Abstract
The reprogramming of cord blood (CB) cells into induced pluripotent stem cells (iPSCs) has potential applications in regenerative medicine by converting CB banks into iPSC banks for allogeneic cell replacement therapy. Therefore, further investigation into novel approaches for efficient reprogramming is necessary. Here, we show that the lentiviral expression of OCT4 together with SOX2 (OS) driven by a strong spleen focus-forming virus (SFFV) promoter in a single vector can convert 2% of CB CD34+ cells into iPSCs without additional reprogramming factors. Reprogramming efficiency was found to be critically dependent upon expression levels of OS. To generate transgene-free iPSCs, we developed an improved episomal vector with a woodchuck post-transcriptional regulatory element (Wpre) that increases transgene expression by 50%. With this vector, we successfully generated transgene-free iPSCs using OS alone. In conclusion, high-level expression of OS alone is sufficient for efficient reprogramming of CB CD34+ cells into iPSCs. This report is the first to describe the generation of transgene-free iPSCs with the use of OCT4 and SOX2 alone. These findings have important implications for the clinical applications of iPSCs.
Introduction
The ability to generate induced pluripotent stem cells (iPSCs) from somatic cells has opened up a new avenue for regenerative medicine. Earlier studies used fibroblasts, such as those derived from a skin biopsy, to generate iPSCs by overexpression of Yamanaka factors (OCT4, SOX2, MYC and KLF4, or OSMK) or Thomson/Yu factors (OCT4, SOX2, NANOG, and LIN28).1,2 However, it takes several weeks to prepare cells from a skin biopsy for reprogramming.1,3 Later, hematopoietic stem/progenitor cells or CD34+ cells from mobilized peripheral blood, bone marrow, or cord blood (CB) captured much attention because blood cells can be used immediately for reprogramming.4,5,6 However, isolation of mobilized peripheral blood and bone marrow is invasive, time consuming and has potential risks for the donor, while harvesting CB cells has none of these limitations. In addition, >400,000 fully characterized and HLA-typed CB units are stored in public banks and are readily available for clinical therapy.7 Moreover, CB has the youngest somatic cells and is expected to carry minimal genetic mutations induced by UV radiation.8,9 Due to its unique advantages as donor cells for the production of clinical-grade human iPSCs, CB is believed to be one of the best sources for reprogramming. An additional advantage is the potential of converting CB banks into iPSC banks for allogeneic cell-based therapy.10
For clinical applications, transgene-free or footprint-free iPSCs need to be used to prevent potential adverse effects due to retroviral or lentiviral integration or due to the interference of residual expression of reprogramming factors on the differentiation of iPSCs into progenies of clinical interest.11,12 Toward this goal, several approaches have been used for obtaining integration or transgene-free iPSCs, including the use of plasmids,13 the Cre/loxP system,14,15 adenoviruses,16,17 piggyBac transposon,18,19 minicircle DNA,20 protein transduction,21,22 Sendai virus,23 and miRNA.24 However, these methods suffer from low efficiency, require repetitive induction or selection, or require virus production. Synthetic modified mRNA might solve the problem,25 but it requires the daily addition of mRNA by lipofection and CB CD34+ cells are among the most difficult to transfect by lipofection.
Several investigators have used the EBNA1-based episomal vector due to its unique features: (i) only one transfection of vector DNA by nucleofection is needed for efficient reprogramming, and (ii) the vector is lost in 5% or more cells after each cell division, leading to depletion of the episomal vector from cells after long-term passage. Recently, several groups have successfully used the pCEP4 episomal vector to generate footprint-free iPSCs.26,27,28 However, in those studies, five to seven factors, including strong oncogenes like MYC and/or simian virus 40 large T antigen (SV40LT) were used, which raises safety concerns for the clinical use of iPSCs.
Earlier studies showed that OCT4 and SOX2 alone can reprogram CB cells into iPSCs, but at a very low efficiency.9 We hypothesized that reprogramming efficiency might depend on expression levels of reprogramming factors, which largely relies on the promoters used. It is well known that the strength of promoters is contextual; several studies have shown that the spleen focus-forming virus (SFFV) promoter is stronger in primary hematopoietic cells or hematopoietic cell lines than many commonly used promoters like human elongation factor 1α (EF1α), cytomegalovirus, and A2UCOE (ubiquitous chromatin opening element).29,30,31,32 Thus, we set out to determine whether iPSCs can be efficiently generated from CB CD34+ cells with the SFFV promoter being used to drive expression of OCT4 and SOX2.
Results
Balanced expression of OCT4 and SOX2 by a lentiviral vector efficiently reprograms CB CD34+ cells into iPSCs
It has been reported that overexpression of OCT4 together with SOX2 (O+S) using a retroviral vector in 2 individual constructs can reprogram CB CD133+ cells into iPSCs.9 However, the efficiency is as low as 0.002–0.005%, making this approach impractical for many applications. We hypothesized that the low efficiency might be due to low-level expression of the reprogramming factors O+S mediated by retroviral vectors. To test this assumption, we cloned reprogramming factors into a lentiviral vector driven by a strong promoter SFFV (Figure 1a).
Figure 1.
