously shown to regulate meristem maintenance, as well as internode, flower, and fruit development (25, 26). Together with the closely related factor POUND-FOOLISH (PNF), it controls floral evocation by regulation of LEAFY (LFY), AP1, and other factors (27). BLR also represses AG in floral and inflorescence meristems, acting synergistically with the general corepressors LUG and SEU (25). Because of the related functions of BLR and AP1 and their coexpression in floral meristems, we used targeted ChIP of BLR on selected AP1 binding sites to test whether AP1 and BLR may regulate flower initiation by binding to common sites in the genome, possibly as part of a protein complex. Indeed, we found that BLR and AP1 binding sites overlap in the regulatory regions of several genes that control floral transition and meristem specification such as the LFY, AP1, AP2, and TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 (TOE1) [at least threefold enrichment of BLR-GFP ChIP in 7 out of 11 tested AP1-bound regions (21)] (Fig. S6C). We also confirmed the interaction of BLR and AP1 by protein complex isolation experiments using BLR as a bait (Fig. S6B).
Next, we analyzed the expression patterns of plants expressing promoter:gene-GFP fusions of several potential MADS interactors. All showed expression in developing flower meristems or at later stages of flower differentiation (Fig. S6D). The nucleosome remodelers BRAHMA (BRM) and CHR17, as well as REF6 and the other chromatin-associated proteins, are broadly expressed throughout floral meristems, suggesting that they achieve their functional specificity through recruitment to target gene promoters by transcription factors, such as MADS-domain proteins.
Biological Roles of Interactions Between MADS-Domain Proteins and Chromatin-Associated Factors.
We identified the H3K27me3 demethylase REF6, as well as nucleosome remodelers, as protein complex partners of floral MADS-domain proteins, suggesting that MADS-domain proteins regulate transcription by modulating chromatin structure and accessibility. We, therefore, tested local H3K27me3 distribution at DNA regions bound by MADS-domain proteins, using the SEP3 genomic locus as an example (Fig. 3B). We studied the H3K27me3 distribution at the SEP3 promoter and genomic loci before and after induction of the AP1-GR fusion protein in ap1 cal background. SEP3 is one of the earliest genes directly activated by AP1, first weakly 8 h after AP1 induction and more strongly after 2 d (21, 28). Surprisingly, no change in H3K27me3 status associated with gene activation is detectable within the first SEP3 intron (Fig. S7), whereas in contrast, we observed a clear reduction in the level of H3K27me3 in the distal enhancer element and less pronounced in the proximal promoter (Fig. 3 C and D). These results suggest that AP1-mediated activation of SEP3 is (initially) associated with removal of H3K27me3 in the SEP3 promoter. Because SEP3 is also a target of the H3K27me3 demethylase REF6 (22), which is an interaction partner of AP1, it is tempting to speculate that AP1 can redirect or enhance REF6 activity at the SEP3 promoter.
The functions of SWI/SNF-type chromatin remodelers BRM and SPLAYED (SYD), as well as the CHD-type remodeler PICKLE (PKL), in the regulation of flower and carpel development have been characterized previously (29–33). In contrast, no flower-specific functions of the ISWI-type nucleosome remodelers CHR11 and CHR17 have been described so far. We, therefore, investigated flower phenotypes of chr11 chr17 double mutants and found pleiotropic phenotypic alterations: sepals were abnormally curled and longer comparing to other organs (Fig. 3E), petals and stamens were replaced by pin-like structures or were significantly reduced in size, and carpels did not fuse completely (Fig. 3 F–H). These floral morphogenetic defects correlate with a function for CHR11 and CHR17 in the MADS complexes.
Discussion
Specificity of DNA binding and mechanisms of gene regulation by transcription factors can depend on recruitment of cofactors to specific regulatory DNA sequences. Here, we showed that a well-known class of transcription factors, the MADS-domain proteins not only interact with each other, as proposed in the “floral quartet” model (5), but also form large complexes with other types of transcriptional regulators in planta, shedding light on mechanisms by which MADS-domain proteins regulate the transcription of their target genes.
According to the current model, SEP proteins are major mediators of higher-order complex formation of MADS-domain proteins. Our complex isolation results suggest some functional diversification within the SEP subfamily, which is partly supported by genetic data (3) and the results of yeast n-hybrid assays (3, 6). The A-class gene AP1 does not only specify the identity of the outer two floral whorls but also plays a role in the switch from inflorescence to floral meristem identity, in a partially redundant fashion with the two related genes CAULIFLOWER (CAL) and FUL (34). The presence of SOC1 and FUL in the AP1 IP may reflect the role of AP1 in Arabidopsis floral meristem specification. AP1 and SOC1 are only transiently coexpressed around stages 2–3 of flower development (17). AP1 has also been shown to repress SOC1 in the two outer floral whorls (35). This supports a role for heterodimers formed by antagonistically acting MADS-domain proteins in the transition from inflorescence to floral meristem identity, as has been suggested previously (36). Several other MADS-domain proteins, such as SHORT VEGETATIVE PHASE (SVP), are also binding partners of AP1 according to Y2H studies (36), but they were not detected in our AP1-GFP IP experiments, perhaps because of the very low abundance of these proteins in the native inflorescence tissues that were used in our analysis and their limited overlap in expression with AP1.
