Abstract
Measles virus (MV) is an enveloped negative strand RNA virus belonging to the family of Paramyxoviridae, genus Morbillivirus, and causes one of the most contagious diseases in humans. Experimentally infected non-human primates are used as animal models for studies of the pathogenesis of human measles. We established a reverse genetics system based on a highly pathogenic wild-type MV. Infection of monkeys with recombinant MV strains generated by reverse genetics enabled analysis of the molecular basis of MV pathogenesis. The essential in vivo function of accessory genes was indicated by infecting monkeys with recombinant MV strains deficient in the expression of accessory genes. Furthermore, recombinant wild-type MV strains expressing enhanced green fluorescent protein enabled visual tracking of MV-infected cells in vitro and in vivo. To date, three different molecules have been identified as receptors for MV. Signaling lymphocyte activation molecule (SLAM, also called CD150), expressed on immune cells, is a major receptor for MV. CD46, ubiquitously expressed in all nucleated cells in humans and monkeys, is a receptor for vaccine and laboratory-adapted strains of MV. The newly identified nectin-4 (also called poliovirus-receptor-like-4) is an epithelial cell receptor for MV. However, recent findings have indicated that CD46 acts as an MV receptor in vitro but not in vivo. The impact of the receptor usage of MV in vivo on the disease outcome is now under investigation.
Keywords: measles virus, monkey, pathogenesis, tropism, reverse genetics, receptor, EGFP
Introduction
Measles is a febrile disease that typically occurs in small children; the incubation period is 10–14 days, after which clinical symptoms such as fever, coughing, and a characteristic rash appears. Since measles is accompanied by immunosuppression, it has a high frequency of complication with secondary bacterial infections, such as otitis media or pneumonia. Although developed countries are eradicating measles by promoting effective vaccination, measles remains an important issue, especially in developing countries (Griffin, 2007).
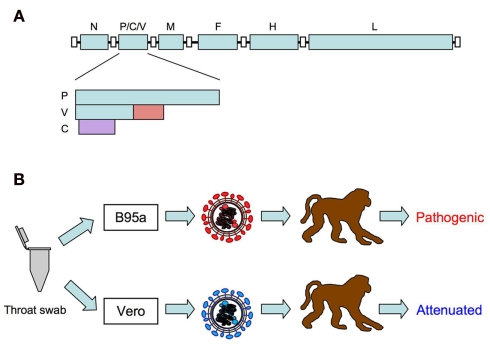
Measles virus (MV), belonging to the genus Morbillivirus of the family Paramyxoviridae, is an enveloped virus with a non-segmented negative strand RNA genome. The MV genome has six genes that encode the nucleocapsid (N), phospho (P), matrix (M), fusion (F), hemagglutinin (H), and large (L) proteins (Figure 1A). MV contains two envelope glycoproteins: the H protein, which is responsible for receptor binding and is important for determining cell tropism of MV; and the F protein, which mediates membrane fusion (Navaratnarajah et al., 2009). The P gene encodes the P protein and the non-structural V and C proteins. The V and C proteins are important for antagonizing the host interferon (IFN) response (Gerlier and Valentin, 2009).
Figure 1.
(A) Schematic diagram of measles virus genome. The P gene encodes the P protein and the non-structural V and C proteins. The V protein is translated from an edited RNA with a non-templated G residue. The V protein-specific region is indicated in red. The C protein is translated from an alternative open reading frame (purple) initiated by the second AUG. (B) Isolation of wild-type MV using B95a cells. Pathogenic wild-type MV strains can be isolated by inoculating throat swabs from measles patients in B95a cells. MV strains isolated in Vero cells lose their pathogenicity.
To date, three different molecules have been identified as receptors for MV. Signaling lymphocyte activation molecule (SLAM, also called CD150), expressed in certain immune system cells including activated B and T lymphocytes, mature dendritic cells, and macrophages, is a receptor for wild-type MV and vaccine and laboratory-adapted strains of MV (Tatsuo et al., 2000). CD46 (also called membrane cofactor protein), expressed in all human and monkey nucleated cells, is a receptor for vaccine and laboratory-adapted strains of MV (Dorig et al., 1993; Naniche et al., 1993). Recently, nectin-4 [also called poliovirus-receptor-like-4 (PVRL4)] has been identified as the epithelial receptor for wild-type MV (Muhlebach et al., 2011; Noyce et al., 2011).
