Abstract
A variety of observations support the hypothesis that deficiency of complex I [reduced nicotinamide-adenine dinucleotide (NADH):ubiquinone oxidoreductase] of the mitochondrial respiratory chain plays a role in the pathophysiology of Parkinson's disease (PD). However, recent data from a study using mice with knockout of the complex I subunit NADH:ubiquinone oxidoreductase iron-sulfur protein 4 (Ndufs4) has challenged this concept as these mice show degeneration of non-dopamine neurons. In addition, primary dopamine (DA) neurons derived from such mice, reported to lack complex I activity, remain sensitive to toxins believed to act through inhibition of complex I. We tissue-specifically disrupted the Ndufs4 gene in mouse heart and found an apparent severe deficiency of complex I activity in disrupted mitochondria, whereas oxidation of substrates that result in entry of electrons at the level of complex I was only mildly reduced in intact isolated heart mitochondria. Further analyses of detergent-solubilized mitochondria showed the mutant complex I to be unstable but capable of forming supercomplexes with complex I enzyme activity. The loss of Ndufs4 thus causes only a mild complex I deficiency in vivo. We proceeded to disrupt Ndufs4 in midbrain DA neurons and found no overt neurodegeneration, no loss of striatal innervation and no symptoms of Parkinsonism in tissue-specific knockout animals. However, DA homeostasis was abnormal with impaired DA release and increased levels of DA metabolites. Furthermore, Ndufs4 DA neuron knockouts were more vulnerable to the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Taken together, these findings lend in vivo support to the hypothesis that complex I deficiency can contribute to the pathophysiology of PD.
INTRODUCTION
Parkinson's disease (PD) is characterized by formation of cytoplasmic inclusions (Lewy bodies) and degeneration of midbrain dopamine (DA) neurons in the substantia nigra pars compacta (SNc), although other neurons are also affected (1). The pathophysiology is unclear and may be complex. One main hypothesis proposes a role of impaired mitochondrial complex I [reduced nicotinamide-adenine dinucleotide (NADH):ubiquinone oxidoreductase]. It originates from findings that the toxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) can cause parkinsonism in humans (2) and laboratory animals (3,4) due to accumulation of its toxic metabolite MPP+ in DA neurons. Within cells, MPP+ accumulates in mitochondria and can act as a specific inhibitor of mitochondrial complex I (5). Decreased mitochondrial complex I activity was also found in the substantia nigra area of postmortem brain tissue from patients with PD (6). Other groups reported reduced complex I activity in the skeletal muscle (7) and platelets (8) of PD patients, suggesting that systemic complex I deficiency may play a role in PD pathophysiology. This idea was further supported by findings that systemic delivery of the complex I inhibitor rotenone caused degeneration of DA neurons and formation of Lewy body-like inclusions (9).
Other lines of studies have instead suggested that age-acquired mutations in mitochondrial DNA (mtDNA) may be of importance. The amount of mtDNA molecules carrying deletions increases with age (10,11) and studies of brain homogenates have shown a higher proportion of such mutations in substantia nigra than what is found in other brain regions (11). A higher proportion of respiratory chain-deficient DA neurons has also been found in brains of PD patients, and laser capture of single cells revealed that these neurons carried a higher proportion of deleted mtDNA molecules (12,13). Vulnerability of DA neurons towards mtDNA deletions is further exemplified by the findings that patients with mutations in the gene encoding the mtDNA polymerase γ accumulate mtDNA deletions and may develop a PD-like phenotype (14). Other genes found to cause recessive Parkinsonism, such as PARKIN, PINK1 and DJ-1, have also been shown to regulate the function of mitochondria (15,16), but how these genes may normally protect against DA cell death is not well understood.
Recently, the importance of complex I in PD has been challenged by studies of mice in which the structural complex I subunit NADH:ubiquinone oxidoreductase iron-sulfur protein 4 (Ndufs4) has been deleted. Ndufs4 knockout mice are viable at birth. During the first postnatal weeks, they develop a fatal neurodegenerative phenotype, reported not to involve DA neurons (17,18). Furthermore, since primary DA neurons cultured from these mice were not resistant to MPP+, paraquat or rotenone (19), while reported to lack detectable complex I activity in vitro, it was concluded that these toxins act through other targets than complex I.
In this study, we have investigated the consequences of tissue-specific loss of Ndufs4 in vivo. We find that complex I lacking Ndufs4 is unstable but only partially impaired, causing only minor effects on ATP production rates in intact mitochondria. In DA neurons, this mild complex I deficiency leads to altered DA levels, impaired DA release and increased vulnerability towards MPTP, thus supporting a role for complex I impairment as a contributing factor in PD pathophysiology.
RESULTS
Loss of Ndufs4 cause a mild complex I deficiency
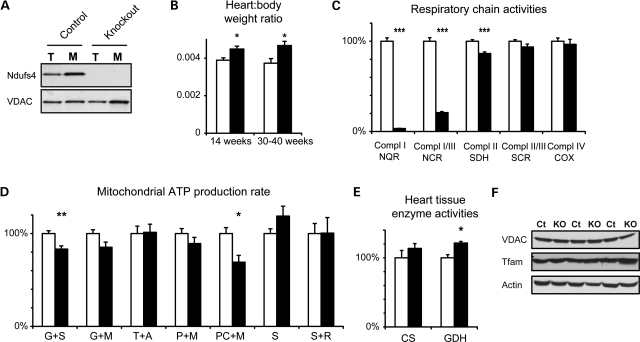
To characterize possible biochemical defects caused by the loss of Ndufs4, we generated heart knockout mice by crossing mice with loxP-flanked Ndufs4 alleles (18) to mice that express the cre-recombinase in heart and skeletal muscle (20). The resulting tissue-specific knockout mice lacked the Ndufs4 protein in heart muscle (Fig. 1A) and had an increased heart-to-body weight ratio (Fig. 1B). Such an increase may indicate cardiomyopathy, but the mice showed no progression to heart failure and appeared to be clinically healthy up to at least 1 year of age.
