Abstract
Preorganizing a ligand in the conformation it adopts upon binding to a protein has long been considered to be an effective way to improve affinity by making the binding entropy more favorable. However, recent thermodynamic studies of a series of complexes of the Grb2 SH2 domain with peptide analogs having constrained and flexible replacements for a phosphotyrosine residue revealed that less favorable binding entropies may result from constraining ligands in their biologically active conformations. Toward probing the origin of this unexpected finding, we examined the complexes of four phosphotyrosine-derived analogs with the Grb2 SH2 domain using molecular dynamics simulations with a polarizable force field. Significantly, the computed values for the relative binding free energies, entropies and enthalpies of two pairs of constrained and unconstrained ligands reproduced the trends that were determined experimentally, although the relative differences were overestimated. These calculations also revealed that a large fraction of the ligands lacking the constraining element exist in solution as compact, macrocyclic-like structures that are stabilized by interactions between the phosphate groups and the amide moieties of the C-terminal pY+2 residues. In contrast, the three-membered ring in the constrained ligands prevents the formation of such macrocyclic structures, leading instead to globally extended, less ordered conformations. Quasiharmonic analysis of these conformational ensembles suggests that the unconstrained ligands possess significantly lower entropies in solution, a finding that is consistent with the experimental observation that the binding entropies for the unconstrained ligands are more favorable than for their constrained counterparts. This study suggests that introducing local constraints in flexible molecules may have unexpected consequences, and a detailed understanding of the conformational preferences of ligands in their unbound states is a critical prerequisite to correlating changes in their chemical structure with protein binding entropies and enthalpies.
Keywords: ligand preorganization, entropy paradox, AMOEBA polarizable force field
INTRODUCTION
Understanding the effects of making chemical modifications to ligands upon their relative binding energetics is a critical step in structure-based drug design. Preorganizing a flexible ligand in the conformation it adopts upon binding has long been considered a useful strategy to achieve more a favorable binding entropy and thus an improved binding affinity.1 Indeed, the program CAVEAT was developed in part to facilitate the design of constrained molecules bearing substituents directed in predefined orientations.2 Awareness of the potential energetic benefits of ligand preorganization dates back to work by Jencks in the 1970s,3 and there are numerous reports of the increased affinities that may accompany the introduction of conformational constraints into flexible ligands.3–9 However, it has recently been reported that the binding entropy of a constrained ligand may actually be less favorable than its flexible control.10–14 Even if ligand preorganization leads to a beneficial entropic contribution to binding, the enhancement to binding affinity may be offset by a compensating enthalpic penalty.7,15–23 Entropy-enthalpy compensation has been widely studied, but its origin is not well understood.5,24–29 While some consider this effect as an intrinsic physical phenomenon, others argue that the entropy-enthalpy compensation is a statistical artifact arising from obtaining entropy and enthalpy based on temperature dependent data from both experiment and theoretical calculations.24,25,27,30–33
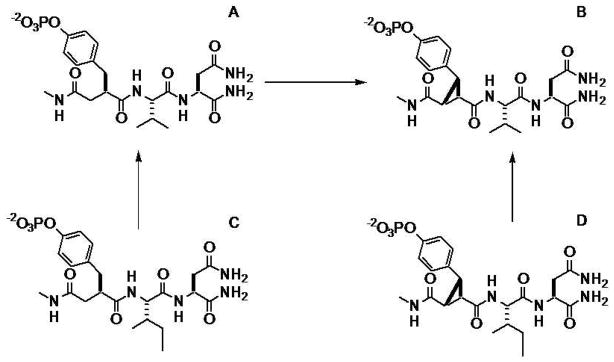
In order to understand the detailed energetic effects of ligand preorganization on protein-ligand interactions, it is essential to perform systematic experimental studies to determine the contributions of entropy and enthalpy to binding free energy and to correlate these with structural and dynamic analyses of the protein-ligand complexes using X-ray crystallographic and NMR spectroscopic methods.34 For example, DeLorbe et al.14 recently examined the binding energetics and structures associated with complexes of a series of constrained and flexible phosphotyrosine-derived peptide analogs with the SH2 domain of growth receptor binding protein 2 (Grb2); Grb2 is a 25 KDa cytosolic adapter protein that is involved in activation of the Ras signal transduction pathway.35 The constrained ligands cpYVN and cpYIN, which were preorganized by incorporating a cyclopropane ring at the pY replacement in the pseudopeptides fpYVN and fpYIN, respectively, (Figure 1), have the same functional groups and the same number and type of heavy atoms as their flexible controls. The thermodynamic binding parameters of these ligands for the Grb2 SH2 domain were determined by isothermal titration calorimetry (ITC),36 and the binding entropies for the constrained ligands were found to be less favorable than for their flexible analogs. This unexpected finding is contrary to the conventional wisdom that ligand preorganization should be accompanied by a more favorable binding entropy. Their less favorable binding entropies notwithstanding, the constrained ligands bound with higher affinities than their more flexible counterparts because of significantly more favorable binding enthalpies. That the measured heat capacity changes on binding of the flexible/constrained ligand pairs were similar suggests that these thermodynamic differences do not arise from desolvation effects.37 Although these experimental studies clearly show that ligand preorganization does not necessarily result in a more favorable binding entropy, the molecular origin of the unanticipated behavior was not examined.
Figure 1.
Chemical structures of the ligands studied and the perturbation scheme. A. fpYVN; B. cpYVN; C. fpYIN; D. cpYIN.
Over the past decades, numerous efforts have been devoted to develop and apply computational approaches to screen and design potent ligands for drug discovery.38–40 Computational methods such as docking and molecular dynamics utilizing continuum and explicit solvent models have long been employed toward predicting protein-ligand binding affinities in silico.38,40–42 Among the various strategies that have been explored, detailed alchemical pathway simulations using explicit solvent show significant promise for providing energetically accurate predictions of protein-ligand binding affinity.34,43,44 The absolute binding free energies calculated from such pathways correlate reasonably well with experimental data, and root mean square (RMS) errors of less than 3 kcal/mol are often reported.41 On the other hand, the relative binding free energies can be calculated more accurately, if there is sufficient sampling of protein-ligand-water configurational space.45 Decomposition of binding free energy into entropic and enthalpic contributions offers important insights into the driving forces for protein-ligand recognition;34 however, quantitative estimation of the binding entropy remains a significant challenge. Common approaches to estimating the binding entropy include quasiharmonic analysis,46,47 normal mode analysis,48–50 and knowledge based scoring functions.51–53 These methods have been applied to several protein-ligand systems,54–60 but the contributions from solvation are typically neglected or approximated due to the computational expense. More physically rigorous alchemical pathway approaches are also applicable to evaluating entropy, but few investigations have reported using such methods in protein-ligand interactions. 61
In the present work, we use molecular modeling to explore how introducing conformational constraints into two closely related phosphotyrosine-derived peptides affects the thermodynamic parameters for their complexation with the Grb2 SH2 domain. Because the phosphate group in these ligands is charged, the AMOEBA polarizable force field,62–65 which accounts for multipole electrostatics and polarization effects, was used to model protein, ligand, and water components in each MD simulation. The AMOEBA force field has been successfully applied to accurately model a number of highly polar molecular systems, including water,63 monovalent and divalent ions,66–69 small molecules,70 peptides,71 and trypsin-benzamidinium binding.72–74 We report herein the calculations of binding free energies, enthalpies and entropies, as well as the results from simulations of the structure and dynamics of the ligands in water and in their complexes with the Grb2 SH2 domain. The results are compared with experimental observations, and a possible molecular origin for the unexpected, unfavorable entropic effect resulting from preorganizing these phosphotyrosine-derived ligands is presented.
METHOD
Entropy Calculation
The change in entropy (ΔS) is related to the change in free energy by75
| (1) |
In this study, we computed the entropy change numerically from the slope of a linear fit to the temperature dependence of the free energy change near room temperature. This approach is essentially identical to the finite difference method that has been used previously.76–78 While other direct or perturbation methods exist for computing enthalpy or entropy, we have found that the numerical approach is the most reliable for calculations of energetics. In addition, because REMD is being used to simulate free ligands in solution, it is possible to take advantage of the temperature dependence of free energy data.
