Abstract
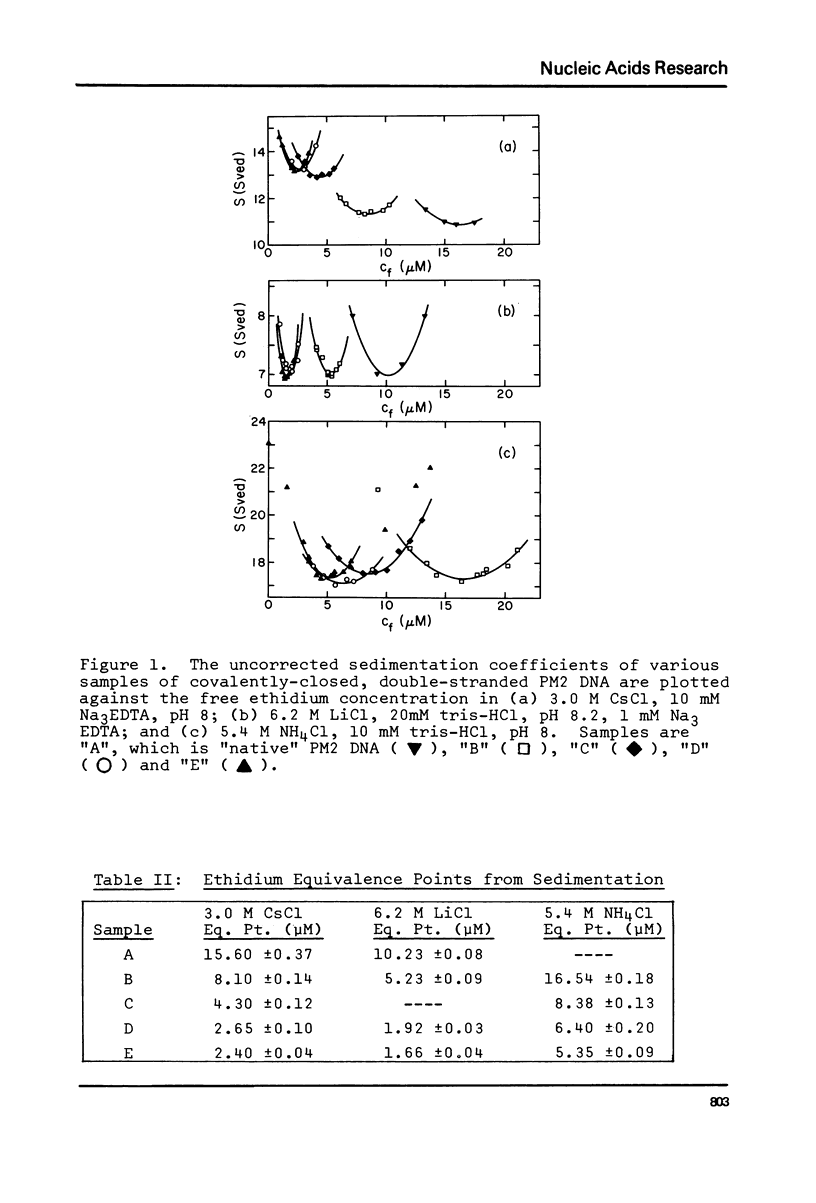
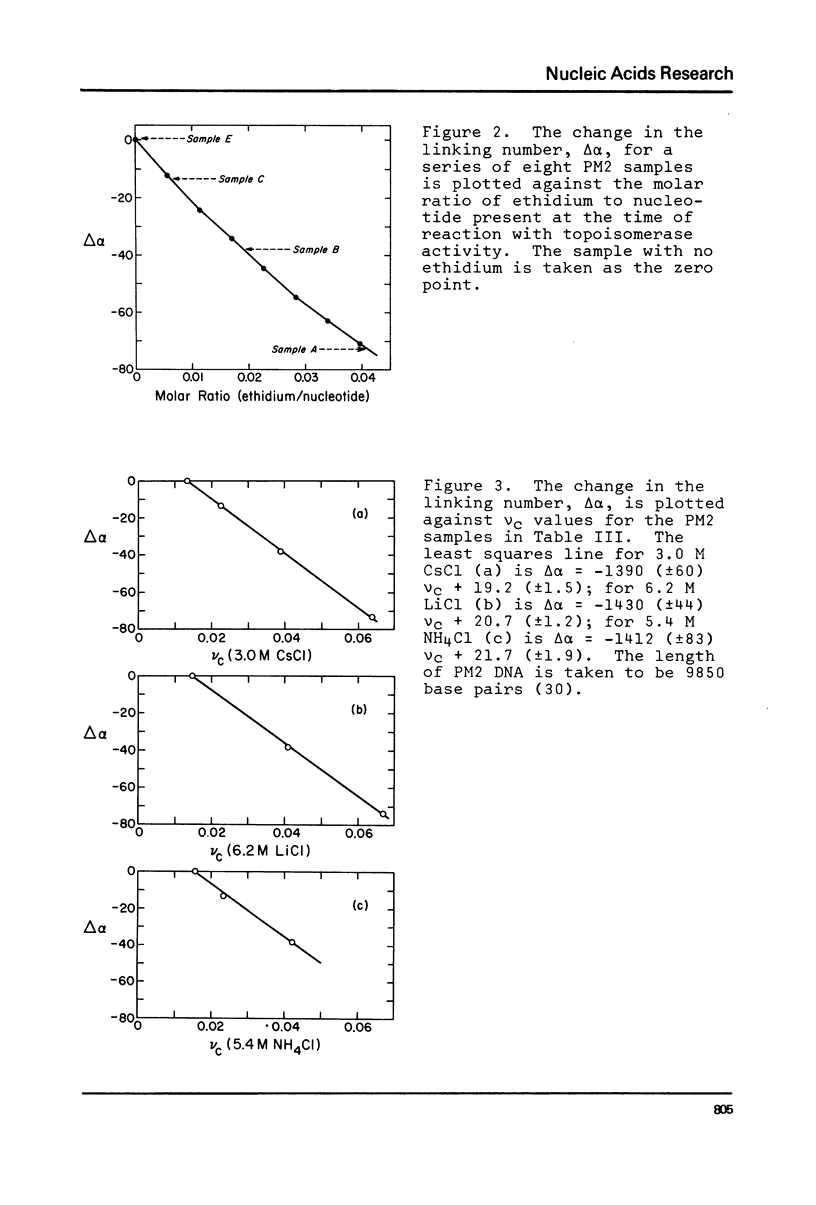
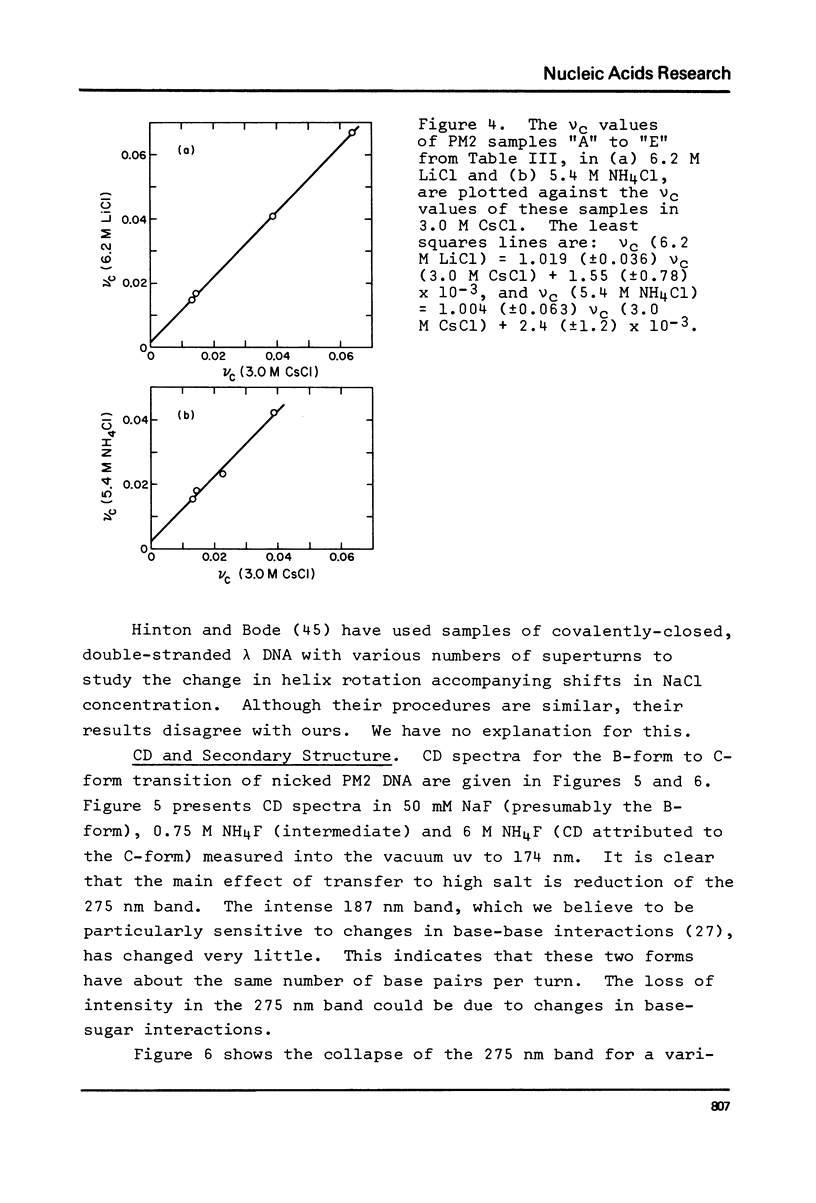
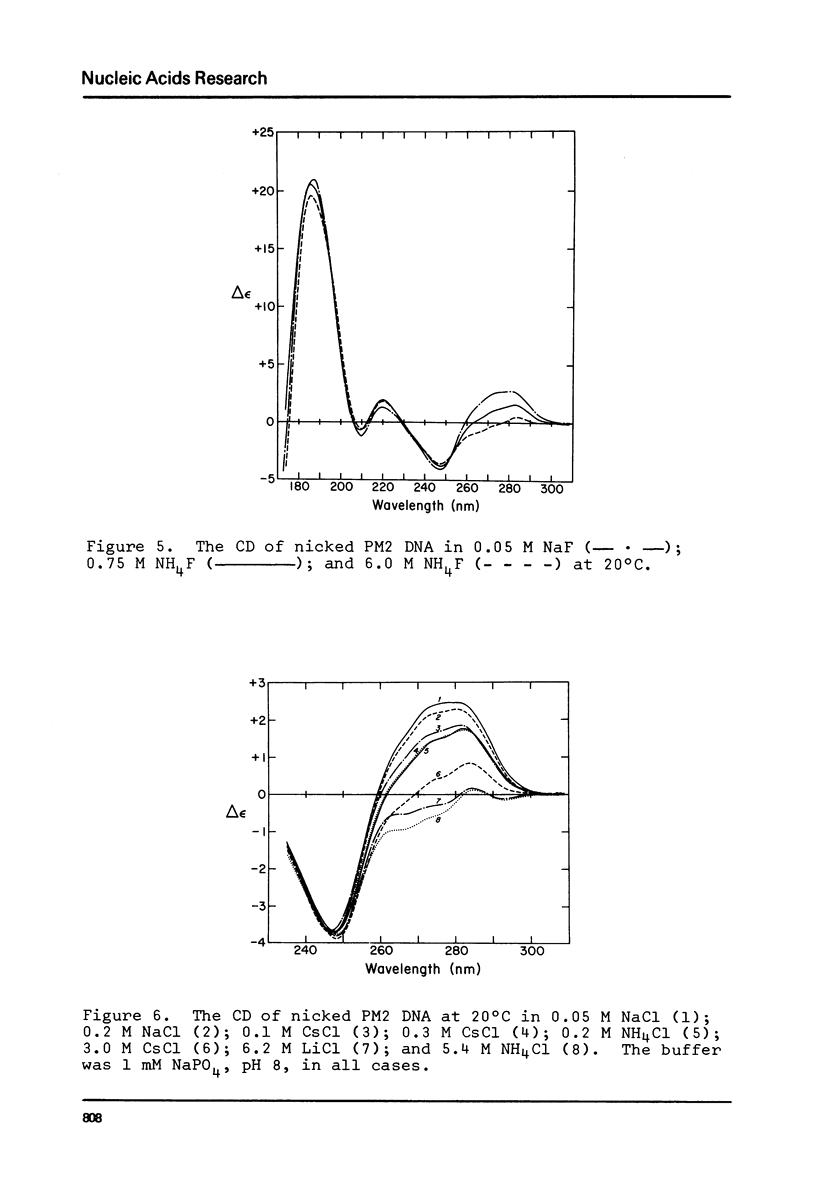
The change in average rotation of the DNA helix has been determined for the transfer from 0.05 M NaCl to 3.0 M CsCl, 6.2 M LiCl and 5.4 M NH4Cl. This work, combined with data at lower salt from other laboratories, allows us to relate the intensity of the CD of DNA at 275 nm directly to the change in the number of base pairs per turn. The change in secondary structure for the transfer of DNA from 0.05 M NaCl (where it is presumably in the B-form) to high salt (where the characteristic CD has been interpreted as corresponding to C-form geometry) is found to be -0.22 (+/- 0.02) base pairs per turn. In the case of mononucleosomes, where the CD indicates the "C-form", the change in secondary structure (including temperature effects) would add -0.31 (+/- 0.03) turns about the histone core to the -1.25 turns estimated from work on SV40 chromatin. Accurate winding angles and molar extinction coefficients were determined for ethidium.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P., Bauer W. Supercoiling in closed circular DNA: dependence upon ion type and concentration. Biochemistry. 1978 Feb 21;17(4):594–601. doi: 10.1021/bi00597a006. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. The conformation of C-DNA. J Mol Biol. 1975 Oct 15;98(1):265–269. doi: 10.1016/s0022-2836(75)80115-x. [DOI] [PubMed] [Google Scholar]
- BRAHMS J., MOMMAERTS W. F. A STUDY OF CONFORMATION OF NUCLEIC ACIDS IN SOLUTION BY MEANS OF CIRCULAR DICHROISM. J Mol Biol. 1964 Oct;10:73–88. doi: 10.1016/s0022-2836(64)80029-2. [DOI] [PubMed] [Google Scholar]
- Chung S. Y., Holzwarth G. Circular dichroism of flow-oriented nucleic acids. I. Experimental results. J Mol Biol. 1975 Mar 5;92(3):449–466. doi: 10.1016/0022-2836(75)90291-0. [DOI] [PubMed] [Google Scholar]
