Abstract
Low efficiencies of gene targeting via homologous recombination (HR) have limited basic research and applications using human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs). Here, we show highly and equally efficient gene knockout and knock-in at both transcriptionally active (HPRT1, KU80, LIG1, LIG3) and inactive (HB9) loci in these cells using high-capacity helper-dependent adenoviral vectors (HDAdVs). Without the necessity of introducing artificial DNA double-strand breaks, 7–81% of drug-resistant colonies were gene-targeted by accurate HR, which were not accompanied with additional ectopic integrations. Even at the motor neuron-specific HB9 locus, the enhanced green fluorescent protein (EGFP) gene was accurately knocked in in 23–57% of drug-resistant colonies. In these clones, induced differentiation into the HB9-positive motor neuron correlated with EGFP expression. Furthermore, HDAdV infection had no detectable adverse effects on the undifferentiated state and pluripotency of hESCs and hiPSCs. These results suggest that HDAdV is one of the best methods for efficient and accurate gene targeting in hESCs and hiPSCs and might be especially useful for therapeutic applications.
Introduction
Increasing the efficiency of gene targeting in human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) is essential to improve both the experimental and therapeutic potential of these cells. Although gene targeting by electroporation has been routinely used in mouse embryonic stem cells (mESCs), its application in hESCs/hiPSCs has been limited.1,2 The ratio of targeted to random chromosomal integration (the “relative” targeting efficiency) in these cell types by electroporation is generally low (0~2%).3,4,5,6,7 Recently, high relative targeting efficiencies were reported in hESCs/hiPSCs by electroporating modified bacterial artificial chromosomes (BACs), encoding long homologous DNA to the target sequences.8 Zinc-finger nucleases (ZFNs) are also used for highly efficient gene targeting at both transcriptionally active and inactive loci.9,10,11 However, the accuracy of homologous recombination (HR) by these methods, which is critical for the therapeutic application of iPSCs, has not yet been proven. Viral vectors have been utilized to overcome the low DNA delivery and gene-targeting efficiencies in hESCs/hiPSCs.9,12,13 A high-capacity helper-dependent adenoviral vector (HDAdV)14,15 recently yielded relative targeting efficiencies of ~45% at the hypoxanthine phosphoribosyltransferase (HPRT1) locus in hESC lines, thus suggesting its potential as a universal gene-targeting vector in human pluripotent stem cells.16
The current study expanded the general applicability of HDAdVs to gene knockout and knock-in at several loci, both transcriptionally active and inactive, in hESCs as well as in hiPSCs. Efficiencies of accurate gene targeting were 7–81% at five loci without detectable effects on the undifferentiated state and pluripotency. Importantly, 75–100% of homologous recombinants were produced by accurate HR without additional vector integration at ectopic sites, suggesting high fidelity and the potential safety of HDAdV-mediated gene targeting.
Results
Gene knockout of HPRT1 locus in hiPSCs
Gene targeting at the HPRT1 locus in hESCs was efficiently achieved using HDAdV (Table 1).16 The current study first targeted the HPRT1 locus in the male hiPSC lines, 246H1 and 246G1, using the same HPRT1-targeting HDAdV (Figure 1a).17 HR, which knocks out the sole allele on the X chromosome, is detected by selecting the infected cells with 6-thioguanine-resistance (6TGR). The cells were also subjected to negative selection with ganciclovir (GANC) to improve the relative gene-targeting efficiency because the vector also encodes the HSVtk gene. HDAdV infection of 246G1 cells resulted in 135 G418-resistant (G418R) colonies, 79 of which were G418R-GANC-resistant (GANCR). Of those G418R-GANCR colonies, 16 were also 6TGR. The resultant gene-targeting efficiency (G418R-GANCR-6TGR colonies)/(G418R-GANCR colonies) was 20% with positive-negative selection (Table 1). A similar efficiency of 7% was observed in the 246H1 cell line (Table 1). The linearized HPRT1-targeting HDAdV plasmid was electroporated into the 246H1 cells as a control, resulting in no HR out of 126 G418R clones. Southern hybridization (Figure 1b) demonstrated that those 6TGR clones had been targeted accurately at the HPRT1 locus via HR, without ectopic vector integration (Supplementary Figure S1a). These results indicate that HDAdV-mediated gene targeting is equally efficient in both hESCs and hiPSCs.
Table 1. Summary of gene-targeting efficiencies in human ESCs and iPSCs.
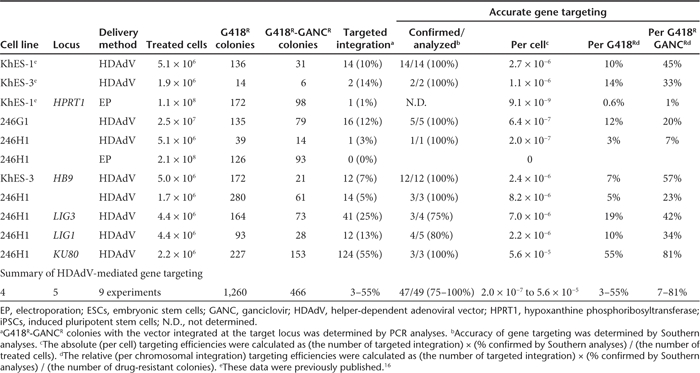
Figure 1.