The relative change in enthalpy is then computed as
| (2) |
The Bennett Acceptance Ratio (BAR) method79 was utilized to calculate the relative binding free energy (ΔA). Not considering finite size effects, this Helmholtz free energy is equal to the Gibbs free energy change at the same pressure. The free energy change between neighboring states (λi and λi+1) is given by the expression
| (3) |
where
| (4) |
and j is the iteration index. Here, Uλi is the potential energy of the system evaluated using the parameters from λi, and <> indicates the ensemble average. ΔA is solved iteratively until the value of (ΔA(j) − ΔA(j−1)) < 0.01 kcal/mol.
In order to avoid the end point singularity during the alchemical transformation, the soft-core buffered 14-7 potential was used for van der Waals (vdW) interactions
| (5) |
where ε is the well depth, λ is the scaling factor, ρij=Rij/Rij0 with Rij as the actual separation between atoms i and j and Rij0 is the minimum energy distance parameter.
Quasiharmonic Analysis
Quasiharmonic analysis was performed to characterize the collective motions of molecules at thermodynamic equilibrium.46,47 The quasiharmonic approximation assumes that the spatial fluctuations in the system follow a multivariate Gaussian distribution. In a quasiharmonic frequency analysis on an n-atom system, the eigenvalues λi (i=1,2,…,3n-6) of the mass weighted covariance matrix of atomic fluctuations are calculated to determine the quasiharmonic frequencies, , which are then used to estimate the conformational entropy, given by
| (6) |
where ħ is the Planck constant, and R is the gas constant. The quasiharmonic method has been successfully applied to simple molecular systems with few energy minima. However, this method will overestimate the configurational entropy for more complex systems having multiple occupied energy wells, particularly when using Cartesian coordinates rather than internal coordinates.80
Force Field and Parameterization
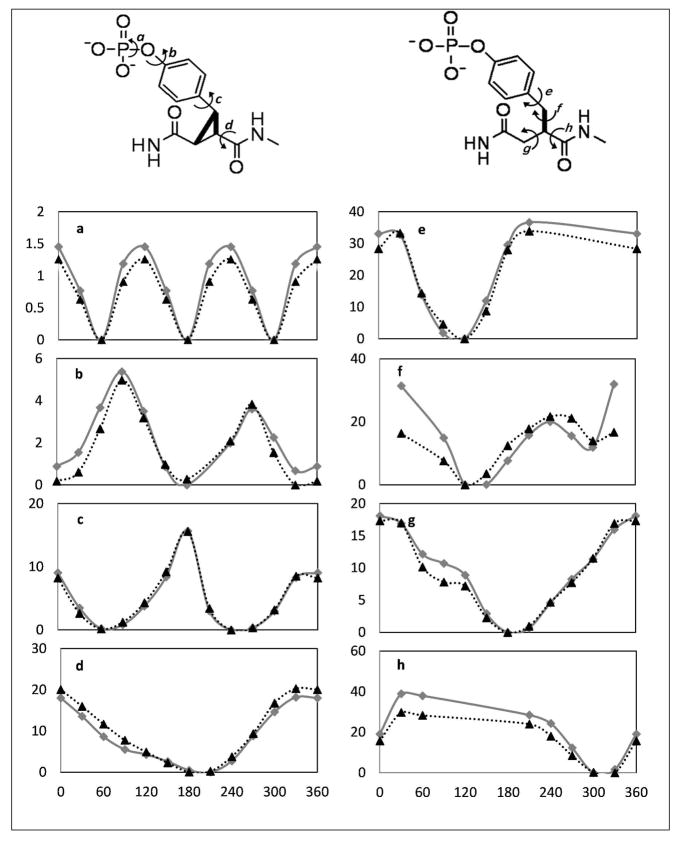
The AMOEBA polarizable force field63,65 was applied to model the protein, water molecules, and the amino acid residues in the peptide-analogs fpYVN, fpYIN, cpYVN and cpYIN (Figure 1). Additional parameterization was necessary for the phosphotyrosine (pY) replacements fpY and cpY. Missing valence parameters for the constrained and unconstrained pY segments were derived from quantum mechanics (QM) calculations by using the “Valence” module in the TINKER software package.81 “Valence” sets the equilibrium bond lengths and angles based on the HF/6-31G* optimized structures of the pY replacements; the force constants and vdW parameters were then transferred from the existing AMOEBA force field parameters for the same atom types. Trimethyl and dimethyl phosphate (TMP and DMP) were used to derive the vdW parameters of the phosphate group by fitting to both the QM structure and energy of TMP/DMP-water dimers. The vdW parameters were further tuned to match the experimental liquid density and heat of vaporization of TMP. The electrostatic parameters for the pY side chain were obtained from QM at the MP2/6-311++G(2d,2p) level by the “original-fit” approach,70,82 wherein the atomic multipoles for the pY segment were initially derived from the MP2/6-311G** density matrix using the original distributed multipole analysis (DMA).83,84 The atomic dipole and quadrupole moments of the pY residue were then optimized to the electrostatic potential (ESP) around the whole ligand computed at the MP2/6-311++G(2d,2p) level (electrostatic parameters of other residues were fixed). The ESP root mean square differences between QM and that from the final atomic multipoles, evaluated over roughly 35000 grid points around the peptide, were 1.55 and 0.92 kcal/mol per unit charge for cpYVN and fpYVN, respectively.
The model compounds of both unconstrained and constrained pY segments and the dihedral angles ah for which parameters were derived are illustrated in Figure 2. Dihedral angle parameters in the pY replacement that were missing were obtained by comparing the QM conformational energy profile to those computed from the corresponding MM (Molecular Mechanics) using all energy terms except for the dihedral angle term. The difference in energy was then fit to a three-term, Fourier series torsional function. The torsional energy in MM serves essentially as an “error” function. Typically, the torsional energy parameters (V1, V2, and V3) are less than 1–3 kcal/mol for rotation about a single bond, whereas the overall conformational energy barriers could be much higher due to intramolecular electrostatic and vdw interactions. (Figure 2)
Figure 2.
Conformational energy profiles for constrained and unconstrained pY segments. The grey lines with squares are QM relative energy, and the back dotted lines with triangles are the MM relative energy. Y-axis is the relative energy in kcal/mol. X-axis is the dihedral angle in degree.
Computational Details
In order to evaluate the thermodynamic driving forces for ligand binding to the SH2 domain, we calculated the relative binding free energy, enthalpy and entropy contributions for constrained and unconstrained ligand pairs, according to the alchemical pathway shown in Figure 1. The relative binding free energies for each pair of constrained and unconstrained ligands were computed from the free energy differences between the ligands in water and in the protein binding pocket. The fpYVN ligand was gradually transformed into cpYVN by performing 26 simulation steps; 10 steps were performed in the alchemical transformations of Val to Ile in the constrained and unconstrained ligands. For the ligand-water systems, replica exchange molecular dynamics (REMD)85–87 simulations were performed with 48 replicas at temperatures between 260 and 620 K (detailed schedule can be found in the Supporting Information).88 For the transformation of fpYVN to cpYVN, the bond forming the cyclopropane ring was introduced by gradually increasing the force constant from 0 to 550 kcal/mol/Å2; while simultaneously transforming two hydrogen atoms into “dummy atoms” by turning off their vdW and electrostatic interactions with all other atoms. Note that the valence contributions from the dummy atoms were canceled between perturbations of the ligand in water and in the solvated complex. Constant-volume and constant-temperature (NVT) simulations of 2.5 ns were performed at each step. Replicas were exchanged every 2 ps, with exchange success rates for all replicas were in the range of 22% to 40%. All the REMD simulations were performed using the parallel SANDER module in AMBER10, and the REMD implemented in AMBER1089 was modified for use with the AMOEBA force field. For the protein-ligand complexes in explicit water, relative free energies for constrained and unconstrained ligands having Val and Ile at the pY+1 position were calculated at 288 K, 298 K and 308 K. For each set of alchemical calculations at each temperature, a total of 65 ns of NVT molecular dynamics was performed over 26 steps, using the PMEMD module in AMBER10. For all the simulations, the vdW cutoff was set at 12 Å and the long-range electrostatics were treated using Particle Mesh Ewald (PME) summation90–92 with a grid spacing of 0.8 Å and a real space cutoff of 7 Å. The induced dipoles, which were also computed with PME, were iterated until the root mean square (RMS) change was below 0.01 Debye. A tighter induced dipole convergence of 10−5 RMS Debye was used in the energy calculation for the post free energy analysis with the Bennett Acceptance Ratio (BAR). By using a bootstrap procedure, the statistical uncertainty was estimated as the standard deviation of the average free energy values that are computed using 100 partial simulation trajectory blocks (1.0 ns). Given that the uncertainty in the entropy calculation was dominated by the free energies at the lowest and highest temperatures (288 K and 308 K), the statistical error for − TΔS was estimated from the upper and lower bound of free energy changes at 288 K and 308 K; thus the statistical error for − TΔS is twice the statistical error of free energy.