Several animal models have been used for studying the pathogenesis of MV (Griffin, 2007). Cotton rats, rats, hamsters, mice, and ferrets can be infected with MV and are commonly used as small animal models for MV pathogenesis. After identification of CD46 and SLAM as MV receptors, numerous transgenic and knock-in mice expressing human CD46 and/or SLAM were established and intensively used to study different aspects of MV infection (Sellin and Horvat, 2009). However, non-human primates are the only animals that exhibit acute disease similar to that seen in humans. In this review, we discuss recent findings regarding tropism and pathogenesis of MV; these findings were obtained by infecting monkeys with recombinant wild-type MV.
Historical Background of Monkey Models
When infected with measles, monkeys exhibit similar symptoms as seen in humans. This was reported as early as 1911, after inoculating monkeys with blood from measles patients (Anderson and Goldberger, 1911). In 1921, it was reported that measles could be transmitted from humans to monkeys by placing a filtered throat swab from a measles patient into the tracheae of monkeys (Blake and Trask, 1921a). These authors also performed histological analysis of infected monkeys (Blake and Trask, 1921b). However, at this time, “MV” had not yet been discovered. MV was first isolated in 1954 from a specimen obtained from a measles patient (Enders and Peebles, 1954). Enders and Peebles (1954) inoculated human and monkey cell cultures with a throat swab taken from a young boy named David Edmonston and isolated MV from these cultures. After this, it was discovered that MV isolated from normal human renal cells caused clinical signs similar to those of human measles in monkeys (Peebles et al., 1957). Since then, numerous studies have been carried out by infecting monkeys with MV, measles vaccines, or specimens from measles patients (Griffin, 2007). In such experiments, two species of monkeys, cynomolgus monkey (Macaca fascicularis) and rhesus monkey (Macaca mulatta), serve as good animal models. New World monkeys are more susceptible to MV than Old World monkeys, and infection of marmoset (Saguinus mystax) with MV results in a fulminant disease (Levy and Mirkovic, 1971; Albrecht et al., 1980).
In the past, it was well known that infection of monkeys with materials from measles patients induced clinical signs similar to those of human measles (Nii et al., 1964; Yamanouchi et al., 1970; Sakaguchi et al., 1986). However, curiously enough, infection of monkeys with MV isolated and propagated in cultured cells did not always induce these clinical signs (Enders et al., 1960; Yamanouchi et al., 1970; van Binnendijk et al., 1994). This riddle was solved by the introduction of B95a cells (a marmoset B-lymphoid cell line) for isolation and propagation of MV (Kobune et al., 1990). Kobune et al. (1990, 1996) found that MV strains could be efficiently isolated in B95a cells using materials from measles patients. More importantly, MV strains isolated from B95a cells retained their original pathogenicity in monkeys. These studies indicated that vaccine and laboratory-adapted strains of MV previously isolated from non-lymphoid cells such as Vero cells were not true MV (Figure 1B). A decade later, it was found that the MV receptor SLAM is highly expressed on B95a cells (Tatsuo et al., 2000), which accounts for the efficient isolation of pathogenic MV from patient samples. Similar to MV strains isolated from B95a cells, MV strains isolated and propagated in monkey mononuclear cells, human cord blood cells, human B lymphoblastoid cell lines, and Vero cells expressing SLAM replicated well in monkeys and induced clinical signs of measles (van Binnendijk et al., 1994; McChesney et al., 1997; Zhu et al., 1997; Auwaerter et al., 1999; El Mubarak et al., 2007; Bankamp et al., 2008). These results suggest that expression of SLAM on cells used for isolation is important for isolation of pathogenic MV.