Gene targeting at HPRT1. (a) Schematic illustration of HPRT1 knockout with HDAdV. The probes for Southern analyses are shown as black bars. a, 5′ probe; b, neo probe; c, 3′ probe. HSVtk, the herpes simplex virus thymidine kinase gene cassette; Neo, the neomycin-resistant gene cassette; Venus, an expression cassette for the Venus (F46L mutant yellow fluorescent protein gene ref. 41); A, AhdI sites; Sb, SbfI sites. (b) Southern analyses of wild type (WT) and the HPRT1-knockout human induced pluripotent stem clones. Genomic DNA was digested with AhdI and SbfI. HDAdV, helper-dependent adenoviral vector.
Heterozygous knockout of KU80, LIG1, and LIG3 genes in hiPSCs
Next, to examine the applicability of HDAdV-mediated gene targeting at other housekeeping loci, the Lupus Ku autoantigen protein p80 (KU80) locus of the nonhomologous end-joining (NHEJ) pathway, and the DNA Ligase I (LIG1) and DNA Ligase III (LIG3) loci of the potential backup pathway of NHEJ were targeted. NHEJ is a major pathway involved in DNA double-strand break (DSB) repair. KU70 and KU80 are the key factors in NHEJ, and the heterozygous mutant cells or RNAi-induced knockdown of Ku resulted in elevated gene-targeting frequencies via HR in the HCT116 and NALM-6 human somatic cell lines.18,19 The 246H1 cells were infected with gene-targeting HDAdVs that were designed to delete exons 3–4 of KU80, exons 3–4 of LIG1, and exons 5–7 of LIG3 (Figures 2a and 3a,b). Detailed Southern analyses of G418R and GANCR clones revealed relative gene-targeting efficiencies of 81% at KU80, 34% at LIG1, and 42% at LIG3 (Figures 2b and 3c, Supplementary Figure S1b–d, Table 1). Importantly, in contrast to ZFN -mediated gene targeting, no ectopic vector integration was observed in any of these targeted clones (Supplementary Figure S1b–d). In these heterozygous hiPS clones, the inserted neomycin-resistant gene (neo) was flanked by the mutant loxP sites (lox71 and lox66). After transfection of pCAGGS-Cre, which encodes the CAG promoter-driven Cre gene, and recloning of the cells, neo was removed in ~25% of the cells (Figures 2c and 3d, Supplementary Figure S1b–d, Supplementary Table S1). Two independent clones from LIG1, LIG3, and KU80 heterozygous mutant iPSCs were subjected to further analyses. The reduction in the copy number of wild-type (WT) allele and of gene expression levels in these heterozygous hiPS clones were confirmed by quantitative genomic Southern analyses, quantitative reverse transcription-PCR (RT-PCR), and western blotting (Figures 2d,e and 3e,f). These results therefore indicate that highly efficient gene targeting can be achieved at various housekeeping loci using HDAdVs.
Figure 2.
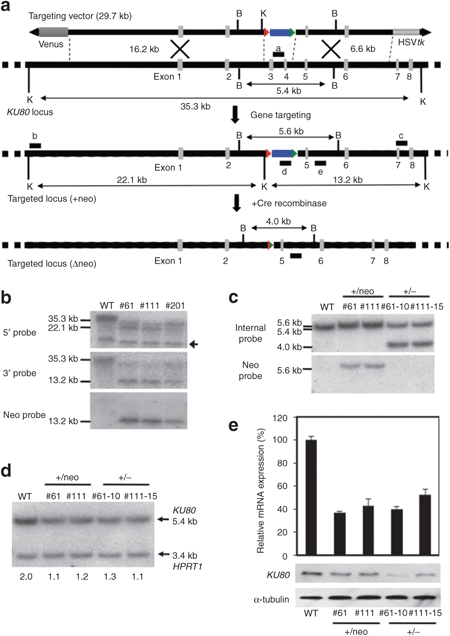
Gene targeting at KU80. (a) Schematic illustration of KU80 heterozygous knockout with HDAdV. The probes for Southern analyses are shown as black bars; a, neo probe; b, 5′ probe; c, 3′ probe; d, deleted-region probe; e, internal probe. HSVtk, the herpes simplex virus thymidine kinase gene cassette; Blue box, the neomycin-resistant gene cassette; Venus, the Venus gene cassette; Triangle, loxP site; B, BglII sites; K, KpnI sites. (b) Southern analyses of WT (246H1) and heterozygous knockout clones (#61 and #111). #201 is an inaccurate recombinant. Genomic DNA was digested with KpnI. An arrow indicates a nonspecific band. (c) Southern hybridization analyses of the KU80 heterozygous knockout clones obtained from 246H1, and the clones from which the PGKneo cassette was removed by transient Cre expression. The genomic DNA was digested with Bg II and hybridized to the 32P-labeled internal probe or neo probe. WT, wild-type parental cells. (d) Quantitative Southern hybridization. Genomic DNA was digested with BglII and hybridized with probe “d.” A DNA fragment from intron 3 of HPRT1 locus was used as a control probe. The copy number of wild-type KU80 allele determined by a densitometric analysis is indicated. (e) Reduction of KU80 gene expression in the heterozygous mutants. The upper panel indicates the mRNA level determined by quantitative RT-PCR. The average of three independent experiments is shown with the standard deviation. The lower panel indicates the protein level determined by Western blot. An anti-α-tubulin antibody was used as a loading control. HDAdV, helper-dependent adenoviral vector. RT-PCR, reverse transcription-PCR.
Figure 3.