Figure 1.
The loss of Ndufs4 causes a mild phenotype in heart. (A) Immunoblots from total (T) and mitochondrial (M) fractions of control and Ndufs4 knockout hearts. (B) Heart-to-body weight ratios in control and Ndufs4 heart knockout mice. (C) Respiratory chain enzyme activities in detergent-permeabilized mitochondrial preparations. Complex I (NADH:coenzyme Q reductase, NQR), complexes I/III (NADH:cytochrome c reductase, NCR), complex II (succinate dehydrogenase, SDH), complexes II/III (succinate:cytochrome c reductase, SCR) and complex IV (cytochrome c oxidase, COX) activities were measured and normalized to activity of citrate synthase (CS) in the mitochondrial suspension. Values are shown as percent of the following absolute values in controls: NQR 0.30, NCR 0.80, SDH 0.32, SCR 0.51 and COX 2.29 units/unit CS. (D) The ATP production rate determined in isolated, intact mitochondria in the presence of substrates glutamate + succinate (G + S), glutamate + malate (G + M), TMPD + ascorbate (T + A), pyruvate + malate (P + M), palmitoyl-l-carnitine + malate (PC + M), succinate (S) and succinate + rotenone (S + R). Activities were normalized to CS activity in the mitochondrial suspension and shown as percent of the following absolute values in controls: G + S 0.19, G + M 0.14, T + A 0.15, P + M 0.11, PC + M 0.095, S + R 0.027 and S 0.023 units/unit CS. (E) CS and GDH activities in heart tissue. Activities were normalized to tissue mass and shown as percent of absolute values in controls: 347 mmol/min/kg for CS and 13.4 mmol/min/kg for GDH. (F) Immunoblots from total extracts of control (Ct) and Ndufs4 knockout (KO) hearts (age ∼12 months). Open bars, controls; black bars, Ndufs4 heart knockouts. Data shown as mean ± s.e.m. (n = 5). *P < 0.05; **P < 0.01; ***P < 0.001 by Student's two-tailed t-test following Levene's test for equality of variance.
We measured the activities of respiratory chain enzyme complexes in detergent-permeabilized mitochondrial extracts from Ndufs4 knockout hearts (Fig. 1C) and found a striking, almost complete absence of isolated complex I activity (<5% residual activity). Coupled complex I/III activity was also reduced, but not as dramatically (21% residual activity). This discrepancy could be explained by different affinity for the artificial electron acceptor coenzyme Q1, which is added to assay isolated complex I activity, while coupled complex I/III activity was measured with only endogenous coenzyme Q10.
Next, we measured mitochondrial ATP production rates in intact, isolated mitochondria (Fig. 1D) and found only slightly reduced ATP production rates using Krebs' cycle substrates that results in delivery of electrons at the level of complex I, e.g. glutamate/malate (85% residual activity; P = 0.065) and glutamate/succinate (83% residual activity; P = 0.0065). The most pronounced reduction was found using β-oxidation substrates palmitoyl-l-carnitine/malate (69% residual activity; P = 0.013). Total extracts from heart tissue had slightly higher mitochondrial citrate synthase (CS; 114% of controls; P = 0.31) and glutamate dehydrogenase (GDH; 121% of controls; P = 0.0031) activities per heart muscle mass (Fig. 1E), suggesting that a compensatory increase in mitochondrial mass maintain near-normal overall tissue ATP production rates. We found no increase in other mitochondrial markers such as mitochondrial transcription factor A (Tfam) or voltage-dependent anion channel (VDAC) when protein levels were analyzed by immunoblotting (Fig. 1F).
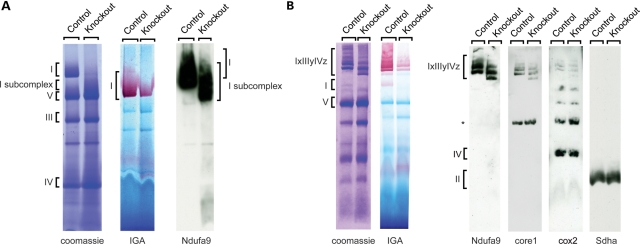
The observed severe reduction in complex I enzyme activity in disrupted mitochondria from Ndufs4 knockout hearts is consistent with published results (18). However, we unexpectedly observed near-normal ATP production with substrates delivering electrons to complex I in isolated intact mitochondria, suggesting that the mutant complex I is intact in mitochondria in vivo. To address this possibility, we performed blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis of heart mitochondria. Following treatment with the more harsh detergent n-dodecyl β-D-maltopyranoside, a majority of complex I in knockout hearts appeared to be a smaller subcomplex (Fig. 2A), as previously reported (21), suggesting impaired assembly and/or stability of the complex. As described by others (21), this subcomplex was enzymatically inactive as assessed by NADH reductase in-gel activity, but at variance with this previous study, we found low levels of NADH dehydrogenase activity corresponding to some remaining fully assembled complex I (Fig. 2A). Next, we looked at supercomplex organization by treating mitochondria with the milder detergent digitonin. We found impaired formation of complex I containing supercomplexes and additional lower molecular weight species that were enzymatically inactive. This again demonstrates a decrease in complex I formation or stability (Fig. 2B). However, residual supercomplex formation with in-gel complex I activity was present, suggesting that this impairment was only partial. In addition to complex I, the supercomplexes found in Ndufs4 knockout hearts also contained complexes III and IV (Fig. 2B).