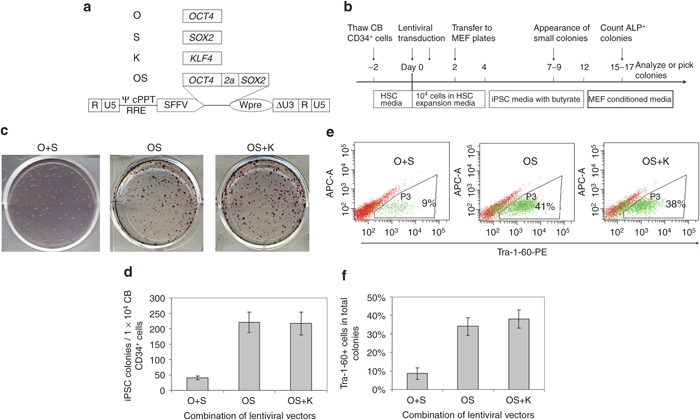
Lentiviral vector-mediated expression of OCT4 and SOX2 efficiently reprogram cord blood (CB) CD34+ cells into induced pluripotent stem cells (iPSCs). (a) Schematic of the self-inactivating (SIN) lentiviral vector backbones for expression of the human reprogramming factor OCT4, SOX2, KLF4. Δ indicates the SIN design with partially deleted U3 of the 3′ long-terminal repeat. cPPT, central polypurine tract; RRE, rev-responsive element; SFFV, spleen focus-forming virus U3 promoter; Wpre, woodchuck post-transcriptional regulatory element; ψ, packaging signal; 2a, a self-cleavage site derived from equine rhinitis A virus. (b) Experimental strategy for reprogramming human CB CD34+ cells using lentiviral vectors. (c) Representative alkaline phosphatase (ALP) staining of iPSC colonies 16 days after lentiviral transduction of 1 × 104 CB CD34+ cells. O, OCT4; S, SOX2; K, KLF4. (d) Numbers of induced pluripotent stem cells (iPSCs) generated from 1 × 104 CB CD34+ cells. n = 3. O+S vs. OS: P < 0.05. OS vs. OS+K: no significant difference. Data shown are presented as mean ± SEM. (e) Representative fluorescence-activated cell sorting (FACS) diagram of TRA-1-60 expression in cells undergoing reprogramming. Cells at day 16 after transduction were harvested and analyzed. (f) Percentages of TRA-1-60 positive cells in reprogramming cultures. O+S vs. OS: P < 0.05; OS vs. OS+K: no significant difference. Data shown are presented as mean ± s.e.m. (n = 3).
As detailed in Figure 1b and the Materials and Methods section, CB CD34+ cells were transduced with lentiviral vectors that express reprogramming factors followed by iPSC generation by culturing transduced cells on mouse embryonic fibroblasts (MEFs). Of interest, in the O+S condition, dozens of small colonies were observed in each well as early as 4–5 days after seeding transduced CB cells onto MEF layers, however, morphologically iPSC-like cells did not appear until a week later (data not shown). Analysis of these non-iPSCs by flow cytometry indicated that many cells expressed mesenchymal markers (data not shown). We also tested the combination of OCT4 and SOX2 (abbreviated as OS for clarity) in a single vector with the use of self-cleavage peptide sequence 2a. In this condition, no colonies were observed in the first week, and the first iPSC-like colonies appeared at 8–10 days after CB transduction. These data suggest that balanced expression of OCT4 and SOX2 may inhibit the outgrowth of non-iPSCs.
In the O+S condition, we routinely observed 300–600 total colonies from 10,000 transduced CB CD34+ cells 2 weeks after transduction. However, the majority of colonies were morphologically non-iPSCs and alkaline phosphatase (ALP) staining showed that ~20% of the colonies were iPSC-like (Figure 1c). In the OS condition, we observed 200–250 colonies in each well, with ~80% of the colonies being morphologically iPSCs, which was further confirmed by ALP staining (Figure 1c,d). In agreement with these results, fluorescence-activated cell sorting (FACS) analysis of the cells in the reprogramming cultures showed that only 9% of the cells in the O+S condition expressed the iPSC marker TRA-1-60, whereas ~40% of the cells in the OS condition were TRA-1-60 positive (Figure 1e,f).
Together, our findings demonstrate that OCT4 and SOX2 alone can efficiently reprogram CB cells into iPSCs and that balanced expression of the two factors that are linked with a 2a self-cleavage peptide sequence can increase reprogramming efficiency and inhibit growth of non-iPSC colonies.
KLF4 does not increase efficiency of lenti SFFV-OS-mediated reprogramming
Because the use of additional factors has been shown to boost reprogramming efficiency, we tested the effects of including other factors like KLF4 in reprogramming. In sharp contrast to expectations, we found that the addition of KLF4 (K) to OS did not increase the reprogramming efficiency. This surprising finding is unlikely to be explained by differential expression levels of reprogramming factors because the same OS vector was used in both conditions, and the expression of KLF4 was confirmed in preliminary studies. In OS conditions with and without K, 2% of transduced CB cells were successfully converted into iPSCs and ~40% of cells in the reprogramming culture expressed the iPSC marker TRA-1-60 (Figure 1c–f). This data suggests that the expression of OS, driven by the SFFV promoter, is sufficient to reprogram CB CD34+ cells at high efficiency and addition of other factors like KLF4 does not significantly increase the reprogramming efficiency.
Efficiency of OS-mediated reprogramming depends on OS expression levels
Having observed up to a 1,000-fold higher efficiency in converting CB cells into iPSCs by OS compared to the previous report,9 we speculated that differences in the expression levels of OS might explain the large difference in reprogramming efficiency. Transgene expression levels are largely determined by the strength of promoters; we thus cloned lentiviral vectors in which green fluorescent protein (GFP) expression is driven by the PGK, EF1, or the SFFV promoter to determine the strength of these promoters in CD34+ cells (Figure 2a). FACS analysis showed that GFP expression driven by the PGK or the EF1 promoters is ~85% or ~60% lower than expression driven by the SFFV promoter in CB CD34+ cells (Figure 2b,c). We reasoned that GFP is more stable than transcription factors; the GFP intensity may not reflect OCT4 or SOX2 expression levels. To address this issue, we cloned OCT4GFP fusion gene-expressing vectors driven by the three promoters. In this system, GFP is fused to the protein of interest. Thus the GFP expression, as measured by fluorescence intensity, can reflect the expression level of its fusion partner.33 Similarly, we observed that the SFFV promoter drove highest level expression of OCT4GFP in CB CD34+ cells, followed by the EF1 and the PGK promoters (Figure 2d). Of note, GFP intensity was decreased by ~20-fold in OCT4GFP-transduced cells, as compared to GFP-transduced cells, and the differences in expression of OCT4GFP were less pronounced than that of GFP, which reflect the rapid turnover of OCT4 in CB CD34+ cells. Together, these data suggest that the SFFV promoter drives significantly higher levels of transgene expression in CB CD34+ cells than the PGK or EF1 promoters.
Figure 2.