- Cowman M. K., Fasman G. D. Circular dichroism analysis of mononucleosome DNA conformation. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4759–4763. doi: 10.1073/pnas.75.10.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman B. P., Maestre M. F. Experimental differential light-scattering correction to the circular dichroism of bacteriophage T2. Proc Natl Acad Sci U S A. 1973 Jan;70(1):255–259. doi: 10.1073/pnas.70.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology. 1968 Apr;34(4):738–747. doi: 10.1016/0042-6822(68)90094-9. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Gennis R. B., Cantor C. R. Optical studies of a conformational change in DNA before melting. J Mol Biol. 1972 Apr 14;65(3):381–399. doi: 10.1016/0022-2836(72)90196-9. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod J. C., Johnson W. C., Jr, Huntington S. K., Maestre M. F. Conformation of deoxyribonucleic acid in alcohol solutions. Biochemistry. 1973 Dec 4;12(25):5092–5096. doi: 10.1021/bi00749a011. [DOI] [PubMed] [Google Scholar]
- Glaubiger D., Hearst J. E. Effect of superhelical structure on the secondary structure of DNA rings. Biopolymers. 1967;5(8):691–696. doi: 10.1002/bip.1967.360050803. [DOI] [PubMed] [Google Scholar]
- Goodwin D. C., Brahms J. Form of DNA and the nature of interactions with proteins in chromatin. Nucleic Acids Res. 1978 Mar;5(3):835–850. doi: 10.1093/nar/5.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. M., Taylor T. N., Lang D. Dehydrated circular DNA: circular dichroism of molecules in ethanolic solutions. Biopolymers. 1978 Jan;17(1):145–157. doi: 10.1002/bip.1978.360170111. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. DNA structure: evidence from electron microscopy. Science. 1978 Aug 11;201(4355):525–527. doi: 10.1126/science.663672. [DOI] [PubMed] [Google Scholar]