Figure 2.
The loss of Ndufs4 leads to complex I subcomplex formation. (A) Blue native polyacrylamide gel electrophoresis analysis of mitochondria from control and Ndufs4 knockout hearts. Isolated mitochondria from hearts were lysed with 1% n-dodecyl β-D-maltopyranoside and further subjected to coomassie staining, in-gel activity (IGA) staining for complex I, or immunoblot anaysis. (B) Mitochondria from control and Ndufs4 knockout hearts were treated as in (A), but lysed with 1% digitonin. Ndufa9, complex I 39 kDa subunit; Sdha, complex II subunit; core 1, complex III subunit; cox2, cytochrome c oxidase subunit 2; IxIIIyIVz; complex I, III, IV supercomplexes; *, unspecific bands.
In summary, we show here that the loss of Ndufs4 results in a subassembled and/or unstable complex I with a complete to near-complete loss of enzyme activity when assayed in disrupted mitochondria, whereas intact mitochondria show only slightly reduced ATP production rates. We also report that interactions with complexes III and IV have a stabilizing effect on the mutant complex I lacking the Ndufs4 subunit, leading to only a moderate decrease of complex I activity and ATP production capacity in vivo.
Generation of DA neuron Ndufs4 knockout mice
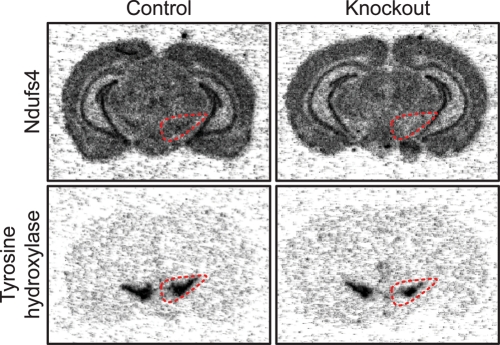
Next, we disrupted Ndufs4 in midbrain DA neurons by crossing Ndufs4loxP and DAT-cre mice, which express cre-recombinase specifically in DA neurons (22). We henceforth refer to these DA-neuron-specific conditional Ndufs4 knockouts (genotype DATcre/+; Ndufs4loxP/loxP) as ‘Ndufs4-DA mice’. The efficiency of the knockout was supported by the observed loss of Ndufs4 transcripts in midbrain DA neurons as observed by in situ hybridization of midbrain sections with an Ndufs4 probe (Fig. 3).
Figure 3.
Ndufs4–DA mice lack Ndufs4 transcript in midbrain DA neurons. In situ hybridization of radiolabeled DNA oligoprobes against TH and Ndufs4 mRNA in two adjacent brain sections of control and Ndufs4–DA mice. The area containing TH-positive DA neurons in SNc and the neighboring ventral tegmental area is outlined.
Ndufs4–DA mice do not show striatal denervation or develop Parkinsonism
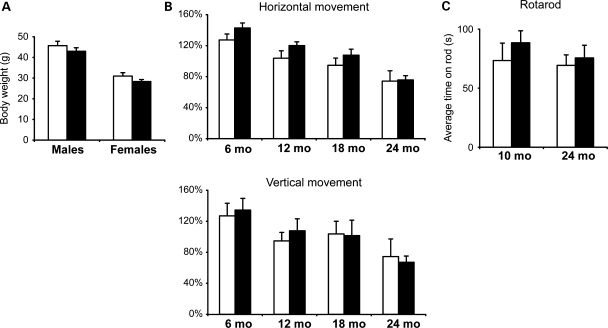
Ndufs4–DA mice appeared healthy and were indistinguishable from control littermates. There was no significant difference in body weight (Fig. 4A). We quantified spontaneous locomotion and exploratory behavior of Ndufs4–DA and control mice (genotype Ndufs4loxP/loxP) using locomotor boxes. We found no significant differences in locomotion (horizontal movement) or rearing (vertical movement) in cohorts of mice at 6, 12, 18 or 24 months of age (Fig. 4B). Balance and coordination were assessed by the accelerating rotarod test. We found no differences in the ability of adult (10 months) or old (24 months) Ndufs4–DA mice to remain on the rod (Fig. 4C).
Figure 4.
Ndufs4–DA mice do not develop Parkinsonism. (A) Body weights of 12-month-old Ndufs4–DA and control mice (n≥ 11). (B) Spontaneous horizontal (locomotion) and vertical (rearing) activity of Ndufs4–DA and control mice monitored over a period of 2 h at ages 6, 12, 18 and 24 months. No significant differences were found in either early exploratory behavior or after 1 h of acclimatization. (C) Performance (latency to fall) of 10- and 24-month-old mice on an accelerating Rotarod. Each mouse was given four trials and the average time was used. Open bars, controls; black bars, Ndufs4–DA knockouts. Data shown as mean ± s.e.m.