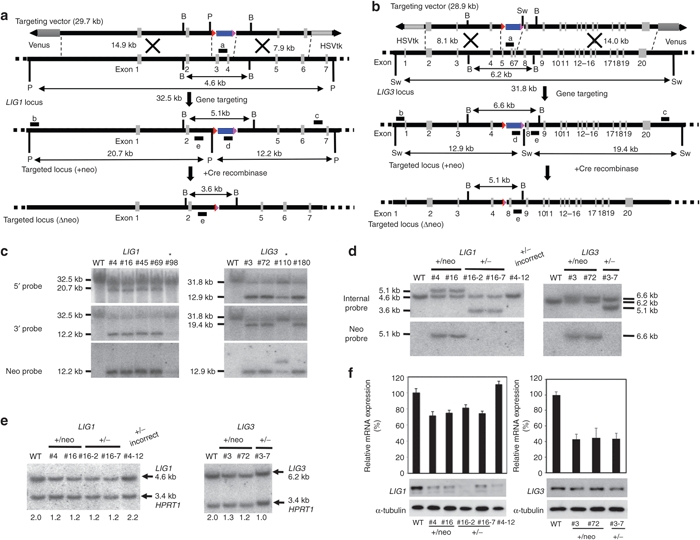
Generation of LIG1 and LIG3 heterozygous mutants in human induced pluripotent stem cells. (a) Schematic illustration of the LIG1 heterozygous knockout with HDAdV. The probes for Southern hybridization analyses are shown as black bars; a, neo probe; b, 5′ probe; c, 3′ probe; d, deleted-region probe; e, internal probe. HSVtk, the herpes simplex virus thymidine kinase gene cassette; Blue box, the neomycin-resistant gene cassette; Venus, the Venus gene cassette; Triangle, loxP site; B, BglII; P, PacI. (b) Schematic illustration of the LIG3 heterozygous knockout in human induced pluripotent stem cells. The probes for Southern hybridization analyses are shown as black bars; a, neo probe; b, 5′ probe; c, 3′ probe; d, deleted-region probe. HSVtk, the herpes simplex virus thymidine kinase gene cassette; Neo, the neomycin-resistant gene cassette; Venus, the Venus gene cassette; Red triangle, loxP site; B, BglII; Sw, SwaI. (c) Southern hybridization analyses of WT, LIG1, and LIG3 heterozygous knockout clones (LIG1: #4, #16, #45, and #69; LIG3: #3, #72, and #180). Genomic DNA was digested with PacI or SwaI. Asterisks indicate inaccurate homologous recombinants. (d) Southern hybridization analyses of the LIG1 and LIG3 heterozygous knockout clones obtained from 246H1 cells and clones in which the PGKEM7neo cassette was removed by transient expression of Cre recombinase to confirm the insertion and removal of the PGKneo cassette. The genomic DNA was digested with BglII and hybridized with the 32P-labeled internal probe or neo probe. #4-12 is an inaccurate recombinant. WT, wild-type parental cells. (e) Quantitative Southern hybridization. Genomic DNA was digested with BglII and hybridized with probe “d.” A DNA fragment from intron 3 of the HPRT1 locus was used as a control probe to normalize the amount of loaded DNA. The copy number of wild-type LIG1 or LIG3 allele determined by a densitometric analysis is indicated. (f) Reduction of LIG1 (left panel) or LIG3 (right panel) gene expression at mRNA and protein levels in the heterozygous LIG1 or LIG3 mutants. The upper panel indicates the mRNA level determined by quantitative RT-PCR. The average of three independent experiments is shown with the standard deviation. The lower panel indicates the protein level determined by Western blotting. An anti-α-tubulin antibody was used as a loading control. HDAdV, helper-dependent adenoviral vector. RT-PCR, reverse transcription-PCR.
Gene knock-in of HB9 locus in hESCs and hiPSCs
Next, to examine the efficiency of HDAdV-mediated gene targeting at transcriptionally inactive loci, reporter hESC and hiPSC lines were established, in which the enhanced green fluorescent protein (EGFP) gene was knocked into the HB9 locus by HR. HB9 is expressed selectively in motor neurons in the developing vertebrate central nervous system and also in pancreatic cells, but not in ES cells.20,21,22 A human HB9-targeting HDAdV was constructed with homology arms of 12.1 kb and 2.0 kb on each side (Figure 4a). The infected hESC line, KhES-3, had 21 GANCR colonies out of 172 G418R (Table 1). Southern hybridization demonstrated that 12 of these 21 colonies (57%) had been accurately targeted at the locus, without ectopic vector integration (Figure 4b, Supplementary Figure S1e,f). In addition to hESCs, the HB9 locus was also targeted in the hiPSC line, 246H1, at similar efficiencies (Table 1, Figure 4b). A karyotype analysis was performed for the knock-in hESC clones G1 and 47, and no abnormality was observed by G-banding (Supplementary Figure S2). We also performed a similar analysis for the HPRT1-knockout hESC clone, which we reported previously, and found no abnormalities (Supplementary Figure S2), HB9-targeted hESCs and hiPSCs maintained an undifferentiated state (Figure 5a) and pluripotency (Figure 5b,c), as we have previously reported for the HPRT1-knockout hESC clones.16 These results indicate that HDAdV-mediated gene targeting is equally efficient regardless of the transcriptional activities of target loci.
Figure 4.