Reverse Genetics of MV
Reverse genetics refers to the methods used for recovering infectious viruses from the cDNA that encodes the viral genome. By using this method, mutations or extra transcription units can be introduced into viral genomes by the modification of cDNA plasmids. Reverse genetics of MV was first established based on the Edmonston vaccine strain (Radecke et al., 1995). However, as previously mentioned, viruses derived from the Edmonston vaccine strain do not induce clinical symptoms of measles in monkeys. Therefore, reverse genetics of pathogenic wild-type MV was needed for the study of MV pathogenesis in monkeys. To this end, we first determined the complete nucleotide sequence of the genome of the pathogenic wild-type IC-B strain (NC_001498/AB016162; Takeuchi et al., 2000), which was isolated in Tokyo in 1984 by using B95a cells (Kobune et al., 1990). Then, we constructed a complete cDNA plasmid of the IC-B strain named p(+)MV323, and successfully recovered infectious MV (IC323 strain) from the p(+)MV323 plasmid (Takeda et al., 2000). Importantly, the IC323 strain induced clinical signs such as rash, Koplik’s spots, and lymphopenia similar to human measles in infected monkeys, indicating that the IC323 strain retains the original pathogenicity of the IC-B strain. Now, infectious MV strains can be easily recovered from cDNA plasmids by using an improved protocol (Takeda et al., 2005).
Reverse genetics of other wild-type MV strains has been reported for the HL strain isolated in Japan (Terao-Muro et al., 2008) and the KS strain isolated in Sudan (Lemon et al., 2011). For vaccine strains of MV, reverse genetics for the Schwarz/Moraten vaccine strain (Combredet et al., 2003; del Valle et al., 2007) and the AIK-C vaccine strain (Nakayama et al., 2001) have also been reported. Reverse genetics for vaccine strains are being used as a platform to generate new multivalent vaccines expressing antigens of other pathogens (Billeter et al., 2009) and oncolytic viruses for cancer therapy (Russell and Peng, 2009).
Function of MV Accessory Proteins in vivo
The P gene of MV encodes two non-structural proteins, namely the C and V proteins. However, the function of the C and V proteins in vivo was not well understood. The C protein is a small (186 amino acid), highly positively charged protein. To study the function of the C protein in the context of the natural course of MV pathogenesis, we generated an IC323 strain deficient in the expression of the C protein, wtMV(C−), by using the reverse genetics of wild-type MV (Takeuchi et al., 2005). Notably, the growth of wtMV(C−) in cynomolgus monkeys was dramatically reduced when compared to the IC323 strain. A similar growth defect of the IC323 strain deficient in the expression of the C protein, Cko, in vivo was observed in rhesus monkeys (Devaux et al., 2008). Interestingly, Cko induced more inflammatory cytokines such as tumor necrosis factor alpha (TNF)-α and interleukin (IL)-6 and interferon (IFN)-α and -β in infected monkeys.
The V protein is translated from an edited mRNA of the P gene (Griffin, 2007). Thus, the amino-terminal domain of the V protein has the same amino acid sequence as the P protein, and the carboxyl-terminal domain of the V protein has a highly conserved amino acid sequence forming a zinc-binding domain, which is important for function as an interferon antagonist (Gerlier and Valentin, 2009). An IC323 strain deficient in the expression of the V protein, Vko, was generated by introducing nucleotide mutations in the RNA-editing signal in the P gene (Devaux et al., 2008). The growth of Vko in infected rhesus monkeys was lower than that of the parental IC323 strain. Vko induced more inflammatory cytokines (TNF-α and IL-6) and IFN-α and -β. An IC323 strain unable to antagonize STAT1 function, STAT1-blind virus, was generated by introducing three amino acid substitutions in the shared domain of the P and V proteins (Devaux et al., 2011). The STAT1-blind virus induced short-lived viremia and no clinical signs in infected rhesus monkeys. This virus induced more inflammatory cytokines (TNF-α and IL-6) and a Th1/Th2 balance cytokine (IL-12) in infected monkeys. Taken together, these findings indicate that the C and V proteins are not non-essential gene products as previously thought, but are stringently required for antagonizing host innate immune and inflammatory responses in vivo.
In contrast, in vitro studies indicated that the V protein blocks IFN-α/β signal transductions in infected cells, inhibits TLR7-mediated IFN-α production in human plasmacytoid dendritic cells, and inhibits IFN induction in infected cells by interacting with MDA5 (Gerlier and Valentin, 2009). Furthermore, the C protein appears to inhibit IFN induction in infected cells by regulating viral RNA synthesis (Nakatsu et al., 2008). Thus, it is necessary to elucidate whether the in vivo phenotypes of C- and V-deficient viruses are similar to the in vitro phenotypes.
