Abstract
CLE peptides, named for the CLV3/ESR-related peptide family, participate in intercellular-signaling pathways. Here we investigated members of the CLE-like (CLEL) gene family that encode peptide precursors recently designated as root growth factors [Matsuzaki Y et al. (2010) Science 329:1065–1067]. CLEL precursors share a similar domain structure with CLE precursors (i.e., they contain a putative N-terminal signal peptide and a C-terminal conserved 13-amino-acid CLEL motif with a variable middle portion). Our evidence shows that, unlike root growth factor, CLEL peptides are (i) unmodified and (ii) function in the regulation of the direction of root growth and lateral root development. Overexpression of several CLEL genes in Arabidopsis resulted in either long roots or long and wavy roots that also showed altered lateral root patterning. Exogenous application of unmodified synthetic 13-amino-acid peptides derived from two CLEL motifs resulted in similar phenotypic changes in roots of wild-type plants. In CLEL peptide-induced long roots, the root apical meristem (RAM) was enlarged and consisted of an increased number of cells, compared with wild-type root apical meristems. The wavy-root phenotype appeared to be independent of other responses of the roots to the environment (e.g., gravitropism, phototropism, and thigmotropism). Results also showed that the inhibition of lateral initiation by CLEL overexpression was not overcome by the application of auxin. These findings establish CLEL as a peptide family with previously unrecognized regulatory functions controlling the pattern of root growth and lateral root development in plants.
Keywords: peptide hormone, root waving, cell division, signal transduction, synthetic peptides
Secreted peptides are important signals in intercellular communication in plants. They elicit signaling pathways that determine the identity and activity of adjacent cells and, in so doing, regulate the structure and function of plant tissues and organs. Secreted peptides are typically derived by proteolytic processing of precursor proteins. They act as signaling ligands, triggering downstream cellular signaling cascade reactions upon binding to a receptor-like kinase (RLK) and/or a receptor-like protein at the plasma membrane (1).
The largest group of such peptides identified so far, CLE, named for the CLV3/ESR-related gene family, is thought to function as a 12- or 13-amino-acid (aa) peptide ligand that regulates meristem activity in shoots and roots as well as in vascular tissues (2–6). In several studies, the 12- and 13-aa peptides encoded by specific CLE genes have been detected in plants, and synthetic forms have been shown to functionally mimic the overexpression of corresponding genes (3–6). Accumulated evidence also indicates that secreted peptides are commonly subjected to a variety of posttranslational modifications, such as proline hydroxylation, hydroxyproline arabinosylation, and tyrosine sulfation (3–7).
All CLE precursors contain an N-terminal signal peptide and a 14-aa conserved C-terminal motif [CLV3/ESR-related (CLE) motif] from which the mature CLE peptide is derived. The sequence between the two termini is highly variable (2). One family member, CLE18, differs from other CLEs and is unique in that its CLE motif is positioned in the variable region, not in the C terminus as is usual. Overexpression of CLE18 in Arabidopsis caused a long-root phenotype (8), whereas a synthetic 12-aa peptide derived from the CLE motif of the variable region of the gene behaved quite differently, suppressing root growth and causing a short-root phenotype (3). In analyzing the CLE18 precursor protein, we identified the 13-aa sequence in its C terminus as a new putative motif that was potentially responsible for the long-root phenotype triggered by overexpressing CLE18 in planta. Homology searches using this C-terminal sequence of CLE18 revealed a putative peptide gene family of 10 members that, for the reasons described below, we have designated the CLE-like (CLEL) family.
While our study of this gene family was in progress, Matsuzaki et al. (7) reported that a secreted 13-aa tyrosine-sulfated peptide, root growth factor 1 (RGF1), restored the root meristem activity of the knockout mutant tpst-1 in the presence of two unrelated sulfated peptides, PSK and PSY1. Tyrosylprotein sulfotransferase (TPST) is responsible for tyrosine sulfation of peptides. In Arabidopsis, the TPST protein is encoded by only one gene (AtTPST). The loss-of-function mutation in mutant tpst-1 resulted in dwarf plants with short, stunted roots owing to a deficiency in tyrosine-sulfated peptides (7, 9). Furthermore, overexpressing RGF1 in Arabidopsis, or treating plants in vitro with the synthetic tyrosine-sulfated RGF1 peptide, caused an enlarged root apical meristem (RAM) (7). Sequence analysis revealed that the RGF1 peptide shared the same sequences with the CLEL8 peptide that we have studied. Notably, the RGFs were reported to be tyrosine-sulfated peptides and to promote RAM activity only when sulfonated (7). However, our findings differed: we observed that (i) unmodified CLEL peptides were active, (ii) they altered the direction of root growth, and (iii) they regulated lateral root development. Taken together, these results demonstrate that members of the CLEL (RGF) family control not only root growth, but also the direction and pattern of growth.