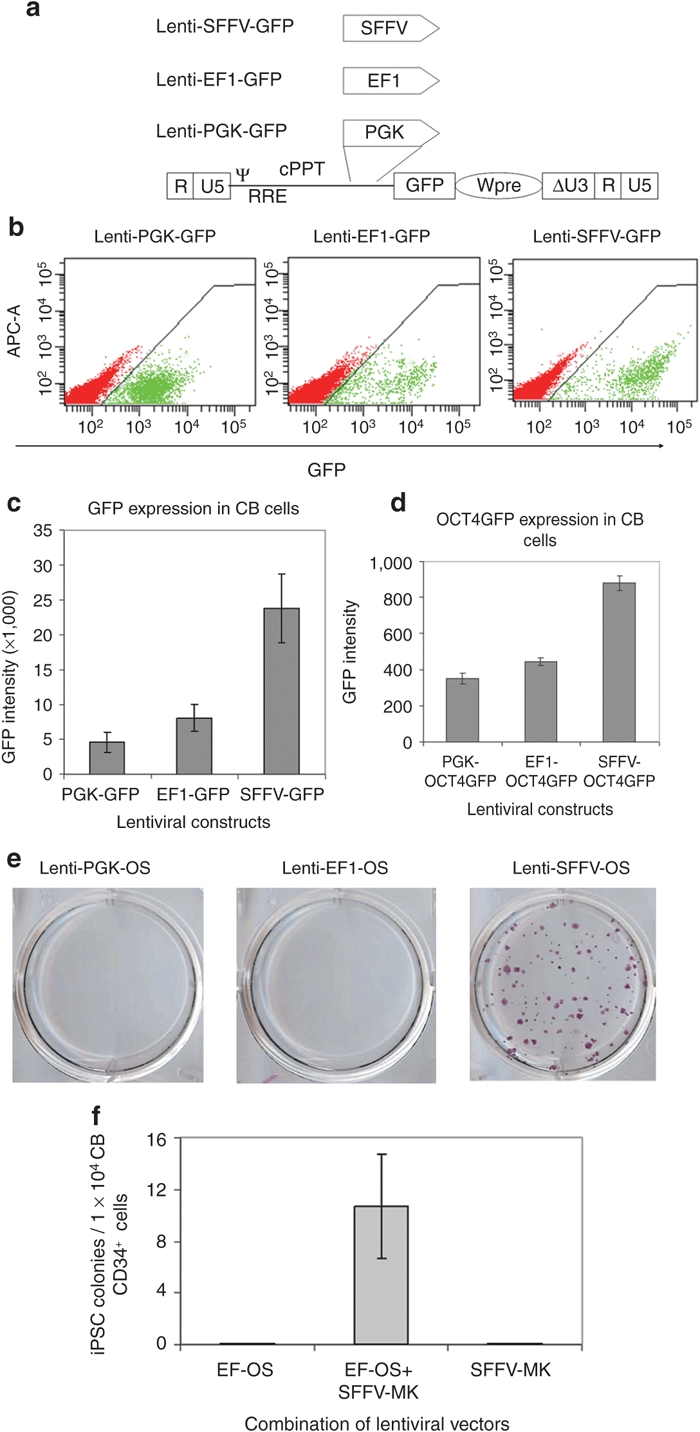
Efficiency of OCT4 and SOX2-mediated reprogramming depends on gene expression levels. (a) Schematic of the self-inactivating (SIN) lentiviral vector backbones for expression of green fluorescent protein (GFP). Δ indicates the SIN design with partially deleted U3 of the 3′ long-terminal repeat. cPPT, central polypurine tract; EF1, elongation factor-1α promoter; PGK, phosphoglycerokinase promoter; RRE, rev-responsive element; SFFV, spleen focus-forming virus U3 promoter; Wpre, post-transcriptional regulatory element; ψ, packaging signal. (b) Representative levels of GFP expression driven by three different promoters in cord blood (CB) CD34+ cells. Fluorescence-activated cell sorting (FACS) analysis was conducted at 3 days post-transduction. (c) Distinct GFP expression levels driven by three different promoters in CB CD34+ cells. n = 3. PGK-GFP vs. EF-GFP: P = 0.05; EF-GFP vs. SFFV-GFP: P < 0.05. (d) Increased expression of OCT4GFP fusion gene driven by SFFV promoter compared to PGK and EF1 in CB CD34+ cells. FACS analysis was conducted at 3 days post-transduction. n = 3. PGK-OCT4GFP vs. EF1-OCT4GFP: P = 0.06; EF1-OCT4GFP vs. SFFV-OCT4GFP: P < 0.01. (e) Alkaline phosphatase (ALP) staining for iPSC cultures from CB cells transduced with PGK-OS, EF1-OS, and SFFV-OS. Note that no colonies were generated in PGK-OS, EF1-OS conditions. (f) Expression of MYC and KLF4 rescues failure of low level OS expression driven by EF1 promoter in generating induced pluripotent stem cells (iPSCs) from CB CD34+ cells. Graphed data are presented as mean ± SEM (n = 3).
To investigate the effects of low OS expression on reprogramming efficiency, we used the weaker PGK and EF1 promoters to drive OS expression. In more than five independent experiments, no iPSC colonies could be generated from 1 × 104 CB CD34+ cells that were transduced with lenti PGK-OS or lenti EF1-OS vectors (Figure 2e). Given that expression of OCT4 is decreased by ~50% when driven by EF1 as compared to the SFFV promoter (Figure 2d), this observation suggests that a 50% decrease in OS expression could lead to reprogramming failure. In hopes of increasing OS expression and thereby reprogramming efficiency, we synthesized an OS gene (synOS) that was codon optimized by DNA 2.0 (Menlo Park, CA). In contrast to our expectation, expression of OS at the protein level by synOS was ~20% lower than the wild-type human OS. Of note, this small decrease in OS expression translated into a fourfold decrease in reprogramming efficiency (data not shown). This observation further supports our conclusion that OS-mediated high-efficiency reprogramming critically depends on OS expression levels, and a slight decrease in OS expression leads to a substantial drop in reprogramming efficiency, whereas a 50% decrease results in reprogramming failure.