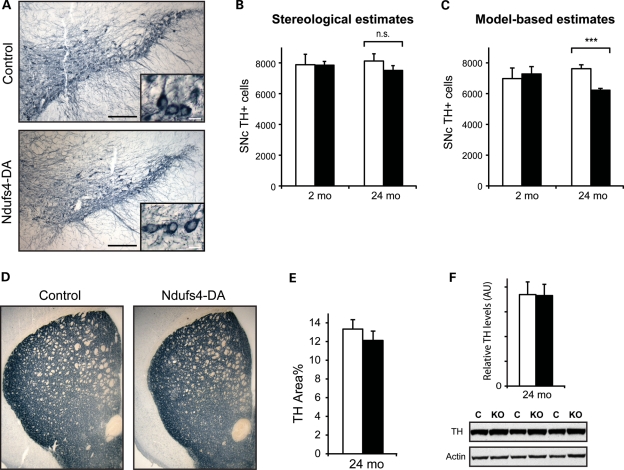
We next undertook a detailed analysis of the midbrain DA system. Tyrosine hydroxylase (TH)-labeled midbrain sections revealed no differences in gross morphology of DA neurons in Ndufs4–DA mice compared with controls (Fig. 5A). We estimated the total number of TH-positive cells in the SNc of Ndufs4–DA mice at 2 and 24 months using both stereology and manual counting followed by Abercrombie's correction (23,24). Using stereology, we found similar numbers of DA neurons at 2 months of age, and a slight (7.5%) decrease at 24 months of age (Fig. 5B). As we expected a possible cell loss to be moderate, we proceeded to directly count all TH-immunoreactive cells on the same series of sections. Such a model-based approach has been found to be accurate under normal conditions and to require fewer samples due to a decreased variance (24). We then found a significant (P = 0.0005) decrease of neurons in the 24-month-old Ndufs4–DA mice (Fig. 5C). At 2 months of age, DA neuron numbers were similar, which is consistent with the results from others showing that Ndufs4 knockouts have normal numbers of TH-positive neurons at 6–7 weeks (17). This indicates that there may be a modest loss of cells between 2 and 24 months of age in Ndufs4–DA mice. No apparent loss of DA nerve terminals was seen by TH immunohistochemistry of striatal sections (Fig. 5D). As small differences in fiber density are difficult to detect in 14 µm sections, we used confocal microscopy to measure TH-immunoreactive areas in thin optical sections. This estimate did not reveal a significant decrease in TH-innervation, although there was ∼10% lower mean levels in Ndufs4–DA mice (Fig. 5E). We also quantified the amounts of TH proteins in striatal homogenates by immunoblotting (Fig. 5F). We found equal levels of TH protein, showing that the loss of Ndufs4 does not affect the striatal TH protein content. We conclude that the loss of Ndufs4 does not lead to overt neurodegeneration, despite some indications of a very low-grade cell loss between 2 and 24 months of age.
Figure 5.
Morphology of the nigrostriatal DA system. (A) TH-positive DA neurons in SNc of control and Ndufs4-DA mice at 24 months of age. Scale bars = 200 µm (overviews); 20 µm (insets). (B) Number of TH-positive cells in SNc of 24-month-old mice as determined by stereology. The left and right SNcs were counted separately (93–152 cells per side with a coefficient of error ≤0.1) and then combined. Total numbers (mean ± s.e.m.) were 7881 ± 677 and 7851 ± 243 at age 2 months (n = 3), and 8120 ± 473 and 7511 ± 312 at age 24 months (n = 4; 6) in controls and Ndufs4–DA mice, respectively. (C) The number of TH-positive cells in SNc of 2- and 24-month-old mice as determined by manual counting of all cells. Absolute numbers were derived using Abercrombie's formula, taking into account diameter of counted objects (nuclei of DA neurons), section thickness and distances between sections, and found to be 6978 ± 689 and 6946 ± 555 at age 2 months, and 7622 ± 254 and 6225 ± 117 at age 24 months in controls and Ndufs4–DA mice, respectively. The nuclear diameter was not found to differ between genotypes, and at age 24 months determined to be 9.86 µm (95% confidence interval 7.54–12.19 µm) in controls and 9.83 µm (6.86–12.8 µm) in Ndufs4–DA. (D) Overview of TH-stained fibers in striatum (age 24 months). (E) TH-density in dorsolateral striatum measured by the area of optical sections (z < 1 µm) occupied by TH-positive fibers. (F) Amounts of TH protein in striatal homogenates determined by semi-quantitative immunoblotting (age 24 months). Levels of TH were normalized to actin in each lane (n= 5). Open bars, controls; black bars, Ndufs4–DA knockouts. Data shown as mean ± s.e.m. ***P < 0.001 by Student's two-tailed t-test.
Ndufs4–DA mice have altered DA homeostasis
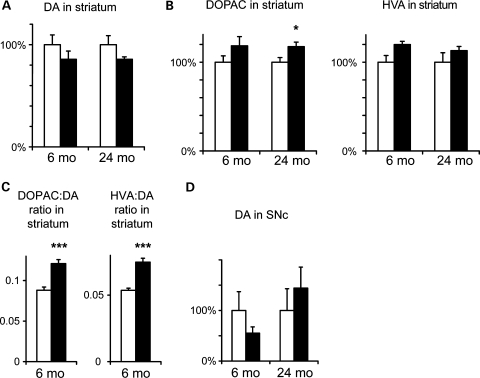
We measured the levels of DA and its major metabolites in homogenates of different brain regions by high-performance liquid chromatography (HPLC). In striatum, we found slight (∼15%) reductions in DA levels in Ndufs4–DA mice compared with littermate controls (Fig. 6A). Similar levels were consistently found in both 6- and 24-month-old mice, and this deficiency was thus stable over time. Consistent with reduced DA levels, we found increased levels of major DA metabolites 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA) in striatum (Fig. 6B). Consequentially, there was a highly significant increase in both DOPAC:DA and HVA:DA ratios (Fig. 6C) as is typically seen in conditions with increased DA turnover. These differences were not caused by introduction of cre in the DA transporter locus (22), as mice only carrying the DAT-cre knockin allele have normal DA levels and metabolite:DA ratios (25). DOPAC and HVA were equally increased, and there was no difference in the ratio between the two metabolites. No significant differences in DA or metabolites were found at the level of SNc (Fig. 6D), possibly due to high variance in the measurements. There were no differences in the levels of noradrenaline or serotonin in SNc or striatum (not shown).