- Hancock R. Interphase chromosomal deoxyribonucleoprotein isolated as a discrete structure from cultured cells. J Mol Biol. 1974 Jul 5;86(3):649–663. doi: 10.1016/0022-2836(74)90187-9. [DOI] [PubMed] [Google Scholar]
- Hanlon S., Brudno S., Wu T. T., Wolf B. Structural transitions of deoxyribonucleic acid in aqueous electrolyte solutions. I. Reference spectra of conformational limits. Biochemistry. 1975 Apr 22;14(8):1648–1660. doi: 10.1021/bi00679a017. [DOI] [PubMed] [Google Scholar]
- Hanlon S., Johnson R. S., Wolf B., Chan A. Mixed conformations of deoxyribonucleic acid in chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3263–3267. doi: 10.1073/pnas.69.11.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton D. M., Bode V. C. Purification of closed circular lambda deoxyribonucleic acid and its sedimentation properties as a function of Sodium chloride concentration and ethidium binding. J Biol Chem. 1975 Feb 10;250(3):1071–1079. [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Minyat E. E., Frank-Kamenetskii M. D., Schyolkina A. K. The B to A transition of DNA in solution. J Mol Biol. 1974 Aug 25;87(4):817–833. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Schyolkina A. K., Poletayev A. I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12(1):89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Jr A circular dichroism spectrometer for the vacuum ultraviolet. Rev Sci Instrum. 1971 Sep;42(9):1283–1286. doi: 10.1063/1.1685367. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Jr, Girod J. C. A novel denaturation of DNA. Biochim Biophys Acta. 1974 Jun 27;353(2):193–199. doi: 10.1016/0005-2787(74)90184-1. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic R. T., van Holde K. E. Sedimentation of homogeneous double-strand DNA molecules. Biochemistry. 1977 Apr 5;16(7):1490–1498. doi: 10.1021/bi00626a038. [DOI] [PubMed] [Google Scholar]