The entropy was extracted from the temperature dependence of the free energies via linear regression (Eq. 1). For ligands in water, the relative free energy ΔAwat was obtained from REMD at 18 temperatures between 260 K and 360 K. A linear fit was used to interpolate ΔAwat at 288 K, 298 K and 308 K. These values were then subtracted from the free energy changes of the ligand in complex (ΔAcomp) at the same temperatures to obtain relative binding free energy ΔΔAbind. The entropic contribution was computed from the slope of the fitted linear temperature dependence of the relative binding free energy. The enthalpy was evaluated via ΔΔA + TΔΔS. The total simulation time for the combined MD simulations of free ligands and complexes at all temperatures was more than 6 μs.
RESULTS AND DISCUSSION
Ligand conformational properties and sampling
One of the essential goals of this study is to obtain an accurate description of the distribution of the different conformational ensembles of the peptide-like ligands in solution. These ligands present a significant challenge for molecular simulations as they each possess a total of more than a dozen rotatable bonds, with four being in the constrained pY replacement (cpY) and six belonging to the unconstrained pY mimic (fpY). At the same time, strong intramolecular interactions between the charged phosphate moiety and other polar groups in the molecule result in certain stable conformations with low potential energies. The conformational energy profile for the pY replacement indicates the energy barrier for escaping these stable conformations can be tens of a kcal/mol (Figure 2). In solution, it is likely that the conformational population will be significantly different because of interactions with water molecules. In fact, it is known that the dipeptide Ala-Ala has distinctly different local minimum energy structures in gas and solution phases.64 We examined each of the main conformational degrees of freedom for the pY replacement that we parameterized. The AMOEBA conformational energy profiles depicted in Figure 2 are in good agreement with high-level ab initio QM results. The average root-mean-square deviation (RMSD) between QM and MM minimized structures for the entire ligand is about 0.25 Å per atom.
Adequate sampling of the configurational space of the system, including the ligand, is critical in order to obtain accurate thermodynamic information from the simulations. When the peptide-like ligands are bound to SH2 domain, they are restricted within the protein pocket, and their structures are well-defined and in good agreement with the X-ray crystallographic structures.14 Since the ensemble of structures of the free ligands in solvent is unknown, we performed MD simulations of the unconstrained fpYVN ligand in a dielectric continuum (ε = 80) at and above room temperature in order to explore the conformational space of these peptide analogs in water and to guide the more detailed, explicit-solvent simulations. The potential energy and torsional RMSD distributions obtained from multiple independent simulation trajectories were compared to examine the convergence of the MD sampling. We found that MD simulations at room temperature do not produce converged distributions after 30 ns. On the other hand, when these simulations are performed at 600 K or above, the distributions quickly converge after a few nanoseconds (see Supporting Information). Based on the information obtained from the simple continuum simulations, replica-exchange molecular dynamics (REMD) simulations in explicit solvent at temperatures ranging from 260 K to 620 K were performed to compute the relative free energy and entropy between different ligands in the solvent environment. Using these simulations, we examined the distribution of the main torsions of fpYVN in solution at 298 K. Overall, we observe a broad sampling in torsional space and a number of torsional transitions. The autocorrelation functions of these torsions exhibit fast decay of only a few ps to reach 1/e, perhaps because of the artificially fast kinetics of REMD; some examples are included in the Supporting Information.
Calculated binding thermodynamics consistent with experimental measurements
We evaluated the relative binding free energy and entropy for complex formation of the constrained ligands cpYVN and cpYIN and their corresponding unconstrained analogs fpYVN and fpYIN with the Grb2 SH2 domain. The free energy and entropy were computed from molecular dynamics simulations using the AMOEBA polarizable force field, and results are summarized and compared with the values that were determined experimentally by ITC in Table 1.14 In order to facilitate the comparison of the calculated and experimental values, we used the fpYVN ligand as the reference and set the calculated values for it to those determined experimentally. The calculated thermodynamic parameters for the other ligands were then obtained by difference according to the relative binding free energies, entropies and enthalpies derived from the simulations.
Table 1.
Calculated and experimental thermodynamics (kcal/mol) for phosphotyrosine (pY)-containing peptide analogs and their constrained counterparts binding to the SH2 domain of Grb2. fpYV(I)N is the unconstrained tri-peptide analog consisting of pY, V (or I) and N residues; cpYV(I)N are the constrained counterparts (see Figure 1). The ΔG°, ΔH° and ΔS° are the absolute binding free energy, enthalpy and entropy differences, respectively. With the calculated values of fpYVN set to experimental values, thermodynamics for the remaining ligands were computed from the relative binding free energy and enthalpy obtained from MD simulations. Statistical errors of the calculated binding free energy are given in the parenthesis.
| Calculation
|
Experiment14 |
|||||
|---|---|---|---|---|---|---|
| ΔG° (kcal/mol) | ΔH° (kcal/mol) | −TΔS (kcal/mol) | ΔG° (kcal/mol) | ΔH° (kcal/mol) | −TΔS° (kcal/mol) | |
| fpYVN | −7.7 | −5.4 | −2.3 | −7.7 | −5.4 | −2.3 |
| cpYVN | −10.0(0.6) | −15.7 | 5.7(1.2) | −8.8 | −7.9 | −0.8 |
| fpYIN | −7.7(0.1) | −3.2 | −4.5(0.2) | −7.7 | −5.5 | −2.2 |
| cpYIN | −9.8(0.4) | −14.0 | 4.3(0.8) | −8.6 | −8.3 | −0.3 |
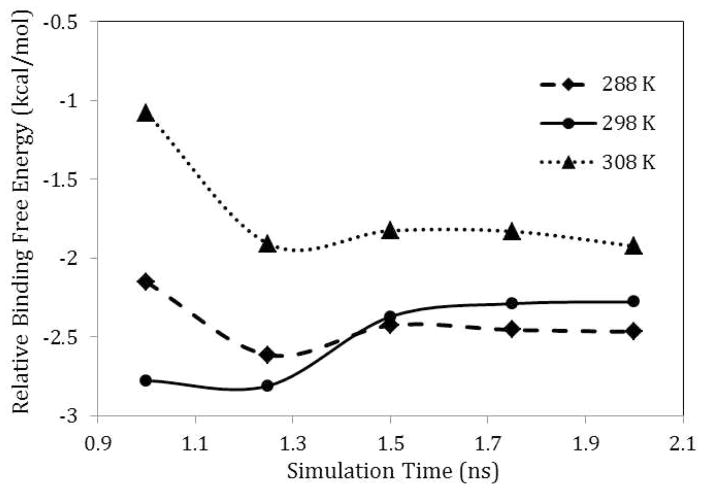
The order of the calculated binding free energies correspond to those determined experimentally with fpYVN ≈ fpYIN < cpYIN < cpYVN. The calculated binding free energy difference between fpYVN and cpYVN was overestimated by about 1.2 kcal/mol compared to experiment, whereas the difference in free energies of the Val and Ile variants match experimental results reasonably well. The transformation of fpYVN into cpYVN requires that a covalent bond be created. A small change in the bond length corresponds to large energy fluctuations due to the stiff bond stretching and angle bending energy term.93,94 During the transformation of fpYVN into cpYVN in water or in their complexes with the Grb2 SH2 domain, 26 intermediate steps are necessary to obtain sufficient overlap between the configurational spaces associated with neighboring steps. As illustrated in Figure 3, the relative free energies converge after 1.5 ns simulations for each intermediate step at different temperatures, with a deviation of less than 0.1 kcal/mol. REMD simulations for the ligands in water at 48 different temperatures were performed to enhance the sampling near room temperature. Accordingly, the relative binding free energies for each pair of ligands are effectively the result of 3 μs of simulation time.
Figure 3.
Convergence of relative binding free energy between fpYVN and cpYVN over simulation time at selected temperatures.