Results and Discussion
CLEL Motif of CLE18 Represents a Family of Peptide Regulators in Arabidopsis.
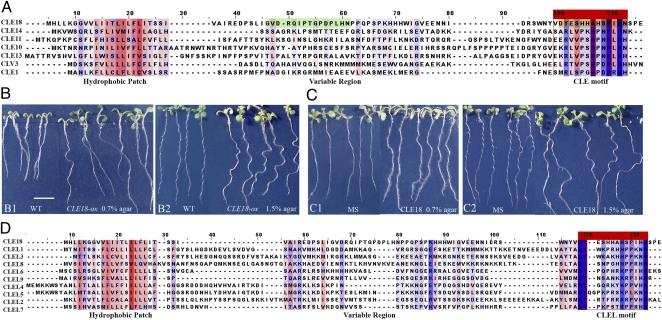
Fig. 1A highlights the unique sequence domain of CLE18 in comparing the complete sequences of several CLE precursors. The CLE motif is located in the variable region of the CLE18 precursor—from residues 35 to 48 (from 47 to 61 in the multiple sequence alignment highlighted in green in Fig. 1A)—rather than in the C terminus as is typical for other members of the family. Furthermore, Strabala et al. (8) reported that overexpressing the full-length CLE18 caused long roots in Arabidopsis, whereas Ito et al. (3) showed that exogenous application of the synthetic 12-aa peptide derived from the CLE18 variable region motif triggered a short-root phenotype. These seemingly contradictory results suggested that the synthetic CLE18 peptide does not represent the native active form of the CLE18 peptide. In other words, either different functional domains in the CLE18 precursor or posttranslational modification of the peptide determines the functionality of the endogenous protein.
Fig. 1.
CLE18 function and sequence analysis of CLE/CLEL families in Arabidopsis. (A) Multiple sequence alignment of CLE18 and several Arabidopsis CLE proteins. (B) Overexpression phenotypes of CLE18 in Arabidopsis plants. (B1) Seven-day-old seedlings of wild type (on left in B1) or plants overexpressing CLE18 grown vertically on 0.7% agar plates (on right in B1). (B2) Nine-day-old seedlings of wild-type plants (on left in B2) or plants overexpressing CLE18 grown on 1.5% agar plates inclined at 45° (on right in B2). (C) In vitro phenotypes after applying the unmodified synthetic 13-aa CLE18 peptide derived from the C-terminal CLEL motif. (C1) Eight-day-old seedlings of wild-type plants grown vertically on 0.7% agar in MS (Murashige and Skoog) medium alone (on left in C1) or in MS medium containing 1.0 μM CLE18 peptide (on right in C1). (C2) Nine-day-old seedlings grown inclined at 45° on 1.5% agar MS plates (on left in C2) or in MS medium containing 1.0 μM CLE18 peptide (on right in C2). (D) Multiple sequence alignment of Arabidopsis CLEL family. In A and D, the red and blue colors identify hydrophobic and hydrophilic residues, respectively. The color intensity corresponds to their relative strength. The CLE motif in the variable region and the CLEL motif at the C terminus of CLE18 are highlighted in green and orange, respectively, in A. (Scale bar, 5 mm.)
To further dissect its function, we overexpressed the full-length CLE18 gene. We observed that, indeed, overexpressing CLE18 caused long roots (Fig. 1, B1), consistent with the results reported by Strabala et al. (8). In addition, roots from these plants showed an enhanced wavy or irregular pattern of growth direction (hereafter we refer to this phenotype as wavy for simplicity) compared with roots from wild-type plants (Fig. 1B). It is known that certain growth conditions can cause the wavy-root phenotype in wild-type plants, a result of a thigmotropic response. One typical assay for this process is to grow the plants on firm (1.5%) agar with a 45° inclination of the growth surface. In this way, we could determine whether the wavy growth pattern elicited by overexpressing CLE18 is related to thigmotropism. As a control, both the wild-type and CLE18-overexpressing plants were grown vertically on soft (0.7%) agar plates for 7 d (Fig. 1, B1). For thigmotropic testing, the plants were vertically grown on firm (1.5%) agar plates for 2 d and then on plates inclined 45 ° for another 7 d (Fig. 1, B2). The CLE18-overexpressing plants showed more wavy roots than did wild type under both conditions (Fig. 1B). The wavy-root phenotype triggered by overexpressing CLE18 therefore appeared to be independent of thigmotropism. To test the possibility that other domains of the CLE18 precursor functioned differently from the CLE motif previously defined in the variable domain, we examined the CLE18 sequence further. Our analysis revealed a 13-aa sequence from residues 75 to 87 in the C terminus (DYESHHAHSPIHN located under the red bar from 101 to 113 in the multiple sequence alignment in Fig. 1A) that appeared to share similar features with CLE peptides. We reasoned that this motif might be responsible for the long and wavy-root phenotype effected by overexpressing the full-length CLE18. To test this possibility, we added the unmodified synthetic 13-aa CLE18 C-terminal peptide to the culture medium. We observed the induction of long and wavy roots (Fig. 1C), suggesting that the C terminus of CLE18 contains a previously undetected motif that functions as a unique regulatory peptide (Fig. 1A).