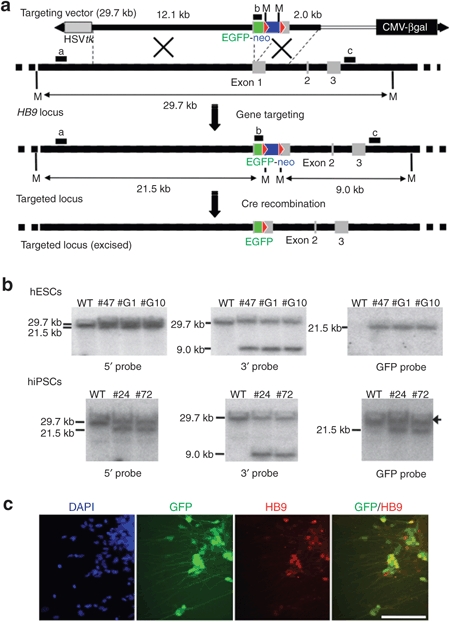
Generation of HB9-EGFP knock-in reporter cell lines from hESCs and hiPSCs. (a) Schematic illustration of HB9-EGFP knock-in with HDAdV. The probes for Southern analyses are shown as black bars. a, 5′ probe; b, EGFP probe; c, 3′ probe. Green box, the EGFP gene. Blue box, the neomycin-resistant gene. Red triangle, loxP site. White bar, stuffer DNA for adjusting the vector size. CMV-βgal, the βgal expression cassette; M, MfeI sites. (b) Southern analyses of wild-type parental and the HB9-EGFP knock-in clones (hESC: #47, #G1, and #G10; hiPSC: #24 and #72). Genomic DNA was digested with MfeI. An arrow indicates a signal from the “endogenous” GFP gene, which was encoded by a retroviral vector used for induction of iPSCs. (c) Immunofluorescence analysis of motor neurons differentiated from the HB9-EGFP knock-in hiPSC clone (#24). Bar, 100 µm. EGFP, enhanced green fluorescent protein; HDAdV, helper-dependent adenoviral vector; hESCs, human embryonic stem cells; hiPSCs, human induced pluripotent stem cells.
Figure 5.
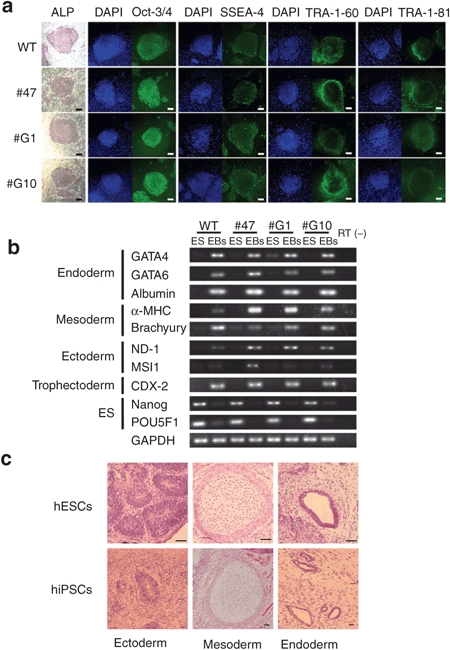
Characterization of HB9-EGFP knock-in hESC and hiPSC clones. (a) Expression of stem cell markers in the HB9-EGFP knock-in hESC clones (#47, #G1, and #G10). ALP, alkaline phosphatase. Bars, 200 mm. (b) Multipotency of the HB9-EGFP knock-in hESC clones. Embryoid bodies (EBs) derived from the hESCs were analyzed by RT-PCR for expression of the lineage-specific markers. GAPDH was used as a control. RT (–), without reverse transcriptase. (c) In vivo differentiation of HB9-EGFP knock-in hESC (#47) and hiPSC (#24) clones. Tissues derived from three germ layers are indicated. Bars, 50 mm. EGFP, enhanced green fluorescent protein; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; RT-PCR, reverse transcription.
The HB9-EGFP knock-in hiPSC line was induced to differentiate into motor neurons to validate differentiation-specific expression of the knocked-in EGFP gene in these reporter cell lines. The neo cassette, which is sandwiched by loxP sites, was excised from the knock-in iPSC clone by transient expression of Cre recombinase (Figure 4a) because the presence of drug-resistant gene may interfere with regulated expression of the reporter gene. After transfection with the pCAGGS-Cre plasmid, followed by analyses of G418 sensitivity and PCR, it was confirmed that the drug-resistant cassette was excised from the HB9 locus in ~50% of the cells (Supplementary Table S1).
The immunofluorescence analysis of HB9-EGFP knock-in hiPSC and hESC lines revealed a correlation between EGFP and HB9 expression (Figure 4c, Supplementary Figure S3). All of the EGFP-positive cells were HB9-positive by immunostaining, suggesting the EGFP gene was precisely knocked into the target chromosomal site by HDAdV-mediated gene targeting. Together, these results demonstrated that HDAdVs are a powerful tool for efficient and accurate gene knockout and knock-in in hESCs and hiPSCs.