Additional analyses were performed in order to scrutinize the convergence and effectiveness of the free energy calculations. To inspect the thermodynamic cycle closure, we reevaluated the relative binding free energy associated with the alchemical transformation fpYVN to cpYVN at 298 K after systematically skipping intermediate perturbation steps (skipping every 4th, 5th … 13th steps). The results indicate that the relative binding free energy converges within 0.3 kcal/mol after 22 to 26 perturbation steps. For neighboring steps during the perturbation, the histograms of potential energy differences from forward and backward perturbation have been obtained and verified to overlap with each other. In addition, statistical inefficiency calculations95 show that ~22 ps and ~45 ps are needed for ligand sampling in water and in the complex to lose “memory” of their previous configurations, respectively. Further details of the thermodynamic cycle, energy overlap, and statistical inefficiency tests are found in the Supporting Information.
The relative binding free energies were then decomposed into enthalpy and entropy contributions. Perturbation methods, including finite difference, single state perturbation, β-perturbation, modified β-perturbation, and the perturbation and correction method, provide a physically rigorous evaluations of changes in entropy and enthalpy.78,96 However, compared with the free energy calculations, the entropy evaluated from computational methods has much greater error. With the exception of the finite difference approach, all of the above methods suffer from numerical underflow problems for systems with large energy fluctuations such as solvated protein-ligand complexes. Recently, Wyczalkowski et al.97 calculated solvation entropy changes based on the analytical temperature derivative of the Bennett Acceptance Ratio and Multistate Bennett Acceptance Ratio (mBAR) estimators. The BAR/mBAR approach to computing entropy is sensitive to energy fluctuations and thus requires long simulation times to achieve convergence. Using the restraint release (RR) approach, Warshel et al.61 reported the binding entropy of three protein-ligand systems with encouraging agreement to experimental measurements. In the RR approach, multiple structures of the ligands must be selected before free energy perturbation and quasiharmonic calculations. Considering the potentially high flexibility of our peptide ligands, a large number of representative configurations are needed to account for the global minima in each perturbation. In the current study, the REMD simulations that were performed on the free ligands in water already provided free energy data at different temperatures. Therefore, we took advantage of the simulations at different temperatures and adopted the finite difference method that has been utilized to calculate entropy change for both small and relatively large systems.60,75,98–100 We used a linear regression to fit the temperature dependence of the relative binding free energies at 288 K, 298 K and 308 K (see Computation Details). For all the systems, the R2 values for the linear fit range from 0.81 to 0.97.
The calculated relative binding entropies and enthalpies for the two ligand pairs show the same trends as experiment. Both calculation and experiment indicate that the constrained ligands bind to the Grb2 SH2 domain with less favorable binding entropy and more favorable binding enthalpy than their more flexible controls (Table 1). Relative to the unconstrained ligands fpYVN and fpYIN, the increased binding affinity, or lower binding free energy, that is observed for the two constrained ligands cpYVN and cpYIN is attributed to an enthalpic advantage rather than an entropic one. Based upon the simulations, preorganization results in an unfavorable binding entropy change (higher −TΔS) of 8 kcal/mol, which is offset by an enthalpic gain of about −10 kcal/mol. Hence, although constraining the ligands fpYVN and fpYIN increases their binding affinities, both computational and experimental results show that this affinity enhancement does not arise from more favorable entropic factors as would normally be expected. The calculated absolute binding entropy −TΔS of the constrained cpYV(I)N ligands is unfavorable (positive), whereas slightly favorable (negative) values were obtained experimentally. This difference is a reflection of our overestimation of the magnitude of the relative binding entropy between constrained and unconstrained ligand pairs, even though the sign of the relative change was predicted correctly. The mutation of Val to Ile in both constrained and unconstrained ligands had an insignificant effect upon both the calculated and the experimental values for binding enthalpies and entropies. Moreover, both the simulated and experimental data seem to suggest that enthalpy/entropy compensation limits the enhancement to the binding affinity, as reflected by the linear relationship between entropy and enthalpy in Figure 4.
Figure 4.
Correlation between binding enthalpy and binding entropy of fpYVN, fpYIN, cpYVN and cpYIN. Black diamonds are calculated values; Grey squares are experimental data. Both calculated and experimental binding enthalpy and entropy of fpYVN are shifted to zero for comparison purpose.
Binding is a process that involves ligand desolvation followed by formation of the protein-ligand complex, so the behavior of unbound ligands in water plays an important role in the overall process. In the current study, the relative binding free energy and entropy was evaluated as the difference between the free energy and entropy changes of the protein and the ligands in solvent and complex environments. The entropy decomposition given in Table 2 suggests that the majority of the unfavorable binding entropy observed for constraining fpYVN and fpYIN arises from differences in the entropies of the free ligands in solvent rather than in their complexes.
Table 2.
Comparison of relative binding energetics (kcal/mol) between unconstrained and constrained ligand pairs fpYVN/cpYVN and fpYIN/cpYIN, and decomposition of the relative binding free energy and entropy into unbound (in solvent) and complex contributions.
| ΔΔG (kcal/mol) | ΔGsolvent* (kcal/mol) | ΔGcomplex* (kcal/mol) | −TΔΔS (kcal/mol) | −TΔSsolvent (kcal/mol) | −TΔScomplex (kcal/mol) | |
|---|---|---|---|---|---|---|
| fpYVN → cpYVN | −2.27 | 26.68 | 24.41 | 8.05 | −6.88 | 1.16 |
| fpYIN → cpYIN | −2.07 | 26.80 | 24.73 | 7.45 | −7.42 | 0.03 |
The free energy contributions from solvent and complex (ΔGsolvent and ΔGcomplex) both include the relevant valence contributions from fpYV(I)N to cpYV(I)N. The valence contributions from water and protein were canceled in the calculation of relative binding free energy.
Effect of a chemical constraint on the organization of unbound ligands
The concept underlying ligand preorganization is that constraining an unbound ligand in the three-dimensional shape that corresponds to its bound structure will lead to a more favorable entropic term for binding. Both computational and structural studies show that the cyclopropane rings in cpYVN and cpYIN preorganize the atoms of the fpY replacement that interact with the protein in the bound conformations of fpYVN and fpYIN. Hence, in order to probe why introducing a conformational constraint into the flexible ligands fpYVN and fpYIN did not result in the expected entropic advantage, we analyzed the simulated structures and dynamics of the bound and unbound forms of the pseudopeptides cpYVN and fpYVN.
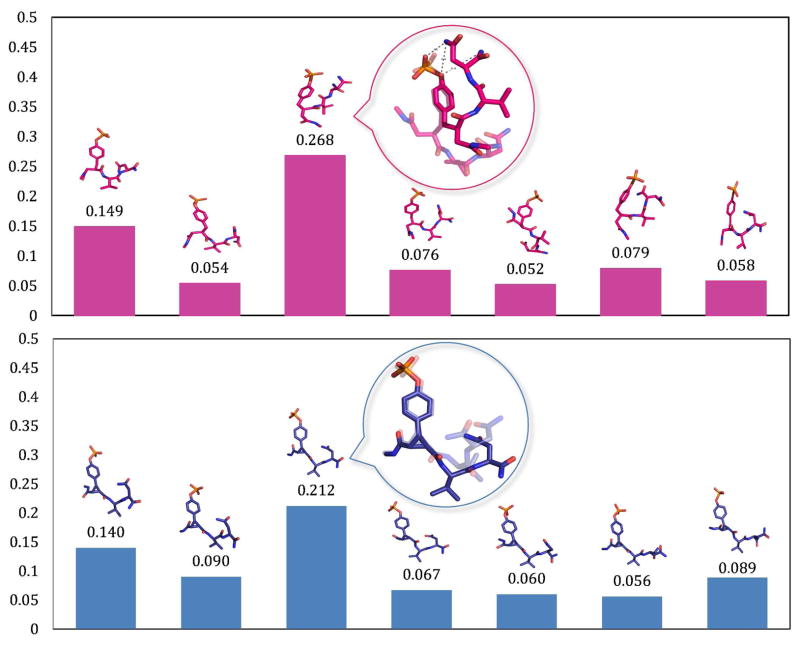
The structures of the ligands extracted from molecular dynamics trajectories were hierarchically clustered based on the all-atom RMSD of the ligands using an average-linkage algorithm101 over 2 ns at 298 K. In an average-linkage algorithm, the distance between one cluster and another cluster is computed as the average distance from any member of one cluster to any member of the other cluster. Recall that REMD simulations with 48 replicas at various temperatures were performed to facilitate the sampling of the ligand configurations at 298 K. This is equivalent to a total simulation time of 100 ns. By using 1.0 Å as the limit for the average distance to centroid, 22 and 20 clusters were obtained for unconstrained and constrained ligand trajectories, respectively. Interestingly, the dominant configuration (35%) of fpYVN is a rather compact, macrocyclic-like structure (Figure 5) with a prominent intramolecular contact between the phosphate oxygen atoms and the amide N–H groups of the pY+2 residue. On the other hand, the cyclopropane ring in the constrained ligands cpYVN and cpYIN prevents these functional groups from interacting with each other, so these residues interact with water and are oriented in opposite directions. Superimposition of the dominant structure of cpYVN with the structure extracted from the complex14 reveals that the cyclopropane ring does preorganize the flexible pY replacement in its biologically active conformation. Thus, even though the cyclopropane ring locally preorganizes the pY replacement, cpY, the simulations suggest that the constrained ligands possess significantly more flexibility at the pY+1 and pY+2 positions relative to their unconstrained counterparts.