Based on our in vitro EMSA studies, we propose that different heteromeric MADS-domain protein complexes can coexist within the nucleus and may compete for partly overlapping sets of DNA-binding sites. The observation that one CArG box is sufficient to recruit a heteromeric higher-order MADS-domain protein complex suggests a mechanism by which these protein complexes might be recruited to DNA target sites in vivo: a preformed higher-order MADS-domain protein complex may first bind to a single, accessible CArG box in a target gene promoter. Then, upon bending of the DNA (37), the second heterodimer present in the complex may bind to another CArG box in the vicinity, which stabilizes the binding of the MADS-domain proteins to DNA. This would suggest a more “active” role of MADS-domain protein complexes in creating DNA loops in native promoters.
Whereas the quaternary complexes that we reconstituted in vitro may represent “core” complexes, we found that floral MADS-domain proteins are part of large complexes or structures in planta. In addition to MADS-domain proteins, we also identified members of other transcription factor families as potential components of MADS-domain protein complexes. This suggests that MADS-domain proteins may also act in a combinatorial fashion with non-MADS transcriptional regulators. Most prominent were members of the homeobox transcription factor family. Homeobox transcription factors form complex, intrafamily interaction networks (24). Therefore, the interaction between MADS-domain protein complexes and individual homeodomain proteins may recruit other members of the family to target gene promoters. Future experiments using more specific plant material for complex isolation might result in a more sensitive detection of additional interactions between MADS-domain proteins and non-MADS transcription factors that may cooperate in the regulation of subsets of target genes.
In the complex isolation experiments, we confirmed a previously identified interaction between AP1 and the transcriptional corepressor SEU (9). We also identified the SEU interaction partner LUH, which acts in a partially redundant manner with LUG (38). In addition, we identified several types of ATP-dependent nucleosome remodelers and their interaction partners in complexes of MADS-domain proteins, possibly as part of larger complexes that are stabilized in the presence of DNA. Chromatin-associated proteins were particularly abundant in the AP1 IP. This could reflect an interaction of AP1 with other proteins to reorganize chromatin structure in target gene promoters during the switch from inflorescence to floral meristem identity. One possible role for the interaction between AP1 and nucleosome remodelers could be in the activation of other floral homeotic genes, because, for example, the SWI/SNF-type chromatin remodeler BRM and PKL have previously been shown to play a role in this process (30, 31). The presence of SYD in the AP1 IP suggests that it can interact with AP1 in activation of LFY and supports the theory of mechanistic control of MADS-domain proteins target genes by modification of the chromatin states (32). The interaction of AP1 and other floral MADS-domain proteins with the H3K27me3 demethylase REF6 also suggests a role in the modification of specific chromatin states, specifically in antagonizing PcG mediated transcriptional repression. This is further supported by the finding of specific reduction of H3K27me3 around MADS DNA-binding sites in the SEP3 promoter upon AP1 induction. The defects in flower development that are observed in mutants of chromatin remodelers support the finding that chromatin-associated factors act together with MADS-domain transcription factors to control flower initiation and differentiation. Examples are phenotypes of brm (31), pkl (29), and chr11 chr17 mutants, as well as phenotypes of overexpression of the H3K27me3 demethylase REF6 and the ref6 curly leaf (clf) double mutant (22). The timed activation of the KNUCKLES (KNU) gene by AG via modification of chromatin states may be another example of interaction between MADS-domain transcription factors and chromatin remodelers (17).
To summarize, our results show that MADS-domain proteins associate with other transcription factors and chromatin-associated proteins into larger structures. Future experiments need to reveal the roles of specific complexes in the selection of target genes and thereby specification of distinct floral organ identities. They also need to reveal how common interactions between DNA sequence-specific transcription factors and the nucleosome remodeling machinery are in plants.