Figure 6.
Altered DA homeostasis in Ndufs4–DA mice. (A) DA levels in striatum in 6- and 24-month-old control and Ndufs4–DA mice (normalized to each control). The slight decrease of DA in Ndufs4–DA mice was significant only when both time points were combined (PGenotype= 0.04 by ANOVA). (B) Relative levels of DOPAC and HVA in striatum of 6- and 24-month-old control and Ndufs4–DA mice. Increased levels of both DOPAC and HVA in Ndufs4-DA mice were significant when both time points were combined (PGenotype< 0.05 by ANOVA). (C) DOPAC:DA and HVA:DA ratios in striatum (age 6 months). (D) Levels of DA in SNc in 6- and 24-month-old control and Ndufs4–DA mice (normalized to each control). Open bars, controls; black bars, Ndufs4-DA knockouts. Data shown as mean ± s.e.m. (n = 7–10). *P < 0.05; ***P < 0.001 by Student's two-tailed t-test following Levene's test for equality of variance.
Impaired DA release but not reuptake in Ndufs4–DA striatum
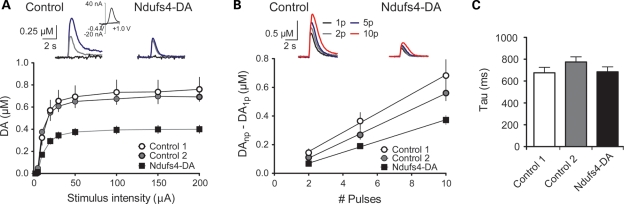
An increase in metabolite:DA ratio is typically seen following partial denervation and viewed as reflecting a compensatory increase in DA turnover in remaining DA neurons. As we found only modest indications of denervation, we proceeded to study striatal DA release by fast scan cyclic voltammetry (FSCV) in brain slices taken from ∼12-month-old control and knockout mice. We first examined release elicited by local, single pulse (1 ms) electrical stimulation, and generated input–output curves by varying stimulation intensity. We found a significant reduction in maximal DA release in slices taken from Ndufs4–DA mice (RM-ANOVA, genotype F(2,78) = 11.81, P < 0.001) relative to controls (Fig. 7A). To ensure that introduction of cre in the DA transporter locus did not affect DA dynamics, we used both normal controls, wild-type for DAT (‘Control 1’; genotype DAT+/+; Ndufs4LoxP/LoxP), and an additional group carrying the DAT-cre allele (‘Control 2’; genotype DATcre/+; Ndufs4+/LoxP). DA release did not significantly differ between these two types of control groups (RM-ANOVA, genotype F(1,44) = 0.08, P = 0.7830). The decay time constants (tau) of the DA signals did not differ significantly among the groups (Fig. 7C; ANOVA, P = 0.2762), suggesting that DAT-mediated reuptake is not affected by the Ndufs4 deletion. As an additional means to verify the changes in DA release, we examined release elicited by 25 Hz stimulation designed to mimic ‘phasic’ activity of DA neurons (26,27). As shown in Figure 7B, an increased DA output was seen in response to an increasing number of pulses delivered at 25 Hz. However, the slope of the curve was significantly reduced in the Ndufs4-DA mice, relative to both control groups (P < 0.05, ANOVA and Neuman–Keuls post-hoc test). Together, these data suggest that mild complex I deficiency, induced by the loss of Ndufs4 in DA neurons, reduces DA release under both tonic and phasic stimulation conditions.
Figure 7.
Reduced DA release in Ndufs4–DA mice. (A) Voltammetric signals elicited by single pulse stimulation at varying stimulus intensities (5, 20, and 200 μA indicated by black, gray and blue traces, respectively) in striatal slices from control and Ndufs4–DA mice. Inset of a control trace showing a typical DA cyclic voltammogram. DA release was significantly reduced across a range of stimulation intensities in Ndufs4–DA mice relative to controls (P < 0.001, two-way RM-ANOVA). Note that controls did not differ from each other, indicating that the presence of the DAT-cre construct alone did not alter DA release. (B) Voltammetric signals elicited by 25 Hz stimulation (1, 2, 5 or 10 pulses). Summary plot demonstrates the relationship between DA release and pulse number. Ndufs4–DA mice showed significantly reduced DA release relative to both control groups (P< 0.01, one-way ANOVA, Neuman–Keuls post hoc). (C) Summary of decay time constants (tau), measured from voltammetric signals. No significant differences were observed among the groups (P = 0.28, one-way ANOVA), indicating that the uptake was not affected by Ndufs4 deletion in DA neurons.