Figure 5.
Clustering of the solvated ligand structures for fpYVN (pink) and cpYVN (blue). The most representative structures are plotted for clusters higher than 5%. The ligands structures from crystal (in transparency) are superposed onto the most dominant configuration.
The entropy of both constrained and unconstrained ligands was estimated from quasiharmonic analysis (Table 3). The absolute entropy calculated for solvated cpYVN is 557 cal/mol/K, which is about 25% greater than the value of 448 cal mol/K−1 that was calculated for fpYVN; this difference corresponds to about 30 kcal/mol in TΔS at room temperature. The same trend is observed for fpYIN and cpYIN ligands, which have essentially the same entropies as fpYVN and cpYVN, respectively. Vibration contributes about 80% of the total entropy, while translation and rotation account for the remainder, so the entropy difference between the constrained and unconstrained ligands arises primarily from the vibrational components. We further decomposed the vibrational entropy into contributions from individual atoms and found that the modes contributing most to the entropy difference arise from the pY+2 residue. This finding is consistent with the structural analysis above. Since the rigid cyclopropane ring separates the pY+2 residue and phosphate group of the pY replacement in the constrained ligands, the pY+2 residue is well exposed in solvent and tends to be mobile. On the other hand, the unconstrained ligands lack this ring, so motion of the pY+2 residue is restrained by hydrogen bonding interactions with the phosphate moiety.
Table 3.
Estimation of absolute configurational entropy by quasiharmonic analysis. Spro(complex), Slig(complex), and Slig(solvent) represent the entropy contributions of α-carbons of the Grb2 SH2 in solvated complex, ligands in solvated complex and unbound ligands in solvent, respectively. All the entropy contributions are in cal/mol/K.
| Spro(complex) | Slig(complex) | Slig(solvent) | |
|---|---|---|---|
| fpYVN | 822.37 | 353.80 | 448.07 |
| fpYIN | 814.78 | 345.58 | 444.43 |
| cpYVN | 831.80 | 358.08 | 557.30 |
| cpYIN | 818.00 | 350.00 | 563.84 |
Water molecules also contribute to the entropy of the system and thus may affect binding thermodynamics. The average number of intermolecular hydrogen-bonds between water and the ligands were computed from the MD simulations at 298.79 K, and the results shown in Table 4 reveal noticeable differences between the constrained and unconstrained ligands. The constrained ligands are solvated by about two more water molecules on average than their unconstrained counterparts. The higher number of contacts with water molecules for constrained ligands is consistent with the observation that constrained ligands are more extended and better exposed to solvent (Table 4). The unconstrained ligands possess more intramolecular hydrogen bonds than their constrained counterparts (2.6 vs. 1.4), so fewer water molecules are associated with the unconstrained ligands. According to our calculation, the entropy contribution from ligand plays a dominant role in the binding energetics. These simulations and quasiharmonic analyses suggest that the unconstrained ligands actually have lower absolute entropies than their constrained counterparts because they are able to form intramolecular hydrogen-bonding interactions between the phosphate group and the amide groups of the terminal pY+2 residue. These computations thus offer a novel insight that accounts for the observation that the constrained ligands bind with less favorable entropies. Namely, even though the cyclopropane ring in cpYVN and cpYIN orients the atoms of the more flexible pY replacements in fpYVN and fpYIN in their bound conformations, this ring precludes the formation of the intramolecular interactions that lower the overall entropy of fpYVN and fpYIN relative to cpYVN and cpYIN.
Table 4.
The average numbers of intermolecular water-ligand hydrogen-bonds around the four solvated ligands, and the average numbers of intramolecular hydrogen bonds within the ligands in solution at 298.79 K.
| fpYVN | fpYIN | cpYVN | cpYIN | |
|---|---|---|---|---|
| Intermolecular H-bond | 33.5 | 33.2 | 35.7 | 35.2 |
| Intramolecular H-bond | 2.7 | 2.4 | 1.4 | 1.4 |
Flexibility of solvated complexes with unconstrained and constrained ligands
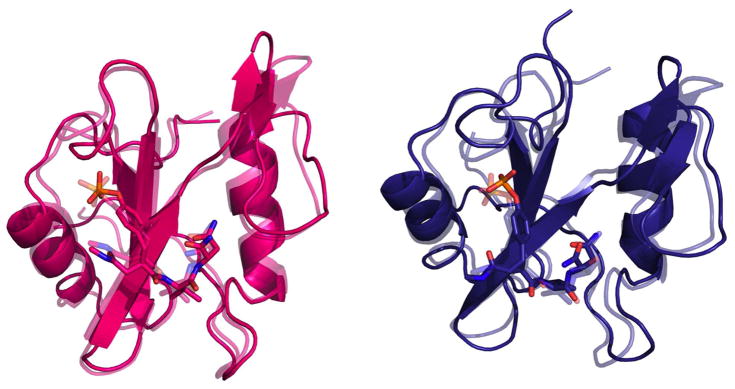
There is the possibility that introducing a conformational constraint into a flexible molecule might affect protein dynamics in the resultant complexes in a manner that could have entropic consequences. In order to probe this question, structures of the four solvated complexes were analyzed using the MD trajectories obtained from the free energy calculations. A comparison of the structures of the solvated complexes of the Grb2 SH2 domain with each of fpYVN, cpYVN, fpYIN and cpYIN derived from simulations and X-ray crystallography yielded all-atom RMSDs of 1.6 Å, 1.6 Å, 1.1 Å and 1.5 Å, respectively, suggesting that the structures obtained computationally are generally in good agreement with those determined by experiment. The RMSDs between the complexes of cpYVN and cpYIN and the complexes of fpYVN and fpYIN are both 1.4 Å. Representative structures of the complexes of fpYVN and cpYVN containing complexes from MD simulations are compared with crystal structures in Figure 7.
Figure 7.