MYC and KLF4 facilitate reprogramming when OS expression levels are low
Having found that low-level OS expression is insufficient to induce CB reprogramming, we further asked whether this can be rescued by MYC and KLF4. As anticipated, in CB CD34+ cells that were transduced with EF1-OS or SFFV-MK alone, no iPSCs were generated. In contrast, after transduction of CB CD34+ cells with both EF1-OS and SFFV-MK, 0.1% cells were converted into iPSCs (Figure 2f). ALP staining and FACS analysis of iPSCs did not show any obvious differences in the expression of pluripotency markers when compared with iPSCs generated with SFFV-OS (data not shown). Of interest, when MYC and KLF4 expression was driven by the EF1 promoter, which leads to lower expression levels, no iPSCs could be generated (data not shown). Together, these findings suggest that high-level expression of OS alone is sufficient for CB reprogramming, whereas reprogramming under low-level OS expression requires other reprogramming factors.
Generation of footprint-free iPSCs using an episomal vector
The successful generation of iPSCs with a lentiviral vector that expresses OCT4 and SOX2 alone prompted us to ask whether this approach would also work in a nonviral system. To test this, we shuttle cloned SFFV-OS from the lentiviral vector construct into a pCEP4 EBNA1/OriP-based episomal vector (Figure 3a). To generate iPSCs, 1 × 105 CB CD34+ cells were cultured in Iscove's modified Dulbecco's medium/10% fetal bovine serum with cytokines SCF, FL, and TPO. After 3 days of culture, the total cell number increased by approximately fivefold and all the cells were harvested for nucleofection with the pCEP-OS (w/o W) plasmid (Figure 3b). In three independent experiments, we failed to generate any iPSCs (left panel of Figure 3c). We reasoned that this failure might be due to the low-level expression of OS mediated by this vector. We then cloned woodchuck post-transcriptional regulatory element (Wpre), a post-transcriptional regulatory element that is commonly used in lentiviral systems to enhance gene expression levels, into the pCEP-OS (w/o W) plasmid (Figure 3a). As expected, the inclusion of Wpre in the episomal vector led to a 50% increase in OCT4 expression and a 55% increase in SOX2 expression (Figure 3d). Using pCEP-OS, we successfully generated ~20 iPSC colonies from the progeny of 1 × 105 freshly thawed CB CD34+ cells (Figure 3c,e).
Figure 3.
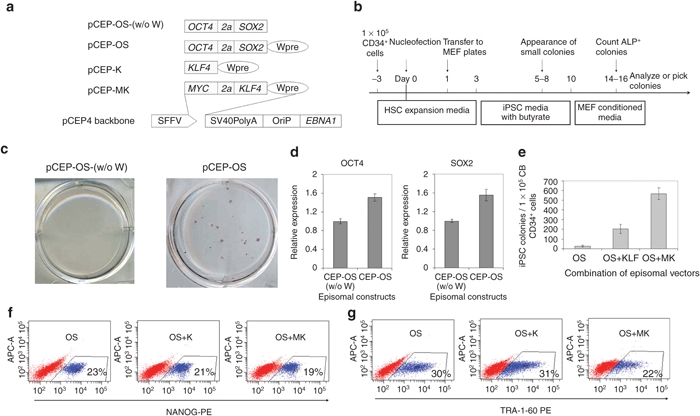
OCT4 and SOX2-mediated reprogramming using episomal vectors. (a) Schematic of episomal vectors used in this study for conversion of cord blood (CB) CD34+ cells into induced pluripotent stem cells (iPSCs). Reprogramming factors were cloned into the pCEP4 backbone; their expression is driven by spleen focus-forming virus U3 promoter (SFFV). 2a is a self-cleavage site derived from equine rhinitis A virus. Wpre, post-transcriptional regulatory element; SV40PolyA, polyadenylation signal from SV40 virus; OriP, EBV origin of replication; EBNA1, Epstein–Barr nuclear antigen 1, which plays essential roles in replication and persistence of episomal plasmid in infected cells. (b) Experimental strategy for reprogramming human CB CD34+ cells using EBNA1-based episomal vectors. (c) Representative alkaline phosphatase (ALP) staining shows that inclusion of the Wpre element in the episomal vector pCEP-OS (w/o W) results in successful reprogramming. n = 3. Colonies are from 1 × 105 CB CD34+ cells. (d) Inclusion of Wpre element in the CEP episomal vector increases gene expression. 293T cells were infected with same amount of plasmids. 3 days after transfection, OCT4 and SOX2 expression was examined by intracellular staining and fluorescence-activated cell sorting (FACS) analysis. n = 3. pCEP-OS (w/o W) vs. pCEP-OS: P < 0.05. (e) Numbers of ALP positive iPSC colonies at 16 days post-transfection of 1 × 105 CB CD34+ cells with pCEP-OS (OS) and pCEP-K (K) or pCEP-MK (MK). n = 3. OS vs. OS+K: P < 0.05; OS+K vs. OS+MK: P < 0.05. Expression of the iPSC markers (f) NANOG and (g) TRA-1-60 in cultures reprogrammed using three different combinations of episomal vectors. Cells were harvested for FACS analysis 20 days after nucleofection.
To better compare our improved vector with published results, we evaluated the effects of KLF4 or MK (MYC and KLF4) together with OS on the efficiency of CB reprogramming. With the addition of KLF4, the reprogramming efficiency increased by eightfold, and further inclusion of MYC led to an additional threefold increase (Figure 3e). Of interest, the appearance of the first iPSC-like colonies was observed at 9–10, 6–7, and 4–5 days after cells were transfected with episomal OS, OS+K, and OS+MK plasmids, respectively. This data suggests that addition of KLF4 and/or MYC accelerates the reprogramming process. Of note, using two episomal vectors that express four factors, we generated up to 600 iPSC colonies from 1 × 105 CB CD34+ cells, compared to 80 colonies from the same amount of CB CD34+ cells even with five factors (OSMK + LIN28).26 These data suggest that our improved episomal vector is substantially more efficient in reprogramming CB cells into iPSCs than previously reported.
We conducted further tests to examine the differences in the expression of pluripotency markers between iPSCs generated with the three different combinations of episomal vectors. Immunostaining and FACS analysis showed that 20–30% of cells expressed the iPSC markers NANOG and TRA-1-60 in all the three combinations, whereas including MYC appeared to decrease the portion of Tra-1-60 positive iPSCs in reprogramming culture (Figure 3f,g).
Taken together, these data demonstrate that we have developed an episomal vector in which increased expression of reprogramming factors leads to efficient reprogramming of CB cells into iPSCs. We show for the first time, that iPSCs can be generated with the episomal vector that expresses only OCT4 and SOX2.