Ndufs4–DA mice are more vulnerable to treatment with MPTP
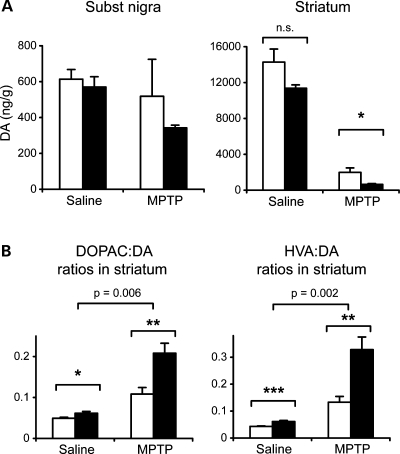
The neurotoxin MPTP has been shown to act by inhibition of complex I. We therefore tested whether Ndufs4–DA mice had altered susceptibility towards MPTP. We challenged ∼12-month-old Ndufs4–DA and littermate controls with MPTP (30 mg/kg for two consecutive days), and assessed the amount of DA and its metabolites 7 days following administration (Fig. 8A). This regimen reduced striatal DA in control mice to 14% (7.0–17.3; 95% confidence interval) of that of saline-treated control mice. In Ndufs4–DA mice, striatal DA levels were reduced to 5.7% (4.2–6.5%) of the levels in saline-treated knockouts. Thus, the relative decrease was more pronounced in Ndufs4–DA mice (P = 0.031, Student's t-test), but factorial ANOVA failed to show a statistical interaction. There was no significant reduction of DA levels in the SNc area. Consistent with our previous findings, levels of DA metabolites in striatum were higher in Ndufs4–DA mice than in controls. Following MPTP-induced denervation, metabolite:DA ratios increased substantially more in Ndufs4–DA than in controls (Fig. 8B). DA neurons of Ndufs4–DA mice thus remain at least as sensitive to MPTP as controls. We conclude that Ndufs4–DA mice are more vulnerable to MPTP, as MPTP treatment aggravates the phenotype with a more pronounced striatal DA loss and metabolite to DA ratios.
Figure 8.
MPTP toxicity in Ndufs4–DA mice. (A) DA levels in SNc and striatum in controls and Ndufs4–DA mice following treatment with MPTP. In striatum, criteria for statistical interaction between genotype and treatment were not reached (P = 0.243; two-way ANOVA). (B) DOPAC:DA and HVA:DA ratios in striatum in control and Ndufs4–DA mice after saline or MPTP administration. Top bracket shows P-value for the statistical interaction between genotype and treatment (two-way ANOVA). Open bars, controls; black bars, Ndufs4–DA knockouts. Data shown as mean ± s.e.m. (n = 7–10). *P < 0.05; **P < 0.01; ***P < 0.001 by Student's two-tailed t-test following Levene's test for equality of variance.
DISCUSSION
Studies of primary mesencephalic neurons lacking Ndufs4 have suggested that complex I activity is dispensable for DA neuron survival and that toxins that kill DA neurons, such as MPP+, rotenone and paraquat, must act by mechanisms other than through inhibition of complex I (19). However, these conclusions are all based on the result that cultured knockout neurons have no detectable complex I activity. Contrary to this, our in vivo data show that loss of Ndufs4 in heart results only in a partial biochemical defect. Although ATP production rates in DA neuron mitochondria were not measured and may vary from that of heart, the fundamental processes governing complex I assembly and stability should be conserved between tissues. Residual complex I activity has also been demonstrated in homogenates of liver (18) and brain (28) from Ndufs4 knockout mice, and may depend on stabilization by complex III in supercomplexes (21). The reason for the discrepancy between measurements of tissues and cultured cells is not known, but may involve the possibility for cultured cells to completely rely on glycolysis for ATP production. However, our data suggest that rather than being in almost complete lack of complex I activity, intact cultured DA neurons from Ndufs4 knockout mice should also possess residual complex I activity, which could explain the reported vulnerability to MPP+, rotenone and paraquat (19). In accordance with such an interpretation, our in vivo studies demonstrated that specific removal of Ndufs4 in DA neurons did not impair sensitivity to MPTP. Rather, DA neurons lacking Ndufs4 appeared to be more sensitive to MPTP than control DA neurons in vivo. Thus it appears that the genetic deletion and MPTP treatment have additive effects. Although this experiment cannot be used to elucidate whether MPTP toxicity acts through complex I inhibition, it illustrates that DA neurons with partial complex I deficiency are more vulnerable to stress.
Full-body Ndufs4 knockouts suffer from early-onset ataxia, lethargy, hypothermia and death within 2 months (18). Almost the entire phenotype can be attributed to loss of Ndufs4 in neurons and glia (28). Degeneration and gliosis are found in cerebellum, the olfactory bulb and vestibular nuclei, whereas DA neurons are spared (17,28). This suggests that DA neurons are not specifically vulnerable to a systemic biochemical defect resulting from loss of Ndufs4. Rather than a systemic defect, however, accumulation of mtDNA deletions or other forms of mitochondrial damage could lead to focal mitochondrial impairment that is more pronounced in DA neurons than elsewhere. As complex I is the largest enzyme complex of the respiratory chain, and heavily depends on mtDNA-encoded subunits, it may be particularly vulnerable to age-acquired damage such as oxidative damage (29) or mtDNA deletions (13).