It has been shown that CLE gene products function as 12- or 13-aa peptide hormones (3, 4, 6). Both the 13-aa proline-hydroxylated CLV3 and the 12-aa proline-hydroxylated CLE2 glycopeptides have been detected in plants overexpressing the CLV3 and CLE2 genes, respectively (4, 6). Accordingly, we determined whether a 12-aa peptide in the CLE18 C terminus (without the “D” residue at the start of the 13-aa motif) functions as well as the 13-aa peptide tested earlier (Fig. S1A). The results indicated that the 12-aa C-terminal peptide of CLE18 elicited root growth effects similar to the 13-aa peptide (Fig. S1B).
On the basis of these results, we suspected that the 12- to 13-aa C-terminal motif of CLE18 represents a family of peptides that function in cell signaling processes. By homology search using this 13-aa motif as a query sequence, we identified a family of 10 proteins, including CLE18. These peptides were conserved only at the C-terminal 13-aa motif, and each contained an N-terminal hydrophobic patch that potentially served as a signal peptide for targeting to the secretory pathway (Fig. 1D). Since this gene family was derived from CLE18 and since the precursor proteins of its members shared a similar domain structure with CLEs, we designated: (i) members of this gene family as CLE-Like (CLEL) genes and (ii) the conserved C-terminal 13-aa sequence as the CLEL motif and the gene products as CLEL peptides. Except for CLE18, the CLEL genes were numbered from CLEL1 to CLEL9 according to the order of their locus numbers in The Arabidopsis Information Resource (Table S1). Of the CLEL proteins in Arabidopsis, CLEL4 (At3g02240) and CLEL5 (At3g02242) share the greatest pairwise amino acid sequence identity at 58%, and the genes are located adjacent to each other on the same chromosome, implying that they are derived from a gene duplication event. The CLEL motif of CLEL5 appears to contain 15 amino acids with a 2-amino-acid (QG) insert (Fig. 1D).
As mentioned above, the peptides under consideration were previously named “root growth factors.” We suggest that a change in nomenclature is justified on several grounds. First, the original name implies that the peptides are confined to roots. On the basis of current information, members of the CLEL family occur not only in roots, but also throughout the plant (Table S2). For example, CLEL6 (RGF6) is especially abundant in stamens and petals (Table S2). The RGF nomenclature gives no information on the nature of the genes. By contrast, the CLEL designation relates these genes to the well-known CLEs, one of which, CLE18, is the founding member of the CLEL family.
CLEL Precursors Are Targeted to the Secretory Pathway.
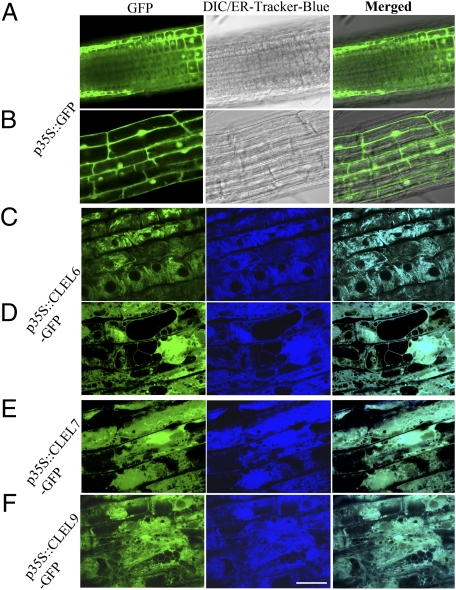
The subcellular localization of three CLEL proteins—CLEL6, CLEL7, and CLEL9—was determined using the GFP-fusion system. The results showed that all three CLEL-GFP fusion proteins (GFP was fused to the C terminus of the full-length CLEL precursors) were located in the endoplasmic reticulum (ER) (Fig. 2 C–F), suggesting targeting to the secretory pathway. The results are further consistent with the presence of an N-terminal signal peptide sequence in the CLEL proteins (Fig. 1D). This analysis suggested that CLEL members, like the CLE peptides, are secreted into the extracellular space via a default pathway following cleavage of their signal peptide in the ER (10). It is worth noting that, due to potential proteolytic cleavage during maturation, all CLEL peptide fusion protein assays, including the GFP fusions described here, should be interpreted with caution. Because of this complexity, GFP distribution may indicate only the subcellular localizations of CLEL precursors, not the mature peptides that may have been separated from the GFP tag after cleavage. Nevertheless, the results demonstrate that CLEL precursors can enter the secretary pathway and that the GFP reporter may indicate the location where the processing of CLEL proteins takes place.