Discussion
Various attempts have been made to improve gene-targeting efficiencies in hESCs/hiPSCs. Four methods have been reported, which routinely achieve high relative targeting efficiencies in hESCs/hiPSCs. The first method is electroporation of BAC-based gene-targeting constructs with large homologies.8 The relative gene-targeting efficiencies were ~33% at three housekeeping loci. However, gene targeting at transcriptionally inactive loci has not yet been reported. The second method utilizes the ZFNs, which are fusion proteins of sequence-specific zinc-finger DNA-binding domains and a nonspecific FokI nuclease domain.23,24 Carefully designed ZFNs introduce a chromosomal DSB at the target site, which greatly enhances the frequency of HR repair near the DSB.25 Several groups have validated the use of ZFNs to target chromosomal loci in hESCs/hiPSCs with high relative targeting efficiencies of 6–100%.9,10,11 However, frequent DSBs at off-target sites, accompanied by inaccurate NHEJ repair and ectopic integration of gene-targeting cassettes, have been observed after ZFN treatment in hESCs/hiPSCs and in other cell types.10,11,26,27,28 The third method utilizes adeno-associated virus (AAV) vectors, with the relative targeting efficiencies of 1–30%.12,13,29 The gene-targeting efficiency can also be evaluated by the efficiency per treated cell (we call this the “absolute” targeting efficiency). The absolute targeting efficiency by the BAC-based method is generally low, at 5 × 10−8 to 5 × 10−7 per cell. In the case of the ZFN-mediated method, the absolute efficiencies were ~7 × 10−5 per cell.11 Homozygous mutant hESCs/hiPSCs could also be obtained by a one-step process.10 The absolute efficiencies by the AAV-based method range from 1.9 × 10−7 to 2.4 × 10−4 per cell.29 While both the relative and absolute targeting efficiencies are high, the AAV vectors have limitations for certain applications, such as the small cloning capacity of −4.7 kb and the relative inefficiency of targeting silent loci.29 In addition to these methods, oligonucleotide-mediated gene modification might be another efficient method, which achieved the absolute efficiencies of −2 × 10−4 per cell in mouse ESCs,30 although its application to hESCs/hiPSCs has not been reported.
The fourth method involves delivering gene-targeting cassettes using HDAdV. In this study, we demonstrated the versatility of HDAdV in a variety of gene knockouts (HPRT1, LIG1, LIG3, and KU80) and knock-ins (HB9) in multiple hESC and hiPSC lines. KU80, LIG1, and LIG3 heterozygous mutant hiPSCs will be useful for analyzing the biological functions of the genes in human pluripotent stem cells and, potentially, to achieve improved efficiencies of gene targeting in these mutant hiPSCs. A knock-in of a fluorescent marker gene into the hapatocyte-specific ALB locus also showed similar targeting efficiencies in hESCs/hiPSCs (manuscript in preparation). Our results and those of a recent report on the LMNA locus31 indicate that, as in the case of ZFN-mediated HR, HDAdV-mediated HR is not affected by the transcriptional activities of the target chromosomal loci. The efficiencies of accurate gene targeting varied depending on the target locus and the cell line but remained consistently high. In this study, to determine the accuracy of HR, the candidate targeted clones were screened by PCR, followed by Southern analyses. Long homology arms hamper detailed Southern analyses to examine the accuracy of HR by BAC-mediated methods. Additional ectopic integrations have been observed in ZFN-mediated targeted hES and hiPS clones.10 HDAdVs yielded 3–55% of chromosomal integration of the vectors (G418R colonies) at the target loci, based on PCR analyses (Table 1). These numbers indicate strong affinity of gene-targeting cassettes delivered by HDAdVs with homologous chromosomal sequences even without artificial DNA DSBs. Detailed Southern analyses with multiple probes and enzymes confirmed that 75–100% of these site-specific integration events were mediated by accurate HR (Supplementary Figure S1). Similar detailed analyses of the structure of integrated gene-targeting cassettes delivered by electroporation into the HPRT locus in mouse ES cells reveal that only 5% of 6TGR colonies are generated by accurate HR,32 indicating high fidelity of HDAdV-mediated gene targeting. Overall, the relative gene-targeting efficiencies were 3–55% and 7–81% of G418R colonies and G418R/GANCR colonies, respectively. Furthermore, gene targeting was equally efficient even at a transcriptionally silent target. Although the cells were maintained under drug (G418 and GANC) selection, deleterious effects were not observed, at least by the analyses of the karyotype, pluripotency, and growth rates of the cells (Figure 5, Supplementary Figure S2, and data not shown). More data from other loci will be needed to elucidate why the gene-targeting efficiency is particularly high at the KU80 locus. Importantly, in contrast to ZFN-mediated gene targeting, no additional ectopic chromosomal integrations of the vectors were detected in any of the 47 HDAdV-mediated homologous recombinants (Table 1). Inaccurate homologous recombinants (Figure 3c,d) might have been produced by the mechanism described by Adair et al.33 The absolute gene-targeting frequencies with HDAdVs were 2.0 × 10−7 to 5.6 × 10−5 per infected cell (9.0 × 10−6 on average). Therefore, depending on the target locus, the absolute efficiency with HDAdVs is as high as that obtained by using ZFNs (see above). This also indicates that a 100-mm dish of starting hiPSCs (~3 × 106 cells) is sufficient to obtain multiple gene-targeted clones. Furthermore, in contrast to electroporation, which requires ten million cells for each attempt, HDAdV-mediated gene targeting is equally efficient even with a smaller number of starting cells,34 which minimizes the number of passages/cell divisions to obtain homologous recombinants.
HR has to be accurate for applications such as establishment of gene knock-in iPSC lines to obtain tissue-specific expression of an inserted marker gene upon differentiation. Furthermore, unintended mutations during chromosomal manipulation are obviously not acceptable for therapeutic applications of hESCs/hiPSCs. The characteristics of highly efficient gene-targeting strategies indicate that BAC-mediated gene targeting might be useful for making heterozygote mutants at housekeeping loci because the procedure is relatively simple. ZFN-mediated gene targeting would be useful for making marker gene knock-ins or homozygous mutants because of its relatively high absolute targeting efficiency. Finally, HDAdVs would be the best suited for manipulation of iPSCs for clinical applications, such as gene repair therapy of inherited disorders, because of the highly efficient and accurate gene targeting with minimal cytotoxicity at both transcriptionally active and inactive loci.