We found that complex I deficiency in DA neurons results in increased metabolite:DA ratios in striatum, indicative of increased turnover rates. Increased DA turnover has also been found in striatum of various rodent models as well as early-stage PD patients (30–32). This has been suggested to reflect a compensatory increase of activity of remaining DA neurons, and helps explain why up to 80% of the striatal DA is lost before the onset of symptoms. Ndufs4–DA mice have only a modest (∼15%) decrease in total striatal DA levels, which can explain the absence of an overt parkinsonian behavioral phenotype. In contrast to the modest decrease in total striatal DA, voltammetric data revealed a more pronounced decrease in release from striatal axon terminals in brain slices. This more profound loss in the releasable pool of DA in Ndufs4–DA mice suggests that more DA is outside of synaptic vesicle storage and therefore more susceptible to enzymatic degradation. Indeed, cytosolic DA levels have been found to increase in primary DA neurons in vitro following treatment with rotenone, and even more so in Ndufs4−/− cells (17,33). In MitoPark mice (22), which have severe combined respiratory chain deficiency in DA neurons, DA release is also selectively impaired during early stages of the degenerative phenotype (26). Taken together, this suggests that impaired DA release is an early consequence of mitochondrial impairment, regardless of whether only complex I or most of the respiratory chain is functionally impaired. How mitochondrial dysfunction leads to impaired DA release is not known, but both vesicular dynamics and/or VMAT2 activity may be particularly susceptible to ATP deficiency or direct oxidative damage (33). DA neurons have numerous axon terminals, but the mitochondria found in these axons are small and often single organelles (34,35), possibly making axon terminals more sensitive to mitochondrial impairment. Toxicity from DA itself has been proposed to play a role in PD (32,36), and increased extravesicular DA may add oxidative stress to neurons with compromised mitochondrial capacity. Consistent with this idea are findings that mice with severe reduction in VMAT2 activity and massively increased metabolite:DA ratios display oxidative damage and late-onset DA cell loss (37). In summary, our results demonstrate that the loss of Ndufs4 causes only a mild complex I deficiency in vivo, which in DA neurons nevertheless leads to alterations that may play a causative or contributing role in the pathophysiology of late onset PD.
MATERIALS AND METHODS
Breeding of mice
DAT-cre (22) and Ndufs4LoxP (18) mice were backcrossed ≥10 generations to C57Bl/6. Ndufs4LoxP/+ were crossed to DATcre/+ mice to generate double heterozygous offspring that were subsequently crossed to homozygous Ndufs4LoxP/LoxP mice. The genes for Ndufs4 and the DA transporter (Slc6a3) are linked on chromosome 13 by a distance of 41.4 Mb. Consequently, once male Ndufs4-DA mice (DATcre/+; Ndufs4LoxP/LoxP) were established, they were further crossed to control females (Ndufs4LoxP/LoxP) to generate littermate knockouts and controls, respectively. Unless otherwise stated, mice with genotype Ndufs4LoxP/LoxP were used as controls.
To generate heart- and skeletal muscle-specific knockouts, Ndufs4Loxp/+ mice were first bred with mice carrying Ckmm-NLS-cre (20). Double heterozygous offspring were crossed to homozygous Ndufs4LoxP/LoxP mice to generate knockouts (Ckmm-NLS-cre; Ndufs4LoxP/LoxP) and littermate controls (genotype Ndufs4+/LoxP or Ndufs4LoxP/LoxP). Experiments were approved by an ethical committee and are in compliance with Swedish law.
Behavior analysis
A cohort of female littermates was analyzed at ages 6 (n= 8), 12 (n= 8), 18 (n= 8) and 24 months (n= 10 controls, 14 knockouts) using an animal activity monitoring system (VersaMax, AccuScan Instruments). Mice were habituated to the dimly lit, low-noise and ventilated experimental room for at least 30 min before initiating experiments. Mice were individually placed in activity cages (40 × 40 cm and 30 cm high) and a grid of infrared light beams at floor level and 7.5 cm above recorded horizontal and vertical locomotor activities, respectively. Spontaneous behavior was measured during a 2 h period between 6 and 8 p.m. at the beginning of the dark phase of the light/dark cycle.
Rotarod experiments were performed during daytime (light phase) using a rotometer (LSi LETICA Scientific Instruments). Mice (n ≥ 8 per genotype and age) were first trained to stay three 1-min periods on the cylinder (rotating at 0, 4 and 4 rpm, respectively). After 1 h, mice were again placed on the rotating cylinder (starting at 4 rpm and accelerating to 40 rpm over 5 min). The time until the mouse fell off was recorded, and each mouse was tested four times with 30 min interval between each trial.
In situ hybridization
Localization of mRNA species in sections using radiolabeled oligo probes followed a published protocol (38). Oligonucleotide probes to detect transcripts of Ndufs4 (5′-AGC TGTGTGTCCCGAGTCTGGTTGTCTGCCAGCTTCCAAGTGGATGTGCT) and TH (5′-GGTGTGCAGCTCATCCTGGACCCCCTCCAAGGAGCGCT) were end-labeled with α-33P-deoxyadenosine 5′-triphosphate using a terminal deoxynucleotidyl transferase kit (GE Healthcare). Coronary cryosections from fresh-frozen brains were hybridized over night at 42°C with radio-labeled probe in 4× saline-sodium citrate (SSC) solution (3.0 m trisodium citrate and 3.0 m NaCl; pH 7.0), 50% formamide, 1× Denhardt's solution (Ficoll, polyvinylpyrrolidone and bovine serum albumin), 1% sarcosyl, 0.02 m sodium phosphate (Na3PO4), 10% dextran sulfate (wt/vol), 0.2 m dithiothreitol and 0.5 μg/μl sheared salmon serum sperm DNA. Slides were washed for 1 h in 60°C 1× SSC buffer. After cooling to room temperature, slides were dehydrated in an ethanol series, air-dried and exposed on phosphoimaging plates (Fujix BAS-3000; Fuji).