- Lawrence J. J., Chan D. C., Piette L. H. Conformational state of DNA in chromatin subunits. Circular dichroism, melting, and ethidium bromide binding analysis. Nucleic Acids Res. 1976 Nov;3(11):2879–2893. doi: 10.1093/nar/3.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- Maestre M. F., Gray D. M., Cook R. B. Magnetic circular dichroism study on synthetic polynucleotides, bacteriophage structure, and DNA's. Biopolymers. 1971;10(12):2537–2553. doi: 10.1002/bip.360101214. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Venable J. H., Jr, Lerman L. S. The structure of psi DNA. J Mol Biol. 1974 Mar 25;84(1):37–64. doi: 10.1016/0022-2836(74)90211-3. [DOI] [PubMed] [Google Scholar]
- Nelson R. G., Johnson W. C., Jr Conformation of DNA in ethylene glycol. Biochem Biophys Res Commun. 1970 Oct 9;41(1):211–216. doi: 10.1016/0006-291x(70)90490-0. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. The structure of the chromatin core particle in solution. Nucleic Acids Res. 1977 Sep;4(9):3199–3214. doi: 10.1093/nar/4.9.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T. Y., Fasman G. D. Conformation of deoxyribonucleic acid in chromatin: a circular dichroism study. J Mol Biol. 1970 Aug 28;52(1):125–129. doi: 10.1016/0022-2836(70)90182-8. [DOI] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell. 1976 Jun;8(2):215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Studdert D. S., Patroni M., Davis R. C. Circular dichroism of DNA: temperature and salt dependence. Biopolymers. 1972;11(4):761–779. doi: 10.1002/bip.1972.360110404. [DOI] [PubMed] [Google Scholar]
- Thomas G. J., Jr, Prescott B., Olins D. E. Secondary structure of histones and DNA in chromatin. Science. 1977 Jul 22;197(4301):385–388. doi: 10.1126/science.560060. [DOI] [PubMed] [Google Scholar]