Materials and Methods
High-resolution LC-MS/MS of protein immunoprecipitates and quantitative data analysis with the MaxQuant software were essentially described before (39). ChIP experiments, in general, were performed as described previously (40). Detailed experimental and data analysis procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Giuseppa Morabito for help with the generation of the BiFC constructs and Frank Wellmer for the 35S:AP1-GR ap1 cal seeds. The authors were supported by grants from the Netherlands Proteomics Centre and Centre for BioSystems Genomics (to C.S.); the European Research Area Network on Plant Genomics Project “The meristematic regulatory network controlling the floral transition” (BLOOM-NET) (K.K. and R.G.H.I.); Netherlands Genomics Initiative Horizon Grant 93519020 (to J.M.M.); Netherlands Organisation for Scientific Research VIDI grant (to K.K.) the French National Agency Young Researcher grant for the ChromFlow project (to C.C.C. and R.B.); and Centre National de la Recherche Scientifique Higher Education Chair Program (to C.C.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112871109/-/DCSupplemental.
References
- 1.Coen ES, Meyerowitz EM. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 2.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 3.Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 4.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 5.Theissen G, Saedler H. Plant biology. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- 6.Immink RG, et al. SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 2009;10:R24. doi: 10.1186/gb-2009-10-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melzer R, Theissen G. Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 2009;37:2723–2736. doi: 10.1093/nar/gkp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melzer R, Verelst W, Theissen G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 2009;37:144–157. doi: 10.1093/nar/gkn900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sridhar VV, Surendrarao A, Liu Z. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development. 2006;133:3159–3166. doi: 10.1242/dev.02498. [DOI] [PubMed] [Google Scholar]
- 10.Brambilla V, et al. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell. 2007;19:2544–2556. doi: 10.1105/tpc.107.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill K, Wang H, Perry SE. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 2008;53:172–185. doi: 10.1111/j.1365-313X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann K, Pajoro A, Angenent GC. Regulation of transcription in plants: Mechanisms controlling developmental switches. Nat Rev Genet. 2010;11:830–842. doi: 10.1038/nrg2885. [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Xi W, Shen L, Tan C, Yu H. Regulation of floral patterning by flowering time genes. Dev Cell. 2009;16:711–722. doi: 10.1016/j.devcel.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Urbanus SL, et al. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 2009;9:5. doi: 10.1186/1471-2229-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Folter S, Urbanus SL, van Zuijlen LG, Kaufmann K, Angenent GC. Tagging of MADS domain proteins for chromatin immunoprecipitation. BMC Plant Biol. 2007;7:47. doi: 10.1186/1471-2229-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombo L, et al. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell. 1995;7:1859–1868. doi: 10.1105/tpc.7.11.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samach A, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 18.Wu X, et al. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130:3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- 19.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann K, et al. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7:e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufmann K, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 22.Lu F, Cui X, Zhang S, Jenuwein T, Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet. 2011;43:715–719. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- 23.Birkenbihl RP, Jach G, Saedler H, Huijser P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J Mol Biol. 2005;352:585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Hackbusch J, Richter K, Müller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA. 2005;102:4908–4912. doi: 10.1073/pnas.0501181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao X, Franks RG, Levin JZ, Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell. 2004;16:1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lal S, Pacis LB, Smith HM. Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 module by the homeodomain proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol Plant. 2011;4:1123–1132. doi: 10.1093/mp/ssr041. [DOI] [PubMed] [Google Scholar]
- 27.Kanrar S, Bhattacharya M, Arthur B, Courtier J, Smith HM. Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 2008;54:924–937. doi: 10.1111/j.1365-313X.2008.03458.x. [DOI] [PubMed] [Google Scholar]
- 28.Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2006;2:e117. doi: 10.1371/journal.pgen.0020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/s0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- 30.Aichinger E, et al. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 2009;5:e1000605. doi: 10.1371/journal.pgen.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurtado L, Farrona S, Reyes JC. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol. 2006;62:291–304. doi: 10.1007/s11103-006-9021-2. [DOI] [PubMed] [Google Scholar]
- 32.Wagner D, Meyerowitz EM. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr Biol. 2002;12:85–94. doi: 10.1016/s0960-9822(01)00651-0. [DOI] [PubMed] [Google Scholar]
- 33.Bezhani S, et al. Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell. 2007;19:403–416. doi: 10.1105/tpc.106.048272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, et al. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development. 2007;134:1901–1910. doi: 10.1242/dev.003103. [DOI] [PubMed] [Google Scholar]
- 36.de Folter S, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West AG, Causier BE, Davies B, Sharrocks AD. DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res. 1998;26:5277–5287. doi: 10.1093/nar/26.23.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sitaraman J, Bui M, Liu Z. LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 2008;147:672–681. doi: 10.1104/pp.108.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC) Methods. 2011;53:453–459. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Kaufmann K, et al. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP) Nat Protoc. 2010;5:457–472. doi: 10.1038/nprot.2009.244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.