MV Tropism in vivo
Recent advances in the study of virus tropism have included the introduction of enhanced green fluorescent protein (EGFP)-expressing viruses. In vivo tropism of Morbillivirus can be visualized with high sensitivity in living animals as well as tissue samples by using EGFP-expressing recombinant canine distemper viruses (von Messling et al., 2004). Similarly, MV target tissues or organs can be visualized with high sensitivity by infecting cynomolgus monkeys with an EGFP-expressing IC323 strain (Figure 2). de Swart et al. (2007) infected rhesus and cynomolgus monkeys with an IC323 strain expressing EGFP (Hashimoto et al., 2002) and examined the tropism of wild-type MV in vivo. They indicated that the major target cells of wild-type MV were B and T lymphocytes and CD11c-positive, major histocompatibility complex (MHC) class-II-positive dendritic cells. This result is consistent with the fact that SLAM is a receptor for wild-type MV. Infection of ciliated epithelial cells in the trachea and lungs was also detected, suggesting the presence of another receptor for MV in epithelial cells.
Figure 2.
Schematic diagram of reverse genetics of wild-type MV expressing EGFP and infection of monkeys. A plasmid carrying the full-genome cDNA of the IC-B strain and the EGFP gene under the control of the T7 promoter is introduced into CHO cells expressing human SLAM (CHO/hSLAM), along with three supporting plasmids expressing N, P, and L proteins, respectively, under the control of the T7 promoter. After infection with vaccinia virus expressing the T7 RNA polymerase, wild-type MV expressing EGFP can be recovered by mixing with B95a cells. EGFP fluorescence in tissues and organs of infected monkeys can be detected using a fluorescence microscope.
Regarding the early target cells of wild-type MV, classical textbooks describe that the primary targets of MV are the epithelial cells of the respiratory tract. However, SLAM is not expressed in these epithelial cells. To examine the early target cells of wild-type and vaccine strains of MV in the lung, de Vries et al. (2010) infected cynomolgus monkeys with EGFP-expressing IC323 or vaccine strains of MV via the intratracheal or aerosol route. They found that CD11c-positive cells, which include alveolar macrophages and dendritic cells, were the major targets of both viruses. Interestingly, although viral replication and cellular tropism in the lungs were similar for the two viruses, only wild-type MV caused significant viremia, suggesting a growth defect of the vaccine strain in lymphocyte cells. Similarly, to examine the early target cells of wild-type MV, Lemon et al. (2011) infected cynomolgus monkeys with an EGFP-expressing wild-type MV based on the KS strain by aerosol infection and found that the early target cells of wild-type MV in monkeys are macrophages and dendritic cells. These studies indicated that alveolar macrophages and dendritic cells but not the epithelial cells of the respiratory tract are the early target cells of wild-type MV.
Nectin-4 is a newly identified epithelial cell receptor (EpR) for MV. To examine the effect of nectin-4-using activity of MV on disease outcomes in monkeys, an IC323 strain recognizing SLAM but not nectin-4 was generated by introducing amino acid mutations in the H protein (Leonard et al., 2008). At that time, nectin-4 had not been identified as a MV receptor, and this strain was called EpR-blind virus. When rhesus monkeys were infected with EpR-blind virus via the conjunctiva and nares, this virus induced viremia and clinical signs in infected monkeys but did not propagate in the lungs. This result indicates the importance of nectin-4 for the propagation of MV in the lungs, which is required for the subsequent exit of MV from the host. Inversely, to examine the impact of the recognition of SLAM by MV, an IC323 strain recognizing nectin-4 but not SLAM, SLAM-blind, was generated (Leonard et al., 2010). When rhesus monkeys were infected with the SLAM-blind virus, it elicited no clinical symptoms. This result indicates that SLAM recognition is necessary for MV virulence and pathogenesis.