Materials and Methods
Construction and preparation of HDAdVs. The human HPRT1-targeting HDAdV was described previously.16 To construct the human HB9-EGFP knock-in HDAdV, 5′ and 3′ homologous arms were amplified from KhES-1 genomic DNA by PCR. The ATG start codon of EGFP gene was fused in-frame with the ATG of HB9 gene. The fragments were then introduced into the pBluescriptII-based vector plasmid encoding the PGK-neo-pA cassette which was sandwiched by loxP sites and then the whole targeting cassette was subcloned into an HDAdV plasmid. To generate the human KU80, LIG1, and LIG3 gene-targeting vectors, BAC clones containing the human KU80, LIG1, and LIG3 loci (BACPAC resources, Oakland, CA) were modified by using the RED/ET recombination technique.35 The loxP71-PGK-EM7-neo-bpA-loxP66 cassette was inserted into the target sites on these BAC clones (Figures 2a and 3a,b). Subsequently, a total of 22–23 kb of homologies including the marker cassette was subcloned into the HDAdV plasmid. A detailed description of these subclonings will be provided on request. The HDAdVs were propagated by serial passages on 293FLPe with addition of FL helper virus (kindly provided by Pedro Lowenstein)36 and purified, as described previously.37 Vector genome titers were determined by quantitative Southern analyses.
Culture of hESCs and hiPSCs. hESC lines, KhES-1 subline 1 and KhES-3,38,39 and hiPSC lines, 246H1 and 246G1 (kindly provided by Shinya Yamanaka),17 were maintained, as previously described.39 246H1 was derived from neonate fibroblast BJ cells with three factors (OCT3/4, SOX2, and KLF4). The hESC lines were used following the hESC research guidelines of the Japanese government.
Isolation of the gene-targeted clones. Clumps of hESCs and hiPSCs were infected with the HDAdVs at a multiplicity of infection of 1,000 vector genomes/cell. Electroporation of plasmid DNA was performed, as reported previously.3 G418 selection (50 µg/ml; Nacalai tesque, Kyoto, Japan) was started 2 days after infection or electroporation. After 3 weeks, surviving colonies were transferred to 96-well plates and GANC selection (2 µmol/l; Invitrogen, Carlsbad, CA) was started. For gene targeting at HRPT1, 6TG (10 mmol/l; Sigma-Aldrich, St Louis, MO) selection was applied after the G418 selection and the 6TG-resistant clones were analyzed. For other loci, G418/GANC double-resistant clones were characterized. Genomic DNA of drug-resistant clones was subject to screening by PCR using primers shown in Table 2 with LA Taq Hot Start Version (TAKARA, Kyoto, Japan) or PrimeSTAR GXL polymerase (TAKARA), following the manufacturer's protocol. To analyze the structure of HB9, KU80, LIG1, and LIG3 loci, DNA was extracted from PCR-positive clones. The accuracy of gene targeting was carefully evaluated by the Southern analyses using restriction enzyme(s), which digested outside the homology arms and also inside the marker gene cassettes, but not within the homology arms. The 5′ and 3′ probes were located between the homology arms and the restriction enzyme sites. Restriction enzymes and probes for Southern hybridization are indicated in Figures 1a, 2a, 3a,b, and 4a. Additional ectopic integration of the vector is therefore expected to produce an extra band with the neo or GFP probe.
Table 2. Primer sequences used in this study.

For the chromosomal analyses, gene knockout and knock-in hES clones were treated with 100 ng/ml colcemid (Invitrogen) for 2 hours, trypsinized, incubated in 0.075 mol/l KCl for 14 minutes, and fixed in Carnoy's fixative. The cells were spread onto glass slides and stained with Giemsa. Chromosome spreads were analyzed by randomly counting 50 cells using the Ikaros Karyotyping System (MetaSystems, Waltham, MA).
Excision of the neomycin-resistant gene cassette in targeted clones. Clumps of hiPSCs were plated feeder-free and transfected with a pCAGGS-Cre plasmid, the CAG promoter-driven Cre gene, using FuGENE HD (Roche, Basel, Switzerland), according to the manufacturer's instructions. Six days later, the cells were sorted into single cells in the presence of Rho-associated coiled kinase (ROCK) inhibitor Y-27632 (10 µmol/l; Wako, Osaka, Japan) onto 96-well plates by using FACS Aria II (BD, Franklin Lakes, NJ). The removal of the neo cassette was confirmed by G418 sensitivity, PCR, and Southern hybridization, and the surviving colonies were used for motor neuron differentiation.