Fig. 2.
Subcellular localization of CLEL-GFP fusion proteins in 5-d-old transgenic Arabidopsis roots. (A and B) Roots expressing the p35S::GFP control construct show a cytosolic localization of the free GFP in less-differentiated cells (A) and in differentiated cells (B). (C–F) Roots expressing CLEL6-GFP (C and D: less-differentiated cells are shown in C and differentiated cells in D), CLEL7-GFP (E), and CLEL9-GFP (F). Left, Center, and Right columns show the GFP, DIC (Differential Interference Contrast) (A and B)/ER Tracker-Blue (C–F), and merged channels, respectively. (Scale bar, 50 μm.)
Overexpression of Several CLEL Genes Causes Long and Wavy Roots.
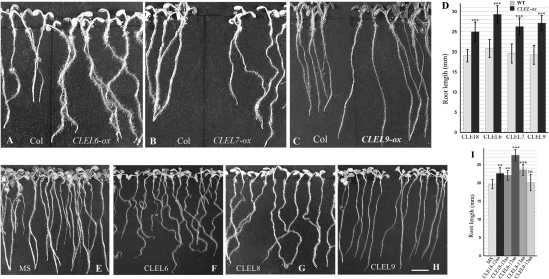
To explore the function of the gene family, we overexpressed several CLEL genes individually in Arabidopsis. The results showed that overexpression of the full-length CLEL genes elicited either long and wavy roots (CLEL6 and CLEL7, Fig. 3 A and B) or long roots exclusively (CLEL9, Fig. 3C). We also sought to determine whether the wavy growth pattern elicited by overexpressing CLEL6 and CLEL7 is related to thigmotropism. To this end, wild-type and CLEL-overexpressing plants were grown vertically on soft (0.7%) agar plates for 7 d (Fig. 3 A and B) or on firm (1.5%) agar plates first vertically for 2 d and then inclined 45° for 7 d (Fig. S2, A1 and A3). Again, the CLEL-overexpressing plants showed more wavy roots compared with wild type under both conditions. We also tested the growth pattern of these plants under light and darkness (Fig. 3 A and B and Fig. S2A) and observed that the CLEL-overexpressing plants formed more wavy roots under both conditions. The results thus indicate that the wavy phenotype elicited by CLEL overexpression is not affected by either thigmotropism or phototropism. The results on CLEL enhancement of root growth (longer-root phenotype) in our study (Fig. 3D) are in general agreement with the reports of Strabala et al. (8) for CLE18 and of Matsuzaki et al. (7) for RGF peptides. However, the wavy-root and altered lateral-root pattern phenotypes described below have not been previously observed. They add another dimension to providing evidence that CLEL peptides play diverse roles in the growth and development of plant roots.
Fig. 3.
Functional analysis of CLELs using overexpression and unmodified synthetic peptide assays. (A–C) Seven-day-old seedlings of wild-type (on left) or plants overexpressing full-length CLEL6 (A), CLEL7 (B), or CLEL9 (C) grown vertically on 0.7% agar plates (on right). (D) Root length in millimeters of 7-d-old seedlings of wild-type plants and plants overexpressing a CLEL gene as shown in A–C. (E–H) Eight-day-old seedlings of wild-type plants grown vertically on 0.7% agar MS medium (E) or on medium containing 1.0 μM of unmodified synthetic CLEL6 peptide (F), CLEL8 peptide (G), or CLEL9 peptide (H). (I) Root length in millimeters of 8-d-old seedlings as shown in E–H. (Scale bar, 5 mm.) Error bars indicate the SD. Two and three asterisks indicate which values are significantly different from the control at P < 0.01 and P < 0.001, respectively. ns, not significant. n = 8–12.
Exogenous Application of Unmodified Synthetic CLEL Peptides Elicits Long and Wavy Roots.