Vaccine and laboratory-adapted strains of MV can utilize both CD46 and SLAM as cellular receptors. However, surprisingly, the impact of the CD46-using activity of vaccine and laboratory-adapted strains of MV on their tissue and organ tropism and attenuation is not well understood. As CD46 is ubiquitously expressed on all nucleated human and monkey cells, vaccine, and laboratory-adapted strains of MV may infect all tissues and organs of humans and monkeys. If so, this tropism shift may have great consequences on vaccine attenuation. In this context, de Vries et al. (2010) indicated that only CD11c-positive cells were infected with the EGFP-expressing vaccine strain via the aerosol route, suggesting that vaccine strains do not use CD46 in vivo. However, when the replication of vaccine and laboratory-adapted strains of MV in monkeys is limited, it will be difficult to identify infected cells in tissues. Furthermore, the infection of vaccine and laboratory-adapted strains of MV may be restricted because of mutations in the P/C/V genes, which are important for antagonizing the host IFN response (Gerlier and Valentin, 2009). Therefore, the tropism shift that solely occurs via the H protein should be evaluated using the wild-type MV expressing EGFP, which bears the H protein of a vaccine strain, such as the IC/EdH strain we developed previously (Takeuchi et al., 2002).
We recently infected cynomolgus monkeys with EGFP-expressing wild-type or IC/EdH strains and found that SLAM-expressing lymphocytes were the main targets of both strains, indicating that CD46 does not act as a receptor for vaccine and laboratory-adapted strains of MV in vivo (Figure 3; Takeuchi et al., 2012). One possible explanation for the limited expansion of the EGFP-expressing IC/EdH strain in vivo is the activation status of lymphocytes. It is known that stimulated lymphocytes are efficiently infected with MV and that stimulated lymphocytes express SLAM (Tatsuo et al., 2000). Thus, lymphocytes expressing SLAM may appear to be infected with both strains. The EGFP-expressing IC/EdH strain that entered into quiescent lymphocytes via the CD46 may not grow well in those cells. Alternatively, the expression level of CD46 in vivo may be too low to allow MV dissemination by cell–cell fusion, as it was reported that high CD46 density is required for MV-induced cell–cell fusion (Anderson et al., 2004). For whatever the reason, the growth of the EGFP-expressing IC/EdH strain was less efficient than that of the EGFP-expressing wild-type strain (Takeuchi et al., 2012), suggesting that the CD46-recognition ability of vaccine and laboratory-adapted MV strains plays a role in the MV attenuation. Further studies are required to elucidate the relationship between CD46-recognition ability and MV attenuation.
Figure 3.
Receptor usage of the wild-type and vaccine and laboratory-adapted strains of MV. Nectin-4 is a recently identified epithelial cell receptor for MV.
Conclusion
As described in this review, our IC323 strain, recovered from a plasmid carrying the full-genome cDNA of the IC-B strain, is now used as a standard wild-type MV strain in MV research. Furthermore, EGFP-expressing IC323 strains are ideal tools for the study of tissue and organ tropism of wild-type MV in vivo. Although the use of monkeys has several limitations, monkey models still provide the only reliable animal model for the study of the pathogenesis of MV in humans. The combination of wild-type and vaccine MV strains generated by reverse genetics and monkey models will provide new insights into the relationship between viral gene functions or individual mutation(s) and the pathogenesis of MV.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This paper was supported by a grant-in-aid (No. 23659227) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Albrecht P., Lorenz D., Klutch M. J., Vickers J. H., Ennis F. A. (1980). Fatal measles infection in marmosets pathogenesis and prophylaxis. Infect. Immun. 27, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson B. D., Nakamura T., Russell S. J., Peng K.-W. (2004). High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 64, 4919–4926 10.1158/0008-5472.CAN-04-0884 [DOI] [PubMed] [Google Scholar]