Characterization of iPSC colonies generated with the pCEP-OS plasmid
To characterize iPSCs, we randomly picked 10 colonies from the pCEP-OS reprogrammed cultures and passaged iPSCs for >3 months. Real-time PCR analysis with two pairs of primers showed that at passage 0, ~0.5 copy of the pCEP-OS plasmid per cell could be detected. After eight passages, the average copy number of residual CEP plasmid decreased to 0.001–0.007/genome and in 2 out of 10 clones, the presence of CEP plasmid was undetectable (Figure 4a). After 12 passages, residual episomal plasmid was disappeared in the majority of clones (data not shown). This finding is consistent with previous reports showing that the presence of episomal vector is undetectable in most iPSC colonies after 10–14 passages.26
Figure 4.
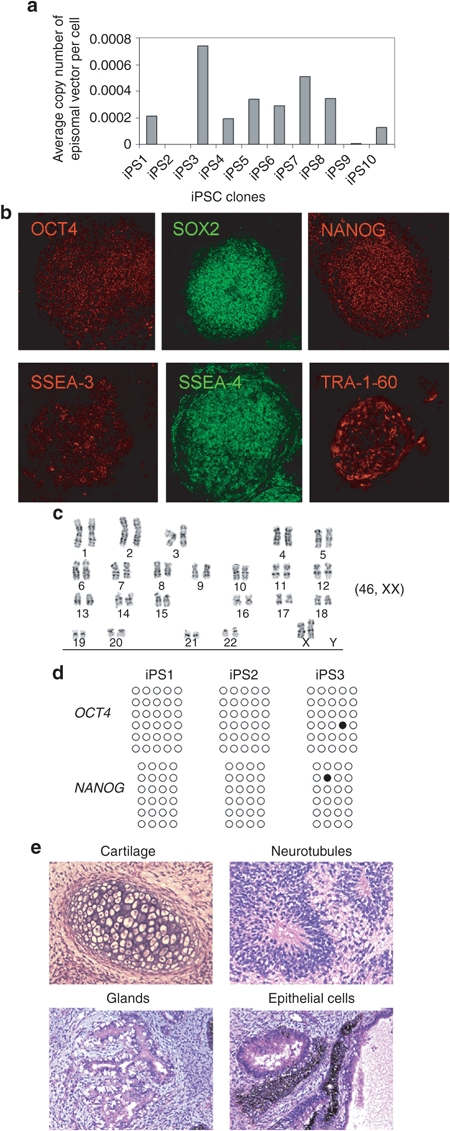
Characterization of induced pluripotent stem cells (iPSCs) generated with pCEP-OS. (a) Copies of residual episomal vectors after eight passages as indicated by real-time PCR. Data shown are from one pair of primers. Similar results were obtained with second pair of primers. (b) Immunohistochemistry analysis of a representative iPSC line showing expression of indicated pluripotency markers. Images were captured using the Zeiss LSM 710 confocal microscope with a ×10 objective. (c) A representative karyogram of an iPSC clone. All analyzed iPSC clones showed a normal karyotype. (d) Bisulphite genomic sequencing of the OCT4 and NANOG promoters indicates demethylation in three independent clones. Each horizontal row of circles represents an individual sequencing reaction of a given amplicon. Open and filled circles represent unmethylated and methylated CpG dinucleotides, respectively. (e) Hematoxylin and eosin (H&E) staining of representative teratoma from pCEP-OS cord blood (CB) iPSCs shows derivatives of three embryonic germ layers. Cartilage (mesoderm); neurotubules with rosettes (ectoderm); glands (endoderm); retina epithelial cells with pigments (ectoderm). Images were acquired using the Olympus microscope with a ×20 objective.
To extensively characterize pCEP-OS generated iPSCs, we selected several clones for a series of tests. Immunostaining of iPSC colonies showed that they expressed typical human iPSC-specific transcription factors OCT4, SOX2, NANOG, and surface markers SSEA-3, SSEA-4, and Tra-1-60 (Figure 4b). Karyotype analysis indicated a normal human karyotype for all the clones tested; one representative is shown in Figure 4c. Sulphite sequencing showed that both the OCT4 and NANOG promoters were demethylated in three randomly picked iPSC clones (Figure 4d). When injected into immunodeficient NSG mice, iPSCs formed teratomas consisting of derivatives of all three embryonic germ layers, demonstrating the pluripotency of these iPSCs (Figure 4e). Together, these data suggest that bona fide transgene-free iPSCs can be generated from human CB CD34+ cells by nucleofection of a pCEP episomal plasmid that expresses OCT4 and SOX2 alone.
Discussion
Here, we report that iPSCs can be generated from human CB CD34+ cells in 2–3 weeks with the use of OCT4 and SOX2 alone. We found that lentiviral vector-mediated transduction of OS is sufficient to reprogram 2% of transduced CB CD34+ cells into iPSCs. This efficiency is up to 1,000-fold higher than previously reported,9 which is attributed to the SFFV promoter-mediated high-level expression of OS. Furthermore, with the use of an improved OS-expressing episomal vector in which the inclusion of Wpre increases transgene expression by 50%, 20 footprint-free iPSCs can be generated from 1 × 105 CB CD34+ cells, an amount that can be purified from ~1 ml of CB. To the best of our knowledge, this is the first report that footprint-free iPSCs can be generated with only two factors.
Striking progress in iPSC reprogramming has been made over the past several years. iPSCs can be generated from almost any kind of mammalian cells. However, recent reports that describe exceedingly high rates of genetic point mutations and gene copy number variations have shifted the research focus from reprogramming efficiency to reprogramming safety.34,35 Two parameters are likely to be the key to the generation of safe iPSCs for clinical use: cell source and reprogramming method. It is widely accepted that CB is one of the best cell sources for reprogramming. However, one of the four transcription factors originally used by Yamanaka and Takahashi for cell reprogramming, MYC, is oncogenic. Overexpression of MYC has been shown to induce malignant transformation,36 Another commonly used reprogramming booster SV40LT is also oncogenic. SV40LT functions by inhibition of the p53 and Rb-family of tumor suppressors and ectopic expression of SV40LT induces in vitro cellular transformation and in vivo tumorigenesis.37 Although expression of reprogramming factors is only required for ~2 weeks, this short-term exposure to MYC may elicit adverse effects on genomic stability.38 Therefore, we propose that an ideal combination of reprogramming factors should be devoid of factors whose overexpression has been demonstrated to induce cellular transformation and in vivo tumorigenesis.