To determine whether peptides derived from the CLEL motif of CLEL proteins are able to function as regulators, we synthesized three 13-aa unmodified peptides derived from the corresponding CLEL motifs of each of the CLEL6, CLEL8, and CLEL9 precursors and applied 1.0 μM of each individually in vitro. The results indicated that the CLEL6 (DYPQPHRKPPIHN) and CLEL8 (DYSNPGHHPPRHN) peptides triggered long and wavy roots (Fig. 3 E–G and Fig. S1 C and D). Moreover, the wavy-root phenotypes elicited by the unmodified synthetic CLEL peptides were independent of thigmotropic or phototropic responses as described above for plants overexpressing the peptides (Fig. S2B). The phenotype resulting from treatment with one of the peptides, CLEL6, was similar to that of plants overexpressing the CLEL6 protein. Thus, the CLEL6 peptide appeared to recapitulate the long- and wavy-root phenotypes triggered by overexpression of the full-length CLEL6 gene (Fig. 3 A and F). In addition, Arabidopsis mutants with an impaired gravitropic response, such as eir1-1 and aux1-7, responded to the CLEL6 peptides by forming long and wavy roots, again suggesting that these phenotypes are not dependent on gravitropism (Fig. S3).
On the other hand, in contrast to the long-root phenotype triggered by overexpression of the full-length CLEL9 gene (Fig. 3C), the 13-aa CLEL9 peptide (DYNSANKKRPIHN) failed to affect root growth significantly (Fig. 3H). It is intriguing to observe such a contrast among different CLEL peptides. In light of the results of Matsuzaki et al. (7), we envisaged that CLEL9 has to be modified by a posttranslational process such as sulfation to become active. By contrast, CLEL6 may not require modification. The observation that CLEL9 promotes longer roots when overexpressed (Fig. 3C) is consistent with the conclusion of Matsuzaki (7) that peptides require sulfation to complement the short-root phenotype as seen in the tpst-1 mutant lacking TPST. In contrast, the previously unreported wavy-root phenotype caused by CLEL6 appears to represent a function mediated by unmodified peptides. In other words, in planta there may be two functional forms of CLEL peptides: a sulfated form and an unmodified form.
Root Apical Meristem of Longer Roots Induced by CLEL Peptides Is Enlarged and Contains More Cells.
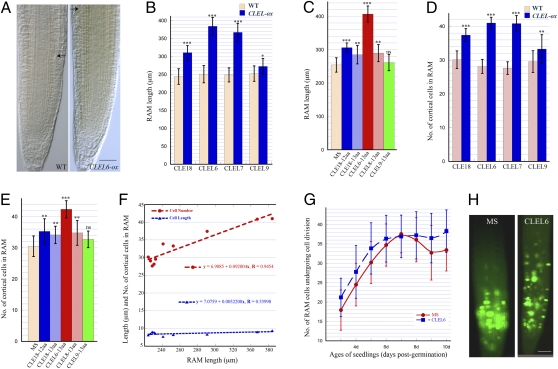
The size of the RAM is an important parameter in root growth and development. To further dissect the long-root phenotype elicited by a CLEL peptide in vivo and in vitro, we measured RAM length in primary roots of 7-d-old seedlings. The results indicated that the long roots observed in plants overexpressing CLE18, CLEL6, CLEL7, and CLE9 contained a significantly longer RAM than did wild-type plants (Fig. 4 A and B). Similar results were observed with the long roots elicited by exogenous application of unmodified synthetic CLEL peptides (Fig. 4C).
Fig. 4.
Root apical meristem size and cell division parameters in wild-type plants and in plants either overexpressing CLEL genes or treated with CLEL peptides. (A) Root tip from a 7-d-old wild type (WT) (Left) and CLEL6-overexpressing plant (CLEL6-ox) (Right). (B–E) Quantitative analysis of RAM in roots of 7-d-old plants overexpressing a CLEL gene (CLEL-ox) (B and D) or in wild-type plants treated with an unmodified synthetic 12- or 13-aa CLEL peptide (C and E). (D and E) Number of cortical cells in the RAM. (F) Regression analysis of RAM length and number of cortical cells in the RAM (red line) and RAM length and length of cortical cells in the RAM (blue line) from 7-d-old wild-type plants and plants overexpressing a CLEL gene. (G) Number of cells undergoing cell division in the RAM of roots grown on MS medium (red line) and in MS medium containing 1.0 μM unmodified synthetic 13-aa CLEL6 peptide (blue line). Age of the seedlings was defined as days post planting. (H) Roots of 7-d-old seedlings of Arabidopsis pCYCB1::CYCB1-GFP marker line grown on MS medium (Left) or on medium containing an 1.0-μM unmodified synthetic 13-aa CLEL6 peptide (Right) to illustrate cells undergoing division. The black arrows in A point to RAM borders in a primary root from wild type (WT) (Left) and a plant overexpressing CLEL6 (CLEL6-ox) (Right), respectively. (Scale bar, 50 μm.) Error bars indicate the SD. One, two, and three asterisks indicate which values are significantly different from the control at P < 0.05, P < 0.01, and P < 0.001, respectively. ns, not significant. n = 8–12.