- Anderson J. F., Goldberger J. (1911). Experimental measles in the monkey: a supplemental note. Public Health Rep. 26, 887–895 10.2307/4566361 [DOI] [PubMed] [Google Scholar]
- Auwaerter P. G., Rota P. A., Elkins W. R., Adams R. J., DeLozier T., Shi Y., Bellini W. J., Murphy B. R., Griffin D. E. (1999). Measles virus infection in rhesus macaques: altered immune responses and comparison of the virulence of six different virus strains. J. Infect. Dis. 180, 950–958 10.1086/314993 [DOI] [PubMed] [Google Scholar]
- Bankamp B., Hodge G., McChesney M. B., Bellini W. J., Rota P. A. (2008). Genetic changes that affect the virulence of measles virus in a rhesus macaque model. Virology 373, 39–50 10.1016/j.virol.2007.11.025 [DOI] [PubMed] [Google Scholar]
- Billeter M. A., Naim H. Y., Udem S. A. (2009). Reverse genetics of measles virus and resulting multivalent recombinant vaccines: applications of recombinant measles viruses. Curr. Top. Microbiol. Immunol. 329, 129–162 10.1007/978-3-540-70523-9_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake F. G., Trask J. D., Jr. (1921a). Studies on measles. I. Susceptibility of monkeys to the virus of measles. J. Exp. Med. 33, 385–412 10.1084/jem.33.3.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake F. G., Trask J. D., Jr. (1921b). Studies on measles. II. Symptomatology and pathology in monkeys experimentally infected. J. Exp. Med. 33, 413–422 10.1084/jem.33.3.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combredet C., Labrousse V., Mollet L., Lorin C., Delebecque F., Hurtrel B., McClure H., Feinberg M. B., Brahic M., Tangy F. (2003). A molecularly cloned Schwarz strain of measles virus vaccine induces strong immune responses in macaques and transgenic mice. J. Virol. 77, 11546–11554 10.1128/JVI.77.21.11546-11554.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Swart R. L., Ludlow M., de Witte L., Yanagi Y., van Amerongen G., McQuaid S., Yuksel S., Geijtenbeek T. B. H., Duprex W. P., Osterhaus A. D. M. E. (2007). Predominant infection of CD150+ lymphocytes and dendritic cells during measles virus infection of macaques. PLoS Pathog. 3, e178. 10.1371/journal.ppat.0030178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. D., Lemon K., Ludlow M., McQuaid S., Yuksel S., van Amerongen G., Rennick L. J., Rima B. K., Osterhaus A. D. M. E., de Swart R. L., Duprex W. P. (2010). In vitro tropism of attenuated and pathogenic measles virus expressing green fluorescent protein in macaques. J. Virol. 84, 4714–4724 10.1128/JVI.02633-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Valle J. R., Devaux P., Hodge G., Wegner N. J., McChesney M. B., Catteneo R. (2007). A vectored measles virus induces hepatitis B surface antigen antibodies while protecting macaques against measles virus challenge. J. Virol. 81, 10597–10605 10.1128/JVI.00923-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P., Hodge G., McChesney M. B., Cattaneo R. (2008). Attenuation of V- or C-defective measles viruses: infection control by the inflammatory and interferon responses of rhesus monkeys. J. Virol. 82, 5359–5367 10.1128/JVI.00169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P., Hudacek A. W., Hodge G., Valle J. R., McChesney M. B., Cattaneo R. (2011). A recombinant measles virus unable to antagonize STAT1 function cannot control inflammation and is attenuated in rhesus monkeys. J. Virol. 85, 348–356 10.1128/JVI.00802-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorig R. E., Marcil A., Chopra A., Richardson C. D. (1993). The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75, 295–305 10.1016/0092-8674(93)80071-L [DOI] [PubMed] [Google Scholar]
- El Mubarak H. S., Yuksel S., van Amerongen G., Mulder P. G. H., Mukhtar M. M., Osterhaus A. D. M. E., de Swart R. L. (2007). Infection of cynomolgus macaques (Macaca fascicularis) and rhesus macaques (Macaca mulatta) with different wild-type measles viruses. J. Gen. Virol. 88, 2028–2034 10.1099/vir.0.82804-0 [DOI] [PubMed] [Google Scholar]
- Enders J. F., Katz S. L., Milovanovic M. V., Holloway A. (1960). Studies on an attenuated measles-virus vaccine I. Development and preparation of the vaccine: techniques for assay of effects of vaccination. N. Engl. J. Med. 263, 153–159 10.1056/NEJM196007282630401 [DOI] [PubMed] [Google Scholar]