Representative structures from MD simulations of fpYVN (pink) and cpYVN (deep blue) binding to Grb2 SH2 domain. The structures are generated by pyMOL.103
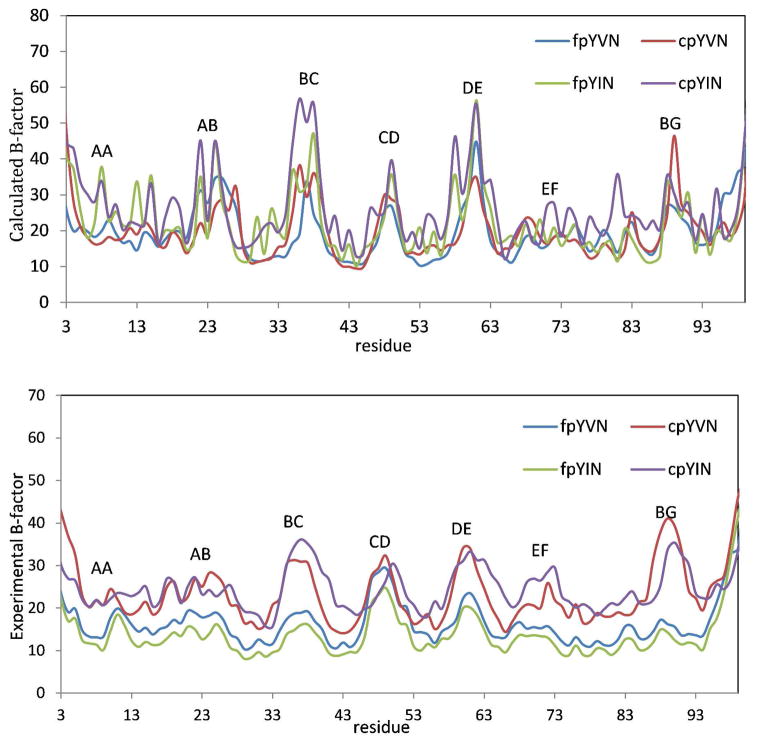
The calculated B-factors about the mean coordinates of the simulations over 2 ns for all Cα atoms are shown in Figure 6. The four complexes share very similar fluctuation modes, and the coordinates of most of the residues vary within only 1 Å, although residues in some of the loops fluctuate by as much as 2.1 Å. Structural variations in the BC loop (GluBC1–GluBC4) were observed upon comparing the crystal structures of complexes of the Grb2 SH2 domain with constrained and unconstrained ligands, although comparable variations in the BC loops were also found in coexisting complexes in the asymmetric unit.11,14,102 In our B-factor calculations, the fluctuations (1.5 to 2.0 Å) in the BC loops of the four complexes are the highest among all other loops. This observation suggests that the variations of the BC loop are more likely to result from the intrinsic flexibility of the loop rather than from differences in the binding modes of the constrained and unconstrained ligands. Based on the simulated B-factors in Figure 6, the B-factors for the protein backbone in the complexes of the constrained ligands are slightly greater than in the complexes of their unconstrained counterparts. Although the trends in the B-factors obtained computationally and experimentally are the same, the differences are more prominent in the experimental values.14
Figure 6.
B-factors of α-carbons calculated from MD trajectories of fpYVN (blue), fpYIN (green), cpYVN (red) and cpYIN (purple) binding to Grb2 SH2 domain (top); B-factor of α-carbons of the four ligands binding to Grb2 SH2 domain from experiment (bottom).
The entropies calculated from quasiharmonic analysis (Table 3) suggest that the Cα-atoms in the Grb2 SH2 domain complexed with each of the four different ligands have similar entropies of about 820 cal/mol/K. The entropies of the domain in the complexes of the constrained ligands cpYVN and cpYIN are slightly higher than in the complexes with their unconstrained controls (about 1%). The higher entropies of the domain in the complexes of the constrained ligands suggest a more favorable contribution to binding free energy, however, the opposite trend is seen by both experiment and calculation. Based on the analyses above, we are unable to find evidence that a change in dynamics of the SH2 domain is responsible for differences in the experimentally observed binding entropies for the various complexes.
The structures of fpYVN, cpYVN, fpYIN, and cpYIN in their respective complexes with the SH2 domain were also examined. The structures of the complexes were clustered based on RMSD of all atoms in the ligands from all MD snapshots using the same method described above for free ligands. For complexes containing fpYVN and cpYVN, there is one dominant conformation of the ligand that forms strong polar interactions with the protein, whereas there are two dominant conformers for fpYIN and cpYIN because of the flexibility of the pY+1 (Ile) residue. Other portions of the pseudopeptides fpYIN and cpYIN are highly similar to each other as well as to fpYVN and cpYVN. The most representative structures of fpYVN and cpYVN are depicted in Figure 7, while the corresponding structures for fpYIN and cpYIN not shown due to their high similarity. The entropies calculated from quasiharmonic analysis of the four ligands in the bound state range from 345.6 to 358.0 cal/mol/K, with the largest entropy difference being only 12 cal/mol/K, which is within statistical error, between cpYVN and fpYIN. Therefore, quasiharmonic analysis of the protein and the ligands in the respective complexes suggests that the entropic advantage for binding of the unconstrained ligands fpYVN and fpYIN does not arise from any differences in the entropy of either the protein or the ligands in bound state.
SUMMARY AND CONCLUSIONS
In order to probe the origin of the unexpected entropic consequences observed for binding of a series of constrained and flexible phosphotyrosine-derived peptide analogs to the Grb2 SH2 domain,14 we performed a series of calculations involving alchemical transformations at different temperatures on two sets of these analogs. Consistent with experimental results, these computations predicted that the binding affinity of the unconstrained peptides fpYV(I)N for the Grb2 SH2 domain is lower than that of the corresponding constrained peptides cpYV(I)N and that the mutation of V to I is not accompanied by significant changes in binding free energy. The experimental observation that unconstrained peptide analogs bind with more favorable entropies but significantly less favorable enthalpies than their constrained counterparts is also well reproduced by our computations, but the differences in the relative binding entropy and enthalpy components are overestimated. Even though these simulations nicely reproduce those trends observed experimentally, further refinements to the method are needed in order to improve the accuracy of predicting differences in the relative binding thermodynamic parameters. Sampling of the conformations of the pseudopeptide ligands in solution is the most demanding calculation in this study. Some 26 perturbation steps were applied to introduce a bond between carbon atoms that are separated by two bonds, and REMD simulations with 48 replicas were performed at each perturbation step. Hence, in order to evaluate the relative solvation free energy/entropy of the two pairs of peptide ligands, about 6 μs MD simulations were performed; however, the statistical uncertainties remain significant. More advanced sampling techniques beyond the first-order scheme adopted in this study are needed in order to compute the solvation and binding free energy more efficiently and precisely.
We analyzed the structures and dynamics of ligands in solution and in their complexes with the SH2 domain in order to probe the molecular origin of the effects of ligand preorganization on binding thermodynamics in this system. Conformational clustering and quasiharmonic analysis of the free ligands in solution suggest that the unconstrained ligands possess significantly lower entropy than their constrained counterparts. This unexpected finding is the consequence of intramolecular hydrogen bonding interactions between the phosphate group of the flexible pY replacement and the C-terminal amide moieties of the pY+2 residue that lead to a more compact and rigid, macrocyclic-like structure. The presence of the cyclopropane ring in the constrained pY replacement prevents this interaction, thereby resulting in more extended conformations in solution. MD simulations of the protein-ligand complexes show that the unconstrained and constrained ligands share similar binding modes; the distribution of conformations of the bound forms of the unconstrained and constrained ligands are comparable as are their nonbonded interactions with the domain. These findings are consistent with the structures determined experimentally by X-ray crystallography.
These simulations reveal an important caveat that has not been previously acknowledged regarding the use of ligand preorganization, which is widely presumed to have a favorable effect upon binding entropy as a general design strategy. Namely, this work demonstrates that introducing a conformational constraint into a flexible ligand does not necessarily lower its entropy in solution, because the flexibility of a ligand in solution is determined by a subtle balance between any intramolecular interactions and the intermolecular interactions between the ligand and its aqueous environment. Comparing the dominant structures of the constrained ligands in their bound and unbound states shows that the cyclopropane ring in the constrained ligands, cpYVN and cpYIN, locally constrains and orients functionality on the flexible phosphotyrosine replacement in fpYVN and fpYIN in the bound conformation as predicted from modeling studies. However, the macrocyclic-like structures of fpYVN and fpYIN in solution, which do not correspond to their bound conformations, reduce the global flexibility of these ligands to an even greater degree than the cyclopropane ring. Because the binding entropies for fpYVN and fpYIN are more favorable than for their constrained derivatives cpYVN and cpYIN, it is now apparent that one cannot think simply in terms of introducing constraints to stabilize the biologically active conformation of a small molecule as a strategy for enhancing ligand binding affinities. Rather, lowering the entropy of a ligand in any way that allows it to adopt its bound conformation can lead to more favorable binding entropies. These studies also reveal that knowing the structures of small molecules in their unbound states is a critical prerequisite to correlating changes in their structures with protein binding entropies and enthalpies.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of General Medical Sciences (R01GM079686) and Teragrid MCB100057. S. F. M. is grateful to the National Institutes of Health (GM 84965), the National Science Foundation (CHE 0750329), the Robert A. Welch Foundation (F-652), and the Norman Hackerman Advanced Research Program for support. The authors thank Prof. Ron Elber for the suggestions in research design and help with technical details. The authors thank Zhen Xia, Dr. Chunli Yan and Dr. John H. Clements for important discussions.