Motor neuron differentiation. Motor neuron induction was performed, as previously described with some modifications.40 Briefly, dissociated iPSC colonies were plated onto culture dishes coated with poly-L-lysine/laminin (PLL/LM; Sigma-Aldrich) in N2B27 neural differentiation medium (1:1 mix of Dulbecco's modified Eagle medium /F12 supplemented with N2 and neurobasal medium supplemented with B27, all from Life Technologies, Carlsbad, CA), supplemented with mouse recombinant Noggin (100 ng/ml; R&D Systems, Minneapolis, MN) for 10 days. Subsequently, colonies were split into small clumps by treatment with Collagenase (200 U/ml; Life Technologies) with 1 mmol/l CaCl2 and cultured on new poly-L-lysine/laminin-coated dishes with N2B27 supplemented with Noggin for 7 additional days. The cells were, then, dissociated by Accutase (Innovative Cell Technologies, San Diego, CA) and cultured on dishes coated with poly-L-lysine/laminin plus human plasma Fibronectin (PL/LM/FN) (Chemicon, Billerica, MA) in N2B27 supplemented with 1 µmol/l of retinoic acid (RA; Sigma-Aldrich) for 4 days, and then cultured in N2B27 supplemented with retinoic acid and human recombinant sonic hedgehog (500 nmol/l; R&D Systems) for additional 7–9 days. The cells were again dissociated by Accutase and cultured on poly-L-lysine/laminin plus human plasma Fibronectin-coated dishes in N2B27 supplemented with brain-derived neurotrophic factor (10 ng/ml; R&D Systems), glial cell-derived neurotrophic factor (10 ng/ml; R&D Systems), and neurotrophin-3 (10 ng/ml; R&D Systems) for another 5 days. Finally, the cells were fixed with 4% paraformaldehyde and subjected to direct visualization of EGFP and immunocytochemistry of HB9 protein, as previously described.40
Quantitative RT-PCR assay and western blot assay. RNA extraction and cDNA synthesis for quantitative RT-PCR were carried out as previously described.16 The ABI PRISM 7000 sequence detection system (Applied Biosystems, Carlsbad, CA) was used for quantitative RT-PCR. Predesigned primers and probe sets for KU80 (Hs00221707_m1), LIG1 (Hs01553527_m1), LIG3 (Hs00242692_m1), and GAPDH (Hs99999905_m1) were obtained from Applied Biosystems. Western blot assays were performed with anti-KU80 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA), anti-LIG1 (10H5; Abcam, Cambridge, MA), anti-LIG3 (1F3; Abcam), or anti-α-tubulin (DM1A; Santa Cruz Biotechnology) antibodies. The membranes were subsequently probed with a horseradish peroxidase-conjugated secondary antibody (anti-goat IgG-HRP or anti-mouse IgG-HRP; Santa Cruz Biotechnology) and developed with ECL PLUS (GE Healthcare, Piscataway, NJ).
Analyses of the stem cell markers and pluripotency. Expression of alkaline phosphatase and surface markers, such as SSEA-4, TRA-1-60, and TRA-1-81, was examined, as previously described.16 Formation of embryoid bodies, RNA extraction, and cDNA synthesis were previously described.16 The sequence of Albumin-specific primers are shown in Table 2. The sequences of other primers were previously described.16 Formation of teratoma and hematoxylin and eosin staining were performed with HB9-EGFP knock-in hESC and hiPSC clones, as previously described.39 Animal protocols were approved by the Institutional Board on Animal Care.
SUPPLEMENTARY MATERIAL Figure S1. Examples of Southern analyses. Figure S2. Karyotype analyses. Figure S3. Motor neurons differentiated from the HB-9-EGFP knock-in hESC clone. Table S1. The number of colonies in which the selection marker was excised.
Acknowledgments
We thank the members of Mitani lab, Haruyoshi Takaki (Teikyo University) and Mizuki Ohno (Kyushu University), for helpful discussions, Ayako Tokumitsu (Saitama Medical University) for technical supports, Tomoaki Hishida (Saitama Medical University) for advice on quantitative RT-PCR and western blotting, Pedro R. Lowenstein (Cedars-Sinai Medical Center) for providing 293FLPe cells and FL helper virus, and Shinya Yamanaka (Kyoto University) for providing human iPSC lines (246G1 and 246H1). This work was supported by national funds from Development of Technology to Create Research Model Cells Project of New Energy and Industrial Technology Development Organization (NEDO) to N.N. and K.M. and, in part, by a grant-in-aid for “Support Project of Strategic Research Center in Private Universities” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to Research Center for Genomic Medicine, Saitama Medical University. The authors declare no conflict of interest.
Supplementary Material
Examples of Southern analyses.
Karyotype analyses.
Motor neurons differentiated from the HB-9-EGFP knock-in hESC clone.
The number of colonies in which the selection marker was excised.