The number and length of cells in the RAM determine its size. To further pinpoint the process whereby CLELs function, we counted the number and calculated the length of cortical cells in the RAM. The results showed a strong positive correlation between RAM length and the number of cortical cells in the RAM (Fig. 4 B–F). In other words, the elongated RAMs contain significantly more cortical cells than the control (Fig. 4 D and E). No significant increase in cell length was observed in the longer RAM brought about by CLEL peptides (Fig. 4F). The results prompt the conclusion that the prolonged RAM results mainly from an increase in RAM cell number.
Next, we monitored the cell division rate in the RAM using a cell-division marker line that contains the pCYCB1::CYCB1-GFP construct (11). We found that the exogenous application of the CLEL6 peptide, which caused a longer RAM (Fig. 4C), did not significantly increase the number of dividing cells (Fig. 4G). However, CLEL6 appeared to alter the distribution of dividing cells in the RAM. On the control medium, cell division in the roots took place in a small niche near QC (Fig. 4H). With CLEL6 treatment, dividing cells were distributed in a much larger region in the RAM (Fig. 4H).
CLEL Peptides Regulate Lateral Root Development Through an Auxin-Independent Pathway.
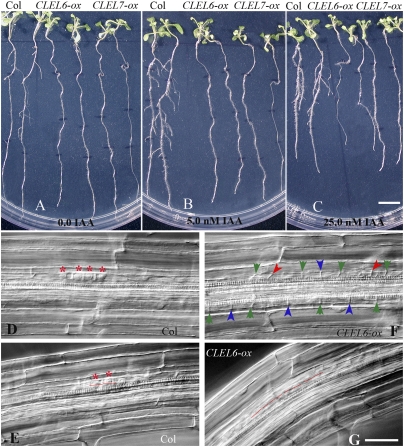
In addition to the above observations, we found that overexpression of CLEL6 or CLEL7 dramatically delayed lateral root development in Arabidopsis. In contrast to the 12-d-old wild-type plants that displayed a well-developed lateral root system, plants overexpressing CLEL6 or CLEL7 had almost no lateral roots (Fig. S4). Although the mechanisms that regulate the distribution and arrangement of lateral roots along the parental root remain largely unknown, auxin has been shown to play a critical role. Lateral roots in Arabidopsis are derived from pericycle founder cells positioned adjacent to the two protoxylem poles (12). Auxin accumulates at the site of lateral root initiation before a primordium starts to form (13). Some auxin-related mutants show impaired lateral root development (14), and, in general, exogenous application of auxin induces cell division and results in lateral root initiation in the entire pericycle at the xylem poles in wild-type plants (15). It is thus proposed that the distribution pattern of lateral roots may be regulated by an auxin-based oscillatory mechanism (16). To determine whether auxin could be responsible for the effect of CLEL on the formation of lateral roots, the 4-d-old transgenic and the wild-type plants grown on the MS (Murashige and Skoog) medium were transferred to the same medium containing 0.0, 5.0, or 25.0 nM IAA (indole-3-acetic acid) (Fig. 5 A–C). Although IAA-containing media promoted lateral root formation in wild-type plants, such treatments failed to rescue lateral root development in plants overexpressing CLEL6 or CLEL7 (Fig. 5 B and C). The results further showed that IAA treatment inhibited root elongation in both wild-type and CLEL-overexpressing plants, indicating that root growth of all plants responded to IAA (Fig. 5 A–C). Taken together, the results suggest that CLEL peptides regulate lateral root development through an auxin-independent pathway.
Fig. 5.