With safety considerations in mind, we initiated experiments to optimize reprogramming conditions using only OS expressed by a lentiviral vector. We found that high-level expression of OS, driven by a strong promoter SFFV, led to the conversion of 2% of transduced cells into iPSCs. This efficiency is up to 1,000-fold higher than previously reported for these factors.9 An ~20% decrease in OS expression levels led to a fourfold decrease in efficiency. Moreover, when OS expression was decreased by 50% or more with the use of promoters like EF1 and PGK, no iPSCs could be generated from CB CD34+ cells. These findings establish that reprogramming of CB cells with OS critically depends on the expression levels of these genes. It is tempting to speculate that high-level expression of OCT4 and SOX2 alone could also reprogram other cells like fibroblasts. However, SFFV is not necessarily a strong promoter in cell types other than hematopoietic cells. For instance, the EF1 promoter drives higher-level expression of transgenes in fibroblasts than the SFFV promoter (data not shown).
To generate footprint-free iPSCs, we used an episomal vector. In the absence of the Wpre element, the OS-expressing pCEP episomal vector was insufficient to reprogram CB cells into iPSCs. However, an improved episomal vector design that included Wpre at the 3′ end of the transgene and in front of the PolyA signal, led to the successful generation of iPSCs. Of note, sodium butyrate was used for ~10 days in our reprogramming culture. Omitting sodium butyrate led to a considerable decrease in reprogramming efficiency (data not shown). This data suggests that sodium butyrate is also crucial for episomal vector-mediated cellular reprogramming. Characterization of iPSC colonies showed no differences in iPSC quality between different combinations of reprogramming factors, as evidenced by a series of in vitro and in vivo tests. Moreover, after 12 passages, no integration or residual episomal plasmid can be identified in most clones by sensitive real-time PCR analysis. However, a caveat is that this does not necessarily mean there is no integration of small fragments in these iPSC clones. Such fragments can only be detected by whole genome sequencing. While the reprogramming efficiency mediated by pCEP-OS is relatively low, this system is capable of generating sufficient numbers (20 iPSCs/ml of CB) of iPSCs for allogeneic cell therapy.
The generation of transgene-free iPSCs from CB cells has recently been reported by several groups. Yu and colleagues found that the use of episomal vectors expressing seven factors can highly efficiently reprogram CB cells; however no iPSCs could be generated in the absence of SV40LT expression.27 Using a 5-in-1 vector (OSMK and LIN28), Cheng and colleagues were able to generate 80 iPSCs from 1 × 105 CB CD34+ cells.26 From the same amount of cells, we can generate ~20 iPSCs with OS alone, and up to 600 iPSCs with OSMK. Considering that the addition of LIN28 increases reprogramming efficiency by three to fivefold,39 our improved vector is at least 20-fold more efficient in reprogramming CB cells than plasmids used in previous studies. Our success is attributed to the inclusion of two features in the vector design: (i) the SFFV promoter, which drives higher levels of transgene expression in hematopoietic cells than PGK, EF1 or other promoters; and (ii) the Wpre element, which increases transgene expression by 50%. Wpre is commonly used in lentiviral vectors to improve transgene expression;30 our findings suggest that Wpre is also functional in episomal plasmids and possibly other DNA vectors such as adenoviral vectors.
In summary, we are the first to report the successful generation of transgene-free human iPSCs with the use of OCT4 and SOX2 alone. All OS-reprogrammed iPSCs examined in our studies showed normal karyotypes. Future studies that compare genetic instability and mutation rates in iPSCs generated with OS alone versus combinations that include oncogenic factors like MYC will be an important next step on the path to clinical application of iPSCs.
Materials and Methods
Cord blood. The use of CB was approved by the institutional review board of Loma Linda University (LLU) and written informed consent was obtained from all participants. CD34+ cells were purified with a CD34+ Microbead Kit (Miltenyi Biotec, Auburn, CA).
Construction of lentiviral and episomal vectors. Human OCT4, SOX2, MYC, and KLF4 cDNAs were purchased from Open Biosystems, Huntsville, AL and cloned into the pRRLSin.cPPT.PGK-GFP.WPRE lentiviral vector that was kindly provided by Luigi Naldini via Addgene, Cambridge, MA (Plasmid 12252).40 Open reading frames of these reprogramming factors and PGK, EF1, or SFFV promoters were inserted into this vector by PCR cloning. For cloning OS or MK vectors, a 2A sequence was used to link OCT4 and SOX2, or MYC and KLF4.41 The EBNA1/OriP-based pCEP4 episomal vector was purchased from Invitrogen (Carlsbad, CA). For cloning pCEP-OS (w/o W), pCEP-OS, pCEP-K, or pCEP-MK vectors, the hygromycin resistance gene element and cytomegalovirus promoter were removed from the pCEP4 vector by digestion with endonucleases NruI and BamHI, and inserts were cut from the counterparts of lentiviral vectors. All the constructs were verified by sequencing. For lentivirus production, a standard calcium phosphate precipitation protocol was used. Titers of 5–10 × 107/ml were routinely achieved in our lab after a 100-fold concentration by centrifugation at 6,000g for 24 hours at 4 °C.42,43
Generation of iPSCs using lentiviral vector. Thawed CB CD34+ cells were cultured in hematopoietic stem cell culture condition: Iscove's modified Dulbecco's medium/10% fetal bovine serum supplemented with TPO, SCF, FL, and G-CSF each at 100 ng/ml, and IL-3 at 10 ng/ml.44 Cytokines were purchased from ProSpec (East Brunswick, NJ). After 2 days prestimulation, 1 × 104 cells/well were seeded into non-TC treated 24-well plates that were precoated with RetroNectin (CH-296; Takara Bio, Shiga, Japan) for lentiviral transduction for 4–5 hours. A second transduction was conducted 24 hours later. One day after transduction, cells were harvested and transferred to 6-well plates, which were preseeded with a mitomycin C-inactivated CF-1 MEF feeder layer (Applied Stemcell, Menlo Park, CA). Passage five MEFs were used in our experiments. Cells were maintained in the hematopoietic stem cell culture condition for 2 more days before being replaced with iPSC media. The iPSC media used in our study is composed of knockout DMEM/F12 medium (Invitrogen) supplemented with 20% knockout serum replacement (Invitrogen), 1 mmol/l GlutaMAX (Invitrogen), 2 mmol/l nonessential amino acids (Invitrogen), 1× penicillin/streptomycin (Invitrogen), 0.1 mmol/l β-mercaptoethanol (Sigma-Aldrich, St Louis, MO), 20 ng/ml FGF2 (ProSpec). To increase reprogramming efficiency, sodium butyrate45 was added at 0.25 mmol/l from day 2–12, and cells were cultured under hypoxia46 by placing culture plates in a Hypoxia Chamber (Stemcell Technologies, Vancouver, British Columbia, Canada) that was flushed with mixed air composed of 92%N2/3%O2/5%CO2. Starting from day 10, MEF-conditioned medium was used. At day 14–16, ALP staining was conducted to quantitate iPSC colonies. Alternatively, all the colonies were harvested by Accutase (Innovative Cell Technologies, San Diego, CA) treatment for FACS analysis.