- Tunis-Schneider M. J., Maestre M. F. Circular dichroism spectra of oriented and unoriented deoxyribonucleic acid films--a preliminary study. J Mol Biol. 1970 Sep 28;52(3):521–541. doi: 10.1016/0022-2836(70)90417-1. [DOI] [PubMed] [Google Scholar]
- VINOGRAD J., BRUNER R., KENT R., WEIGLE J. Band-centrifugation of macromolecules and viruses in self-generating density gradients. Proc Natl Acad Sci U S A. 1963 Jun;49:902–910. doi: 10.1073/pnas.49.6.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J. Physical and topological properties of circular DNA. J Gen Physiol. 1966 Jul;49(6):103–125. doi: 10.1085/jgp.49.6.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H. P., Grossman L. I., Vinograd J. Isolation and partial characterisation of the relaxation protein from nuclei of cultured mouse and human cells. Eur J Biochem. 1975 Jun 16;55(1):79–93. doi: 10.1111/j.1432-1033.1975.tb02140.x. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Degree of superhelicity of covalently closed cyclic DNA's from Escherichia coli. J Mol Biol. 1969 Jul 28;43(2):263–272. doi: 10.1016/0022-2836(69)90266-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Variation of the average rotation angle of the DNA helix and the superhelical turns of covalently closed cyclic lambda DNA. J Mol Biol. 1969 Jul 14;43(1):25–39. doi: 10.1016/0022-2836(69)90076-x. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Complex formation between ethidium bromide and nucleic acids. J Mol Biol. 1965 Aug;13(1):269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- Young L. S., Champoux J. J. Interaction of the DNA untwisting enzyme with the SV40 nucleoprotein complex. Nucleic Acids Res. 1978 Feb;5(2):623–635. doi: 10.1093/nar/5.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]