Defects in lateral root development caused by CLEL overexpression in Arabidopsis. (A–C) Inhibition of lateral root development by CLEL peptides is not rescued by auxin. Wild-type (Col-0) plants (on left) and plants overexpressing CLEL6 (CLEL6-ox) (center) or CLEL7 (CLEL7-ox) (on right) were grown on MS medium for 4 d after planting and then transferred to MS medium containing 0.0 nM (A), 5. 0 nM (B), or 25.0 nM (C) IAA and grown for another 7 d before taking the photograph. (D–G) Root segments from 8-d-old wild type (D and E) and plants overexpressing CLEL6 (F and G) during lateral root initiation at stage I (D and F) or at stage II (E–G). Asterisks indicate the small cells in the central core of the new lateral root at stage I in D or the central cells of the central core of the lateral root undergoing periclinal cell division at stage II in E. The arrowheads in F indicate the new cell walls from asymmetric (red arrowheads) or symmetric cell divisions (blue arrowheads), and the green arrowheads denote the old cell walls. (Scale bar: 5 mm in A–C; 50 μm in D–G.)
CLEL Peptides Disturb the Restrictive Division of Pericycle Cells During Lateral Root Initiation.
At stage I of lateral root initiation, the two adjacent pericycle founder cells undergo two successive rounds of anticlinal asymmetric cell divisions, producing a center of four small cells and two larger flanking cells (Fig. 5D). Thereafter, anticlinal cell division halts, and the two cells in the center of four small cells divide periclinally to produce two layers of cell files at stage II of the lateral root initiation (Fig. 5E) (17). Compared with lateral root initiation in wild-type roots (Fig. 5 D and E), the anticlinal division of pericycle cells was strikingly irregular in roots overexpressing CLEL6 or CLEL7. Here, the pericycle cells appeared to divide both asymmetrically and symmetrically and to lack the formation of the central core of small pericycle cells at stage I (Fig. 5F). Among the 20 seedlings examined for each group of plants, the wild type had 13 central cores of pericycle cells at stage I, whereas the CLEL6- and CLEL7-overexpressing lines had only 2 and 1, respectively. On the other hand, the pericycle cells dividing abnormally at stage I (Fig. 5F) appeared to reach stage II (Fig. 5G), but no further development was observed. The results suggest that CLEL peptides disturb the restrictive division of pericycle cells involved in lateral root initiation.
An exciting question concerns how CLEL peptides act in the development of lateral roots. Here, it is worth noting that a receptor-like kinase, ACR4 (ARABIDOPSIS CRINKLY4), was shown to function in specifying the fate of pericycle cells during lateral root initiation (17). ACR4 appears to be critical for repressing the division of nearby pericycle cells and in this way controls the patterning of lateral roots. ACR4 was also reported to regulate stem cell fate in the RAM via the CLE40-ACR4-WOX5 signaling loop (18). Because LRR-RLKs, for example, CLV1 and ACR4 (4, 18), serve as receptors for CLE peptides, it seems possible that CLEL peptides may interact with unknown receptor-like kinase(s) to initiate signaling cascades that regulate asymmetric division of pericycle cells and thus lateral root initiation.
Concluding Remarks
In this study we have identified previously unrecognized functions of peptides designated RGFs. Overexpression of several CLEL genes individually in Arabidopsis promoted either long roots or, unexpectedly, long and wavy roots with altered lateral root patterns. Exogenous application of unmodified synthetic 13-aa peptides, derived from the CLEL motifs, reproduced both the wavy- and long-root phenotypes. This finding suggests the presence of at least two forms of endogenous peptides derived from CLEL genes: one, the tyrosine-sulfated RGFs, described earlier, which promote RAM activity and lead to a long-root phenotype (7), and another, the presently described nonmodified CLELs, which regulate the growth and developmental patterns of roots. The wavy-root phenotype was seemingly independent of known environmental responses. The evidence supports the view that CLEL peptides participate in previously unrecognized mechanisms that regulate root growth pattern and pericycle cell division, leading to lateral root initiation. These findings open an avenue for studying peptides and their function in intercellular signaling and in regulating root growth and lateral root development.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana ecotype Columbia (Col-0) and designated lines of transgenic plants with the same genetic background were used in this study. The cell-division marker line BJ3 in the Arabidopsis Col-0, which contains a pCYCB1::CYCB1-GFP construct, was provided by Peter Doerner (University of Edinburgh, Edinburgh, UK). CLEL peptides (12- and 13-amino acids derived from CLEL motifs) were synthesized at the Howard Hughes Medical Institute Mass Spectrometry Laboratory, Department of Molecular and Cell Biology, University of California, Berkeley.
For typical agar plate assays, seeds were sterilized for 5–10 min in 50% (vol/vol) bleach (Chlorox) containing a final concentration of 3% sodium hypochlorite, washed three times with sterilized, distilled H2O, and then planted on solid MS medium containing: 0.5-X Murashige and Skoog + Gamborg B5 vitamins, 1.5% (wt/vol) sucrose, and 0.7% (wt/vol) Bacto agar, pH 5.7–5.8. After 3 d at 4 °C, plates were positioned vertically in growth chambers (16 h light, 8 h dark) at 23 °C. Transgenic plants were screened by growth on MS medium containing 50 μg/L kanamycin. Fifteen-day-old kanamycin-resistant seedlings were transferred to soil and grown in a greenhouse under a 16-h light, 8-h dark cycle to harvest seeds for further assays.