REFERENCES
- Nieminen M, Tuuri T., and, Savilahti H. Genetic recombination pathways and their application for genome modification of human embryonic stem cells. Exp Cell Res. 2010;316:2578–2586. doi: 10.1016/j.yexcr.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Zembowicz F., and, Cowan CA. Genome modification in human embryonic stem cells. J Cell Physiol. 2010;222:278–281. doi: 10.1002/jcp.21948. [DOI] [PubMed] [Google Scholar]
- Zwaka TP., and, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M., and, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- Di Domenico AI, Christodoulou I, Pells SC, McWhir J., and, Thomson AJ. Sequential genetic modification of the hprt locus in human ESCs combining gene targeting and recombinase-mediated cassette exchange. Cloning Stem Cells. 2008;10:217–230. doi: 10.1089/clo.2008.0016. [DOI] [PubMed] [Google Scholar]
- Ruby KM., and, Zheng B. Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Shimoji M, Tahimic CG, Aiba K, Kawase E, Hasegawa K.et al. (2010Efficient integration of transgenes into a defined locus in human embryonic stem cells Nucleic Acids Res 38e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Chung SK., and, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA.et al. (2007Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery Nat Biotechnol 251298–1306. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC.et al. (2009Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases Nat Biotechnol 27851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK.et al. (2009Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells Cell Stem Cell 597–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Suzuki K, Aizawa E, Kawase E, Suemori H, Nakatsuji N.et al. (2009Gene targeting in human pluripotent stem cells with adeno-associated virus vectors Biochem Biophys Res Commun 388711–717. [DOI] [PubMed] [Google Scholar]
- Khan IF, Hirata RK, Wang PR, Li Y, Kho J, Nelson A.et al. (2010Engineering of human pluripotent stem cells by AAV-mediated gene targeting Mol Ther 181192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani K, Graham FL, Caskey CT., and, Kochanek S. Rescue, propagation, and partial purification of a helper virus-dependent adenovirus vector. Proc Natl Acad Sci USA. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DJ., and, Ng P. Helper-dependent adenoviral vectors for gene therapy. Hum Gene Ther. 2005;16:1–16. doi: 10.1089/hum.2005.16.1. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T.et al. (2008Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors Proc Natl Acad Sci USA 10513781–13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K.et al. (2007Induction of pluripotent stem cells from adult human fibroblasts by defined factors Cell 131861–872. [DOI] [PubMed] [Google Scholar]
- Bertolini LR, Bertolini M, Maga EA, Madden KR., and, Murray JD. Increased gene targeting in Ku70 and Xrcc4 transiently deficient human somatic cells. Mol Biotechnol. 2009;41:106–114. doi: 10.1007/s12033-008-9098-8. [DOI] [PubMed] [Google Scholar]
- Fattah FJ, Lichter NF, Fattah KR, Oh S., and, Hendrickson EA. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc Natl Acad Sci USA. 2008;105:8703–8708. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison KA, Druey KM, Deguchi Y, Tuscano JM., and, Kehrl JH. A novel human homeobox gene distantly related to proboscipedia is expressed in lymphoid and pancreatic tissues. J Biol Chem. 1994;269:19968–19975. [PubMed] [Google Scholar]
- Arber S, Han B, Mendelsohn M, Smith M, Jessell TM., and, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- Thaler J, Harrison K, Sharma K, Lettieri K, Kehrl J., and, Pfaff SL. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH., and, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH., and, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Händel EM., and, Cathomen T. Zinc-finger nuclease based genome surgery: it's all about specificity. Curr Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- Gupta A, Meng X, Zhu LJ, Lawson ND., and, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PA, Gelazauskaite M, Randøl M., and, Krauss S. Analysis of illegitimate genomic integration mediated by zinc-finger nucleases: implications for specificity of targeted gene correction. BMC Mol Biol. 2010;11:35. doi: 10.1186/1471-2199-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radecke S, Radecke F, Cathomen T., and, Schwarz K. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol Ther. 2010;18:743–753. doi: 10.1038/mt.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IF, Hirata RK., and, Russell DW. AAV-mediated gene targeting methods for human cells. Nat Protoc. 2011;6:482–501. doi: 10.1038/nprot.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarts M., and, te Riele H. Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Ther. 2011;18:213–219. doi: 10.1038/gt.2010.161. [DOI] [PubMed] [Google Scholar]
- Liu GH, Suzuki K, Qu J, Sancho-Martinez I, Yi F, Li M.et al. (2011Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs Cell Stem Cell 8688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Rivera-Pérez J, Chang C., and, Bradley A. Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol Cell Biol. 1991;11:4509–4517. doi: 10.1128/mcb.11.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair GM, Nairn RS, Wilson JH, Seidman MM, Brotherman KA, MacKinnon C.et al. (1989Targeted homologous recombination at the endogenous adenine phosphoribosyltransferase locus in Chinese hamster cells Proc Natl Acad Sci USA 864574–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi F, Balamotis MA, Kishimoto A, Aizawa E, Diaz A, Hasty P.et al. (2005Correction of chromosomal mutation and random integration in embryonic stem cells with helper-dependent adenoviral vectors Proc Natl Acad Sci USA 10213628–13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA., and, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umaña P, Gerdes CA, Stone D, Davis JR, Ward D, Castro MG.et al. (2001Efficient FLPe recombinase enables scalable production of helper-dependent adenoviral vectors with negligible helper-virus contamination Nat Biotechnol 19582–585. [DOI] [PubMed] [Google Scholar]
- Palmer DJ., and, Ng P. Physical and infectious titers of helper-dependent adenoviral vectors: a method of direct comparison to the adenovirus reference material. Mol Ther. 2004;10:792–798. doi: 10.1016/j.ymthe.2004.06.1013. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Fujioka T, Nakamura Y, Nakatsuji N., and, Suemori H. A method for the selection of human embryonic stem cell sublines with high replating efficiency after single-cell dissociation. Stem Cells. 2006;24:2649–2660. doi: 10.1634/stemcells.2005-0657. [DOI] [PubMed] [Google Scholar]
- Suemori H, Yasuchika K, Hasegawa K, Fujioka T, Tsuneyoshi N., and, Nakatsuji N. Efficient establishment of human embryonic stem cell lines and long-term maintenance with stable karyotype by enzymatic bulk passage. Biochem Biophys Res Commun. 2006;345:926–932. doi: 10.1016/j.bbrc.2006.04.135. [DOI] [PubMed] [Google Scholar]
- Wada T, Honda M, Minami I, Tooi N, Amagai Y, Nakatsuji N.et al. (2009Highly efficient differentiation and enrichment of spinal motor neurons derived from human and monkey embryonic stem cells PLoS ONE 4e6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K., and, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of Southern analyses.
Karyotype analyses.
Motor neurons differentiated from the HB-9-EGFP knock-in hESC clone.
The number of colonies in which the selection marker was excised.