Immunostaining and flow cytometry. Staining for ALP was carried out using an ALP-staining kit (Stemgent, San Diego, CA) to quantitate iPSC colonies.
For intracellular staining, cells were fixed for 30 minutes at room temperature in fixation buffer and permeabilization buffer (eBiosciences, San Diego, CA). After washing, cells were stained at room temperature for 2 hours with NANOG-PE (BD Pharmingen, San Diego, CA), followed by washing twice with permeabilization buffer. For staining of cell surface marker TRA-1-60-PE (Stemgent), cells were incubated with the antibody for 30 minutes at room temperature. Flow cytometric analysis was performed using FACS Aria II (BD Biosciences, San Jose, CA) with a 488-nm laser. Thirty thousand events were collected for each sample.
Episomal vector and nucleofection. Fresh or thawed 1 × 105 CB CD34+ cells were cultured in Iscove's modified Dulbecco's medium/10% fetal bovine serum supplemented with TPO, SCF, and FL at 100 ng/ml. Three days later, cells were harvested for nucleofection with a total of 12 µg CEP plasmid DNAs. Human CD34 Cell Nucleofector Kit (Lonza, Walkersville, MD) was used. Nucleofection was performed with Amaxa Nucleofector II using program U-008. Immediately after nucleofection, cells were cultured in a CH-296 pretreated well plate to facilitate the CB cell recovery. The next day, half of the cells were transferred to each well of MEF-coated 6-well plates. Cells were cultured the same way as for reprogramming with lentiviral vector. The total number of iPSC colonies was counted on day 16 post-transfection after ALP staining. At day 14–17, colonies were picked for further culture or harvested for FACS analysis.
Confocal imaging. For immunostaining of iPSC colonies, iPSCs were cultured in chamber slides for 4–5 days. Cells were treated with fixation buffer and permeabilization buffer (eBiosciences) for 30 minutes before being stained overnight with PE or FITC-conjugated antibodies OCT4 (eBiosciences), SOX2 (BD Pharmingen), NANOG (BD Pharmingen), SSEA-3 (eBiosciences), SSEA-4 (eBiosciences), and TRA-1-60 (Stemgent). The samples were washed twice with permeabilization buffer, counterstained with 4′,6-diamidino-2-phenylindole and coverslipped before being imaged. Imaging was performed using the Zeiss LSM 710 NLO laser scanning confocal microscope with a ×10 objective at the LLU Advanced Imaging and Microscopy Core. High resolution monochrome image was captured using a Zeiss HRm CCD camera (Thornwood, NY).
Teratoma assay. The use of NOD/SCID/IL2RG−/− (NSG) immunodeficient mice for the teratoma formation assay was approved by the Institutional Animal Care and Use Committee at LLU. NSG mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and maintained at the LLU animal facility. Approximately 1 × 106 iPSCs were harvested by Dispase (Invitrogen) digestion, washed with culture medium and resuspended in 200 µl DMEM/F12 diluted (1:1) Matrigel solution (BD, San Jose, CA). Cells were injected into the subcutaneous tissue above the rear haunch of NSG mice. At 6–8 weeks after injection of iPSCs, teratomas were dissected and fixed in 10% formalin. After sectioning, samples were embedded in paraffin and stained with hematoxylin and eosin and analyzed by a board certified pathologist.
Bisulphite sequencing. Bisulphite sequencing of genomic DNA from iPSC clones was used to assess methylation status of OCT4 and NANOG promoter. Genomic DNA was purified from human iPSCs by DNeasy Kit (Qiagen, Valencia, CA). The conversion of unmethylated cytosines to uracil was carried out using EZ DNA Methylation-Gold Kit (ZYMO Research, Irvine, CA). Approximately 1 µg genomic DNA was treated in each reaction, and 4 µl of elution was used for each PCR. PCR with primers OCT4-mF3/R3 and NANOG-mF3/R3, which were used by other investigators,47 was carried out using Titanium Taq polymerase (Clontech Laboratories, Mountain View, CA): The cycling conditions were 95°C 10 minutes, followed by 40 cycles of 95 °C for 30 seconds, 60 °C for 30 seconds, 72 °C for 30 seconds, and finally 72 °C for 7 minutes. The PCR products were cloned into a pJET1.2 vector (Fermentas, Glen Burnie, MD) and sequenced by MCLAB (San Francisco, CA).
Karyotyping and G-banding. GTG-banding chromosome analysis was carried out in the LLU Radiation Research Laboratories. Standard DNA spectral karyotyping procedures were followed and a HiSKY Complete Cytogenetic System was used (Applied Spectral Imaging, Vista, CA). For each clone, 10 metaphases were analyzed and karyotyped. The data were interpreted by a certified cytogenetic technologist.