- Enders J. F., Peebles T. C. (1954). Propagation in tissue cultures of cytopathic agents from patients with measles. Proc. Soc. Exp. Biol. Med. 86, 277–286 [DOI] [PubMed] [Google Scholar]
- Gerlier D., Valentin H. (2009). Measles virus interaction with host cells and impact on innate immunity. Curr. Top. Microbiol. Immunol. 329, 163–191 10.1007/978-3-540-70523-9_8 [DOI] [PubMed] [Google Scholar]
- Griffin D. E. (2007). “Measles virus,” in Fields Virology, 5th Edn, eds Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (Philadelphia, PA: Lippincott Williams & Wilkins; ), 1551–1585 [Google Scholar]
- Hashimoto K., Ono N., Tatsuo H., Minagawa H., Takeda M., Takeuchi K., Yanagi Y. (2002). SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76, 6743–6749 10.1128/JVI.76.13.6743-6749.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobune F., Sakata H., Sugiura A. (1990). Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64, 700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobune F., Takahashi H., Terao K., Ohkawa T., Ami Y., Suzaki Y., Nagata N., Sakata H., Yamanouchi K., Kai C. (1996). Nonhuman primate models of measles. Lab. Anim. Sci. 46, 315–320 [PubMed] [Google Scholar]
- Lemon K., de Vries R. D., Mesman A. W., McQuaid S., van Amerongen G., Yuksel S., Ludlow M., Rennick L. J., Kuiken T., Rima B. K., Geijtenbeek T. B. H., Osterhaus A. D. M. E., Duprex W. P., de Swart R. L. (2011). Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathog. 7, e1001263. 10.1371/journal.ppat.1001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard V. H. J., Hodge G., Valle J. R., McChesney M. B., Cattaneo R. (2010). Measles virus selectively blind to signaling lymphocytes activation molecule (SLAM; CD150) is attenuated and induces strong adaptive immune responses in rhesus monkeys. J. Virol. 84, 3413–3420 10.1128/JVI.02304-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard V. H. J., Sinn P. L., Hodge G., Miest T., Devaux P., Oezguen N., Braun W., McCray P. B., Jr., McChesney M. B., Cattaneo R. (2008). Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 118, 2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B. M., Mirkovic R. R. (1971). An epizootic of measles in a marmoset colony. Lab. Anim. Sci. 21, 33–39 [PubMed] [Google Scholar]
- McChesney M. B., Miller C. J., Rota P. A., Zhu Y., Antipa L., Lerche N. W., Ahmed R., Bellini W. J. (1997). Experimental measles. I. Pathogenesis in the normal and the immunized host. Virology 233, 74–84 10.1006/viro.1997.8576 [DOI] [PubMed] [Google Scholar]
- Muhlebach M. D., Mateo M., Sinn P. L., Prufer S., Uhlig K. M., Leonard V. H., Navaratnarajah C. K., Frenzke M., Wong X. X., Sawatsky B., Ramachandran S., McCray P. B., Jr., Cichutek K., von Messling V., Lopez M., Cattaneo R. (2011). Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480, 530–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y., Takeda M., Ohno S., Shirogane Y., Iwasaki M., Yanagi Y. (2008). Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 82, 8296–8306 10.1128/JVI.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Komase K., Uzuka R., Hoshi A., Okafuji T. (2001). Leucine at position 278 of the AIK-C measles virus vaccine strain fusion protein is responsible for reduced syncytium formation. J. Gen. Virol. 82, 2143–2150 [DOI] [PubMed] [Google Scholar]
- Naniche D., Varior-Krishnan G., Cervoni F., Wild T. F., Rossi B., Rabourdin-Combe C., Gerlier D. (1993). Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67, 6025–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnarajah C. K., Leonard V. H. J., Cattaneo R. (2009). Measles virus glycoprotein complex assembly, receptor attachment, and cell entry. Curr. Top. Microbiol. Immunol. 329, 59–76 10.1007/978-3-540-70523-9_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nii S., Kamahora J., Mori Y., Takahashi M., Nishimura S., Okuno Y. (1964). Experimental pathology of measles in monkeys. Biken J. 6, 271–297 [PubMed] [Google Scholar]
- Noyce R. S., Bondre D. G., Ha M. N., Lin L. T., Sisson G., Tsao M. S., Richardson C. D. (2011). Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7, e1002240. 10.1371/journal.ppat.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles T. C., McCarthy K., Enders J. F., Holloway A. (1957). Behavior of monkeys after inoculation of virus derived from patients with measles and propagated in tissue culture together with observations on spontaneous infections of these animals by an agent exhibiting similar antigenic properties. J. Immunol. 78, 63–74 [PubMed] [Google Scholar]