Constructs and Plant Transformation.
Genomic DNA was used to amplify the full-length coding sequences for constructing the CLEL-expressing constructs because CLEL genes do not contain introns. For GFP fusion constructs, the full-length CLEL ORFs were amplified by PCR from the genomic DNA of the Arabidopsis wild-type Col-0 plants using appropriate primers (Table S3). The XhoI and BamHI cloning sites were added to the 5′ ends of the forward and reverse primers, respectively. PCR products were cloned into the pEZS-NL vector (S. Cutler and D. Ehrhardt, Carnegie Institute of Washington, Stanford, CA) at the XhoI and BamHI sites and fused individually in frame to GFP at the C terminus of the CLEL ORF. For overexpressing CLEL plasmids, the amplified CLEL ORFs were cloned into the pEZS-CL vector through the XhoI and BamHI sites. A NotI fragment, containing the 35S promoter, the cloned full-length CLEL ORF, and the OCS (octopine synthase) 3′ terminator, was isolated from the pEZS-NL or pEZS-CL plasmid after NotI digestion and inserted into the binary vector pART27 (19) at the NotI site. The cloned CLEL genes in the pART27 binary vector were introduced into Agrobacterium tumefaciens strain GV3101 using the freeze–thaw method (20). Transgenic plants were generated via the floral-dip method (21).
Microscopy, Photography, and Root Growth Assays.
Primary roots from 5-d-old transgenic T2 plants expressing one of the CLEL-GFP fusion proteins were examined for subcellular localization of the GFP fluorescence using a Zeiss LSM 710 confocal microscope with excitation (nm)/emission (nm) of 488/530. The ER localization of the fusion proteins was confirmed by the ER-Tracker-Blue-White DPX (Molecular Probes, Invitrogen) using the same microscope at excitation (nm)/emission (nm) of 364/470.
Petri-dish–grown seedlings were used for various root growth assays. Seedlings were photographed with a digital camera (Nikon CoolPIX990). The root apex was visualized using a Zeiss AxioImager M1 fluorescence microscope (Carl Zeiss). RAM length was measured as the distance between the QC and the elongation zone of the root (Fig. 4A). The length of cortical cells in the RAM region was calculated by dividing the RAM length by the number of the cortical cells in the RAM. The measurement was conducted on the high-resolution images of the root tips using ImageJ software.
Statistical Analysis.
Statistical analyses of the data were performed using two-tailed Student's t test. Error bars indicate the SD. One, two, and three asterisks in the figures (Figs. 3 and 4) indicate significant differences from the control at P < 0.05, P < 0.01, and P < 0.001, respectively (ns, not significant; sample size, n = 8–12).
Supplementary Material
Acknowledgments
We thank Dr. David King for synthesis of the CLEL peptides. We are grateful to Dr. Peter Doerner for providing the pCYCB1::CYCB1-GFP transgenic lines. We thank Drs. S. Ruzin and D. Schichnes of the University of California, Berkeley, Biological Imaging Facility for assistance.This work was supported by a grant to S.L. from the National Science Foundation. L.M. was partially supported by an award from the Agricultural and Environmental Chemistry Graduate Program at the University of California, Berkeley.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119864109/-/DCSupplemental.
References
- 1.Diévart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131:251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- 2.Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126:939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito Y, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 4.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 5.Ohyama K, Ogawa M, Matsubayashi Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008;55:152–160. doi: 10.1111/j.1365-313X.2008.03464.x. [DOI] [PubMed] [Google Scholar]
- 6.Kondo T, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 7.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 8.Strabala TJ, et al. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006;140:1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:15067–15072. doi: 10.1073/pnas.0902801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol. 2003;51:415–425. doi: 10.1023/a:1022038932376. [DOI] [PubMed] [Google Scholar]
- 11.Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 12.Blakely LM, Durham M, Evans TA, Blakely RM. Experimental studies on lateral root formation in radish seedling roots: I. General methods, developmental stages, and spontaneous formation of laterals. Bot Gaz. 1982;143:341–352. [Google Scholar]
- 13.Casimiro I, et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- 15.Himanen K, et al. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 17.De Smet I, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322:594–597. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- 18.Stahl Y, Wink RH, Ingram GC, Simon RA. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19:909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 19.Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 20.Höfgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.