Real-time PCR. To determine the average copy numbers of residual or integrated CEP vector in iPSC clones, real-time PCR analysis was performed. Total DNA (genomic and episomal) was extracted from iPSCs using the DNeasy kit from Qiagen. Equal amounts of DNA (100 ng) isolated from naive cells (before nucleofection) were used as negative control, while a manual mixture of 1 copy pCEP-OS vector per genome was used as a positive control to calculate the average copy numbers of residual episomal vector in each iPSC after multiple passages. Real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on 7500 Fast Real-Time PCR System (Applied Biosystems). Two sets of primers were used to detect CEP plasmid DNA (in either episomal or integrated form): EBNA1-F: 5′-TTTAATACGATTGAGGGCGTCT-3′, EBNA1-R: 5′-GGTTTTGAAGGATGCGATTAAG-3′; OSW-F: 5′- GGATTACAAGG ATGACGACGA-3′, OSW-R: 5′- AAGCCATACGGGAAGCAATA-3′. The amplification program consisted of 50 °C for 2 minutes and 95 °C for 10 minutes, and was followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute.
Statistical analysis. Data are presented as mean ± s.e. of the mean (s.e.m.). Two-tailed Student t-test was performed. P value of <0.05 was considered statistically significant.
Acknowledgments
Imaging was performed in the LLUSM Advanced Imaging and Microscopy Core that is supported by NSF Grant No. MRI-DBI 0923559 (Sean M Wilson) and the Loma Linda University School of Medicine. The authors thank Monica Rubalcava for technical support in confocal imaging. This work was supported by Loma Linda University Department of Medicine (X.-B.Z.), Loma Linda University GRASP Award (X.-B.Z.), and DOD Basic Award W81XWH-11-1-0607 (X.-B.Z.), USAMRAA Grant W81XWH-08-1-0697 (D.J.B), and the Division of Anatomy, the Department of Basic Sciences, the Center for Health Disparities and Molecular Medicine at Loma Linda University (K.J.P. and R-J.S.) and Radiation Research Laboratories in the Department of Radiation Medicine at Loma Linda University (L.R. and D.S.G). The authors declared no conflict of interest.
REFERENCES
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K.et al. (2007Induction of pluripotent stem cells from adult human fibroblasts by defined factors Cell 131861–872. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S.et al. (2007Induced pluripotent stem cell lines derived from human somatic cells Science 3181917–1920. [DOI] [PubMed] [Google Scholar]
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F.et al. (2008Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes Nat Biotechnol 261276–1284. [DOI] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC.et al. (2009Generation of induced pluripotent stem cells from human blood Blood 1135476–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY.et al. (2009Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders Blood 1145473–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisato A, Wakatsuki M, Kodama Y, Shinba H, Ishida I., and, Nagao K. Generation of induced pluripotent stem cells by efficient reprogramming of adult bone marrow cells. Stem Cells Dev. 2010;19:229–238. doi: 10.1089/scd.2009.0149. [DOI] [PubMed] [Google Scholar]
- Butler MG., and, Menitove JE. Umbilical cord blood banking: an update. J Assist Reprod Genet. 2011;28:669–676. doi: 10.1007/s10815-011-9577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C.et al. (2009Generation of induced pluripotent stem cells from human cord blood Cell Stem Cell 5434–441. [DOI] [PubMed] [Google Scholar]
- Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodríguez-Pizà I, Vassena R.et al. (2009Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2 Cell Stem Cell 5353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer HE. Will iPS cells enhance therapeutic applicability of cord blood cells and banking. Cell Stem Cell. 2010;6:21–24. doi: 10.1016/j.stem.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA.et al. (2010Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency Proc Natl Acad Sci USA 1074335–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y.et al. (2010Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence Stem Cells 28704–712. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T., and, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN., and, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C.et al. (2009Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells Stem Cells 271042–1049. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G., and, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., and, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R.et al. (2009piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells Nature 458766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P., and, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z.et al. (2010A nonviral minicircle vector for deriving human iPS cells Nat Methods 7197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS.et al. (2009Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins Cell Stem Cell 4472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T.et al. (2009Generation of induced pluripotent stem cells using recombinant proteins Cell Stem Cell 4381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M.et al. (2011Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors Proc Natl Acad Sci USA 10814234–14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y.et al. (2011Reprogramming of mouse and human cells to pluripotency using mature microRNAs Cell Stem Cell 8633–638. [DOI] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F.et al. (2010Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA Cell Stem Cell 7618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM.et al. (2011Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures Cell Res 21518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chau KF, Vodyanik MA, Jiang J., and, Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS ONE. 2011;6:e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Yu J, Suknuntha K, Tian S, Montgomery K, Choi KD.et al. (2011Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells Blood 117e109–e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C.et al. (2002High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter Hum Gene Ther 13803–813. [DOI] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS., and, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- Yam PY, Li S, Wu J, Hu J, Zaia JA., and, Yee JK. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther. 2002;5:479–484. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
- Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J.et al. (2007Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells Blood 1101448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Xu Q, Chou DM, Zhao Z., and, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J.et al. (2011Somatic coding mutations in human induced pluripotent stem cells Nature 47163–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E.et al. (2011Copy number variation and selection during reprogramming to pluripotency Nature 47158–62. [DOI] [PubMed] [Google Scholar]
- Adhikary S., and, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Ahuja D, Sáenz-Robles MT., and, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Takizawa N, Narita M, Ichisaka T., and, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci USA. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Wu Z, Wang Y, Cheng L, Cui C, Gao Y.et al. (2008Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors Cell Res 18600–603. [DOI] [PubMed] [Google Scholar]
- De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E.et al. (2005Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells Blood 1052307–2315. [DOI] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M.et al. (2009Reprogramming of murine and human somatic cells using a single polycistronic vector Proc Natl Acad Sci USA 106157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer I, Li Z, Persson J, Liu Y, van Rensburg R, Yumul R.et al. (2011Controlled extracellular matrix degradation in breast cancer tumors improves therapy by trastuzumab Mol Ther 19479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li ZY, Liu Y, Persson J, Beyer I, Möller T.et al. (2011Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14 Nat Med 1796–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XB, Beard BC, Beebe K, Storer B, Humphries RK., and, Kiem HP. Differential effects of HOXB4 on nonhuman primate short- and long-term repopulating cells. PLoS Med. 2006;3:e173. doi: 10.1371/journal.pmed.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S.et al. (2010Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes Stem Cells 28713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T., and, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II.et al. (2009Human induced pluripotent stem cells free of vector and transgene sequences Science 324797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]