- Radecke F., Spielhofer P., Schneider H., Kaelin K., Huber M., Dotsch C., Christiansen G., Billeter M. A. (1995). Rescue of measles virus from cloned DNA. EMBO J. 14, 5773–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. J., Peng K. W. (2009). Measles virus for cancer therapy. Curr. Top. Microbiol. Immunol. 330, 213–241 10.1007/978-3-540-70617-5_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Yoshikawa Y., Yamanouchi K., Sata T., Nagashima K., Takeda K. (1986). Growth of measles virus in epithelial and lymphoid tissues of cynomolgus monkeys. Microbiol. Immunol. 30, 1067–1073 [DOI] [PubMed] [Google Scholar]
- Sellin C. I., Horvat B. (2009). Current animal models: transgenic animal models for study of measles pathogenesis. Curr. Top. Microbiol. Immunol. 330, 111–127 10.1007/978-3-540-70617-5_6 [DOI] [PubMed] [Google Scholar]
- Takeda M., Ohno S., Seki F., Hashimoto K., Miyajima N., Takeuchi K., Yanagi Y. (2005). Efficient rescue of measles virus from cloned cDNA using SLAM-expressing Chinese hamster ovary cells. Virus Res. 108, 161–165 10.1016/j.virusres.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Takeda M., Takeuchi K., Miyajima N., Kobune F., Ami Y., Nagata N., Suzaki Y., Nagai Y., Tashiro M. (2000). Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74, 6643–6647 10.1128/JVI.74.14.6643-6647.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Miyajima N., Kobune F., Tashiro M. (2000). Comparative nucleotide sequence analyses of the entire genomes of B95a cell-isolated and Vero cell-isolated measles viruses from the same patient. Virus Genes 20, 253–257 10.1023/A:1008196729676 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Nagata N., Kato S., Ami Y., Suzaki Y., Suzuki T., Sato Y., Tsunetsugu-Yokota Y., Mori K., Nguyen V. N., Kimura H., Nagata K. (2012). Wild-type measles virus with the hemagglutinin protein of the Edmonston vaccine strain retains wild-type tropism in macaques. J. Virol. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Takeda M., Miyajima N., Ami Y., Nagata N., Suzaki Y., Shahnewaz J., Kadota S., Nagata K. (2005). Stringent requirement for the C protein of wild-type measles virus for growth in vitro and in macaques. J. Virol. 79, 7838–7844 10.1128/JVI.79.12.7838-7844.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Takeda M., Miyajima N., Kobune F., Tanabayashi K., Tashiro M. (2002). Recombinant wild-type and Edmonston strain measles viruses bearing heterologous H proteins: role of H protein in cell fusion and host cell specificity. J. Virol. 76, 4891–4900 10.1128/JVI.76.10.4891-4900.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuo H., Ono N., Tanaka K., Yanagi Y. (2000). SLAM (CDw150) is a cellular receptor for measles virus. Nature 406, 893–897 10.1038/35022579 [DOI] [PubMed] [Google Scholar]
- Terao-Muro Y., Yoneda M., Seki T., Watanabe A., Tsukiyama-Kohara K., Fujita K., Kai C. (2008). Heparin-like glycosaminoglycans prevent the infection of measles virus in SLAM-negative cell lines. Antiviral Res. 80, 370–376 10.1016/j.antiviral.2008.08.006 [DOI] [PubMed] [Google Scholar]
- van Binnendijk R. S., van der Heijden R. W., van Amerongen G., UytdeHaag F. G., Osterhaus A. D. (1994). Viral replication and development of specific immunity in macaques after infection with different measles virus strains. J. Infect. Dis. 170, 443–448 10.1093/infdis/170.2.443 [DOI] [PubMed] [Google Scholar]
- von Messling V., Milosevic D., Cattaneo R. (2004). Tropism illuminated: lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proc. Natl. Acad. Sci. U.S.A. 101, 14216–14221 10.1073/pnas.0403597101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi K., Egashira Y., Uchida N., Kodama H., Kobune F., Hayami M., Fukuda A., Shishido A. (1970). Giant cell formation in lymphoid tissue of monkeys inoculated with various strains of measles virus. Jpn. J. Med. Sci. Biol. 23, 131–145 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Heath J., Collins J., Greene T., Antipa L., Rota P., Bellini W., McChesney M. (1997). Experimental measles. II. Infection and immunity in the rhesus macaque. Virology 233, 85–92 10.1006/viro.1997.8575 [DOI] [PubMed] [Google Scholar]