Footnotes
Supporting Information Available:
Temperatures in Replica Exchange Molecular Dynamics Simulations (in K); Figure S1. Potential energy distributions of configurations sampled at different temperatures (300 K, 600 K and 1000 K) and with different simulation length (10 ns and 30 ns). QM optimized structure and the structure taken from complex crystal structure are as the starting points for repeated simulations. The frames are sampled every 2 ps, and then minimized with a convergence of 0.1 kcal/mol/Å. Thus the conformational distributions we are examining are for 0 K, even though the simulations have been done at high temperatures to accelerate the sampling process. The dihedral angles are measured from 0~360 degree (instead of 0~180 degree) in order to capture all the different orientations. The QM optimized structure is selected as the reference to calculate torsional RMSDs for all snapshots; Figure S2. Atom types of model compounds used for parameterization; Figure S3. Autocorrelation of dihedral angle C407-C350-C352-C354 and C350-C352-C354-C428 of fpYVN in solvent from REMD simulation at 298 K; Figure S4. Conformational distribution of dihedral angle C407-C350-C352-C354 and C350-C352-C354-C428 of fpYVN in solvent from REMD simulation at 298 K; Figure S5. Overlap of potential energy differences calculated forward (blue) and backward (red) of perturbation from fpYVN to cpYVN in water. The X-axis represents the potential energy difference, and the Y-axis shows the probability; Figure S6. Overlap of potential energy differences calculated forward (blue) and backward (red) of perturbation from fpYVN to cpYVN in solvated complex. The X-axis represents the potential energy difference, and the Y-axis shows the probability; Figure S7. Statistical inefficiency of the free energy calculations for fpYVN->cpYVN in water (upper panel) and fpYVN in solvated complex (lower panel) at 298K; Table S1. The dependence of relative binding free energy (fpYVN->cpYVN) at 298K on the number of intermediate steps in the alchemical calculation (energy in kcal/mol). Every 2nd (13 steps total), 4th (20 steps), 5th (21 steps), 6th (22 steps), 7th (23 steps), 10th (24 steps), 13th (23 steps with the middle step skipped) step is skipped respectively. Parameters for C428, O430, N426 and H429 are transferred from AMOEBA PRO; Force field parameters of the model compounds in Figure S2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mann A. In The Practice of Medicinal Chemistry. 2. Academic Press; London: 2003. [Google Scholar]
- 2.Lauri G, Bartlett PA. J Comput Aid Mol Des. 1994;8:51–66. doi: 10.1007/BF00124349. [DOI] [PubMed] [Google Scholar]
- 3.Page MI, Jencks WP. P Natl Acad Sci USA. 1971;68:1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan AR, Parrish JC, Fraser ME, Smith WW, Bartlett PA, James MNG. Biochemistry. 1998;37:16839–16845. doi: 10.1021/bi9821364. [DOI] [PubMed] [Google Scholar]
- 5.Widlanski T, Bender SL, Knowles JR. J Am Chem Soc. 1989;111:2299–2300. [Google Scholar]
- 6.Bartlett PA, Smith WW. J Am Chem Soc. 1998;120:4622–4628. [Google Scholar]
- 7.Davidson JP, Lubman O, Rose T, Waksman G, Martin SF. J Am Chem Soc. 2002;124:205–215. doi: 10.1021/ja011746f. [DOI] [PubMed] [Google Scholar]
- 8.Davidson JP, Martin SF. Tetrahedron Lett. 2000;41:9459–9464. [Google Scholar]
- 9.Verdine GL, Harrison BA, Gierasch TM, Neilan C, Pasternak GW. J Am Chem Soc. 2002;124:13352–13353. doi: 10.1021/ja027150p. [DOI] [PubMed] [Google Scholar]
- 10.Udugamasooriya DG, Spaller MR. Biopolymers. 2008;89:653–667. doi: 10.1002/bip.20983. [DOI] [PubMed] [Google Scholar]
- 11.DeLorbe JE, Clements JH, Whiddon BB, Martin SF. Acs Med Chem Lett. 2010;1:448–452. doi: 10.1021/ml100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin SF. Pure Appl Chem. 2007;79:193–200. [Google Scholar]
- 13.Martin SF, Benfield AP, Teresk MG, Plake HR, DeLorbe JE, Millspaugh LE. Angew Chem Int Edit. 2006;45:6830–6835. doi: 10.1002/anie.200600844. [DOI] [PubMed] [Google Scholar]
- 14.DeLorbe JE, Clements JH, Teresk MG, Benfield AP, Plake HR, Millspaugh LE, Martin SF. J Am Chem Soc. 2009;131:16758–16770. doi: 10.1021/ja904698q. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Guo QX. Chem Rev. 2001;101:673–695. doi: 10.1021/cr990416z. [DOI] [PubMed] [Google Scholar]
- 16.Gilli P, Ferretti V, Gilli G, Borea PA. J Phys Chem. 1994;98:1515–1518. [Google Scholar]
- 17.Dunitz JD. Chem Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 18.Cooper A, Johnson CM, Lakey JH, Nollmann M. Biophys Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 19.Breslauer KJ, Remeta DP, Chou WY, Ferrante R, Curry J, Zaunczkowski D, Snyder JG, Marky LA. P Natl Acad Sci USA. 1987;84:8922–8926. doi: 10.1073/pnas.84.24.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunwald E, Steel C. J Am Chem Soc. 1995;117:5687–5692. [Google Scholar]
- 21.Rekharsky M, Inoue Y. J Am Chem Soc. 2000;122:4418–4435. [Google Scholar]
- 22.Chen W, Chang CE, Gilson MK. Biophys J. 2004;87:3035–3049. doi: 10.1529/biophysj.104.049494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al Omari MM, Zughul MB, Davies JED, Badwan AA. J Incl Phenom Macro. 2007;57:379–384. [Google Scholar]
- 24.Cornish-Bowden A. J Biosciences. 2002;27:121–126. doi: 10.1007/BF02703768. [DOI] [PubMed] [Google Scholar]
- 25.Krug RR, Hunter WG, Grieger RA. J Phys Chem. 1976;80:2335–2341. [Google Scholar]
- 26.Leung DH, Bergman RG, Raymond KN. J Am Chem Soc. 2008;130:2798–2805. doi: 10.1021/ja075975z. [DOI] [PubMed] [Google Scholar]
- 27.Sharp K. Protein Sci. 2001;10:661–667. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds CH, Holloway MK. Medicinal Chemistry Letters. 2011;2:433. doi: 10.1021/ml200010k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams DH, Stephens E, O’Brien DP, Zhou M. Angew Chem Int Edit. 2004;43:6596–6616. doi: 10.1002/anie.200300644. [DOI] [PubMed] [Google Scholar]
- 30.Exner O. Collect Czech Chem C. 1975;40:2762–2780. [Google Scholar]
- 31.Exner O, Beranek V. Collect Czech Chem C. 1973;38:781–798. [Google Scholar]
- 32.Schowen RL. J Pharm Sci. 1967;56:931. doi: 10.1002/jps.2600560802. [DOI] [PubMed] [Google Scholar]
- 33.Linert W, Jameson RF. Chem Soc Rev. 1989;18:477–505. [Google Scholar]
- 34.Zhou HX, Gilson MK. Chem Rev. 2009;109:4092–4107. doi: 10.1021/cr800551w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradshaw JM, Waksman G. Adv Protein Chem. 2002;61:161–210. doi: 10.1016/s0065-3233(02)61005-8. [DOI] [PubMed] [Google Scholar]
- 36.Ladbury JE. Thermochim Acta. 2001;380:209–215. [Google Scholar]
- 37.Sturtevant JM. P Natl Acad Sci USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen WL. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 39.Gohlke H, Klebe G. Angew Chem Int Edit. 2002;41:2645–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 40.Brandsdal BO, Osterberg F, Almlof M, Feierberg I, Luzhkov VB, Aqvist J. Adv Protein Chem. 2003;66:123. doi: 10.1016/s0065-3233(03)66004-3. [DOI] [PubMed] [Google Scholar]
- 41.Mobley DL, Dill KA. Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamb ML, Jorgensen WL. Curr Opin Chem Biol. 1997;1:449–457. doi: 10.1016/s1367-5931(97)80038-5. [DOI] [PubMed] [Google Scholar]
- 43.Gilson MK, Zhou HX. Annu Rev Bioph Biom. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 44.Pande VS, Chodera JD, Mobley DL, Shirts MR, Dixon RW, Branson K. Curr Opin Struc Biol. 2011;21:150–160. doi: 10.1016/j.sbi.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirts MR, Mobley DL, Chodera JD. Annu Rep Comput Chem. 2007;3:41–57. [Google Scholar]
- 46.Andricioaei I, Karplus M. J Chem Phys. 2001;115:6289–6292. [Google Scholar]
- 47.Brooks BR, Janezic D, Karplus M. J Comput Chem. 1995;16:1522–1542. [Google Scholar]
- 48.Gohlke H, Case DA. J Comput Chem. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 49.Tidor B, Karplus M. J Mol Biol. 1994;238:405–414. doi: 10.1006/jmbi.1994.1300. [DOI] [PubMed] [Google Scholar]
- 50.Case DA. Curr Opin Struc Biol. 1994;4:285–290. [Google Scholar]
- 51.Bohm HJ. J Comput Aid Mol Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- 52.Jackson RN, Kulharia M, Goody RS. J Chem Inf Model. 2008;48:1990–1998. doi: 10.1021/ci800125k. [DOI] [PubMed] [Google Scholar]
- 53.Muegge I, Martin YC. J Med Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 54.Carlsson J, Aqvist J. Abstr Pap Am Chem S. 2005;229:U783–U784. [Google Scholar]
- 55.Hermans J, Wang L. J Am Chem Soc. 1997;119:2707–2714. [Google Scholar]
- 56.Luo HB, Sharp K. P Natl Acad Sci USA. 2002;99:10399–10404. doi: 10.1073/pnas.162365999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo R, Gilson MK. J Am Chem Soc. 2000;122:2934–2937. [Google Scholar]
- 58.Swanson JMJ, Henchman RH, McCammon JA. Biophys J. 2004;86:67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baginski M, Fogolari F, Briggs JM. J Mol Biol. 1997;274:253–267. doi: 10.1006/jmbi.1997.1399. [DOI] [PubMed] [Google Scholar]
- 60.Olano LR, Rick SW. J Am Chem Soc. 2004;126:7991–8000. doi: 10.1021/ja049701c. [DOI] [PubMed] [Google Scholar]
- 61.Warshel A, Singh N. Proteins. 2010;78:1724–1735. doi: 10.1002/prot.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren PY, Ponder JW. J Comput Chem. 2002;23:1497–1506. doi: 10.1002/jcc.10127. [DOI] [PubMed] [Google Scholar]
- 63.Ren PY, Ponder JW. J Phys Chem B. 2003;107:5933–5947. [Google Scholar]
- 64.Ponder JW, Case DA. Adv Protein Chem. Vol. 66. Academic Press Inc; San Diego: 2003. Force fields for protein simulations; p. 27. [DOI] [PubMed] [Google Scholar]
- 65.Ponder JW, Wu CJ, Ren PY, Pande VS, Chodera JD, Schnieders MJ, Haque I, Mobley DL, Lambrecht DS, DiStasio RA, Head-Gordon M, Clark GNI, Johnson ME, Head-Gordon T. J Phys Chem B. 2010;114:2549–2564. doi: 10.1021/jp910674d. [PubMed - in process] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grossfield A, Ren PY, Ponder JW. J Am Chem Soc. 2003;125:15671–15682. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- 67.Grossfield A, Ren PY, Ponder JW. Biophys J. 2003;84:94A–94A. [Google Scholar]
- 68.Jiao D, King C, Grossfield A, Darden TA, Ren PY. J Phys Chem B. 2006;110:18553–18559. doi: 10.1021/jp062230r. [DOI] [PubMed] [Google Scholar]
- 69.Wu JC, Piquemal J-P, Chaudret R, Reinhardt P, Ren P. J Chem Theory Comput. 2010;6:2059–2070. doi: 10.1021/ct100091j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y, Wu CJ, Ponder JW, Ren PY. J Comput Chem. 2011;32:967–977. doi: 10.1002/jcc.21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pappu RV, Drozdov AN, Grossfield A. J Am Chem Soc. 2004;126:2574–2581. doi: 10.1021/ja039051x. [DOI] [PubMed] [Google Scholar]
- 72.Jiao D, Golubkov PA, Darden TA, Ren P. P Natl Acad Sci USA. 2008;105:6290–6295. doi: 10.1073/pnas.0711686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiao D, Zhang JJ, Duke RE, Li GH, Schnieders MJ, Ren PY. J Comput Chem. 2009;30:1701–1711. doi: 10.1002/jcc.21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi Y, Jiao D, Schnieders MJ, Ren P. Engineering in Medicine and Biology Society, 2009. EMBC 2009; Annual International Conference of the IEEE; 2009. pp. 2328–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kubo MM, Gallicchio E, Levy RM. J Phys Chem B. 1997;101:10527–10534. [Google Scholar]
- 76.Smith DE, Haymet ADJ. J Chem Phys. 1993;98:6445–6454. [Google Scholar]
- 77.Tsunekawa N, Miyagawa H, Kitamura K, Hiwatari Y. J Chem Phys. 2002;116:6725–6730. [Google Scholar]
- 78.Lu N, Kofke DA, Woolf TB. J Phys Chem B. 2003;107:5598–5611. [Google Scholar]
- 79.Bennett CH. Journal of Computational Physics. 1976;22:245–268. [Google Scholar]
- 80.Chang CE, Chen W, Gilson MK. J Chem Theory Comput. 2005;1:1017–1028. doi: 10.1021/ct0500904. [DOI] [PubMed] [Google Scholar]
- 81.Ponder JW. Washington University Medical School. 2010 http://dasher.wustl.edu/tinker/
- 82.Ren P, Wu C, Ponder JW. J Chem Theory Comput. 2011;7:3027–3034. doi: 10.1021/ct200304d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stone AJ, Alderton M. Molecular Physics. 1985;56:1047–1064. [Google Scholar]
- 84.Stone AJ, Alderton M. Molecular Physics. 2002;100:221–233. [Google Scholar]
- 85.Sugita Y, Okamoto Y. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 86.Yoda T, Sugita Y, Okamoto Y. Chem Phys Lett. 2004;386:460–467. [Google Scholar]
- 87.Zhou RH. P Natl Acad Sci USA. 2003;100:13280–13285. doi: 10.1073/pnas.2233312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patriksson A, van der Spoel D. Phys Chem Chem Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- 89.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, erguson D, Seibel G, Kollman P. Comput Phys Commun. 1995;91:1–41. [Google Scholar]
- 90.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 91.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 92.Sagui C, Darden TA. Annu Rev Bioph Biom. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- 93.Boresch S, Karplus M. J Phys Chem A. 1999;103:103–118. [Google Scholar]
- 94.Boresch S, Karplus M. J Phys Chem A. 1999;103:119–136. [Google Scholar]
- 95.Allen MP, Tildesley DJ. Computer simulation of liquids. Clarendon Press; Oxford: 1989. [Google Scholar]
- 96.Chipot C, Pohorille A. Theory and Applications in Chemistry and Biology. Springer; 2007. Free Energy Calculations. [Google Scholar]
- 97.Wyczalkowski MA, Vitalis A, Pappu RV. J Phys Chem B. 2010;114:8166–8180. doi: 10.1021/jp103050u. [DOI] [PubMed] [Google Scholar]
- 98.Syme NR, Dennis C, Bronowska A, Paesen GC, Homans SW. J Am Chem Soc. 2010;132:8682–8689. doi: 10.1021/ja101362u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peter C, Oostenbrink C, van Dorp A, van Gunsteren WF. J Chem Phys. 2004;120:2652–2661. doi: 10.1063/1.1636153. [DOI] [PubMed] [Google Scholar]
- 100.Aqvist J, Carlsson J. J Phys Chem B. 2009;113:10255–10260. doi: 10.1021/jp900818z. [DOI] [PubMed] [Google Scholar]
- 101.Lee JK. Statistical bioinformatics: a guide for life and biomedical science researchers. John Wiley & Sons, Inc; Hoboken, New Jersey: 2010. [Google Scholar]
- 102.Martin SF, Clements JH, DeLorbe JE, Benfield AP. Acta Crystallogr D. 2010;66:1101–1115. doi: 10.1107/S0907444910035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.DeLano WL. DeLano Scientific. San Carlos, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.