Biochemical measurements
Heart tissue samples from 14-week-old heart knockouts and littermate controls (n= 5) were collected, weighed and mitochondria were isolated. Respiratory chain enzyme activities and mitochondrial ATP production rates were measured in coded samples as previously described (39). Activities of complex I (NADH:coenzyme Q reductase), complexes I/III (NADH:cytochrome c reductase), complexes II/III (succinate:cytochrome c reductase) and complex IV (cytochrome c oxidase) as well as citrate synthase (CS) activity were determined at 35°C using a Thermo T20xti automated photometer (Thermo Fisher Scientific). ATP production rates in intact mitochondria were determined at 25°C using a firefly luciferase and a Victor3 Multi label counter 1420 (Perkin Elmer) in the presence of the following different substrate combinations: glutamate + succinate (G + S), glutamate + malate (G + M), TMPD + ascorbate (T + A), pyruvate + malate (P + M), palmitoyl-l-carnitine + malate (PC + M), succinate (S) and succinate + rotenone (S + R). Activities were expressed as units per unit of CS activity in the mitochondrial suspension. CS and GDH activities were determined in heart tissue samples as described previously (39) and used as mitochondrial markers.
Western blot
Heart tissue was homogenized in a sample buffer (0.1 m NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 10 mm Tris; pH 7.6) with protease inhibitors (Complete; Roche). For analysis of striatal TH-content, protein pellets from homogenates used for HPLC analysis were washed in ice-cold acetone and then resuspended in the sample buffer. Samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a nylon membrane (GE Healthcare). Antibodies against Ndufs4 (Mitosciences; 1:1000), VDAC (VDAC 31HL, Calbiochem; 1:2000), Actin (Abcam; 1:5000), Ndufa9 (Invitrogen; 1:2000), Sdha (Invitrogen; 1:1000), core1 (Invitrogen; 1:1000) and TH (TH-16, Sigma; 1:10000) or antisera against COX2 or Tfam (40) (1:5000) were used to detect the corresponding antigens.
Blue native gel electrophoresis
Mitochondria (75 µg) were solubilized in a solubilization buffer [20 mm Tris, pH 7.4, 0.1 mm EDTA, 50 mm NaCl, 10% (v/v) glycerol] containing either 1% (w/v) digitonin (Calbiochem) or 1% (w/v) dodecylmaltosyde (Sigma). Following 15 min of incubation on ice, unsolubilized material was removed by centrifugation and the supernatant was mixed with loading dye [5% (w/v) Coomassie Brilliant Blue G-250 (Serva), 100 mm Tris pH 7 and 500 mm 6-aminocaproic acid]. Samples were resolved on 4–10% gradient BN-PAGE gels (41). BN gels were further subjected to western blot analysis or in gel catalytic activity staining assay.
In gel, complex I activity was assessed by incubating the BN-PAGE gels in 2 mm Tris–HCl, pH 7.4, 0.1 mg/ml NADH (Roche) and 2.5 mg/ml iodonitrozolium (Sigma) at room temperature.
Tissue processing, immunohistochemistry and microscopy
Mice were deeply anesthetized with pentobarbital and subjected to transcardial perfusion with heparinized Ca2+-/Mg2+-free Tyrode's solution followed by fixative (4% paraformaldehyde with 0.4% picric acid in 0.16 m phosphate buffer). Brains were dissected, postfixed and thereafter equilibrated with 10% sucrose in phosphate buffer. Brains were frozen on CO2 ice and cryosectioned to obtain 14 or 20 µm sections.
Rehydrated sections were immunolabeled by incubation with an antibody against TH (1:500 dilution; Pel-Freez) overnight followed by a biotinylated secondary antibody (1:200 dilution; Vector Laboratories), which in turn was detected using a peroxidase substrate (Vector SG, Vector Laboratories). Images were acquired using a Zeiss axiophot 2 and ×5/0.45NA, ×10/0.45NA and ×40/0.65NA objectives and Axioplan 2 Imaging Software (Zeiss).
For confocal imaging, Cy3-conjugated secondary antibodies (1:400 dilution; Jackson Biolabs) were used. For measurements of TH density, systematically chosen areas of dorsolateral striatum (18 per brain; each 54 × 54 µm; z < 1.0 µm) were imaged by a confocal microscope (LSM510, Zeiss) using a ×100/1.45NA oil objective. The area of each image occupied by TH-immunoreactive fibers was measured using ImageJ (NIH).
Cell counts
Coronary midbrain cryosections (20 µm thick) were thawed onto coded slides and every sixth section was immunolabeled for TH. SNc was outlined as described previously (24) and nuclei of TH-positive neurons were counted in 11 sections encompassing all of SNc. A microscope with a motorized stage (Nikkon E600) controlled by stereology software (Stereologer, SRC; version 2001) was used for design-based stereology. SNc was outlined using a ×4 objective and cells counted using a ×60/1.4NA oil objective with counting frames spaced 200 µm apart. SNcs on the two sides were counted separately and then combined. Subsequently, all TH-positive cells on the same sections were counted manually using a ×20 objective (Nikkon). Nuclear diameter was measured as the maximal diameter in the x–y-plane of randomly selected cells. All cell counts were performed with the genotype blinded to the investigator.
High-performance liquid chromatography
Monoamine and metabolite levels were determined using HPLC. Mice were sacrificed by cervical dislocation, brains were rapidly removed, chilled in ice-cold saline and bilateral pieces of striatum, midbrain and frontal cortex were dissected (n= 7–10 for each genotype and age), frozen on dry ice and stored at −70°C until analysis. All tissues were weighed and homogenized by sonication in 0.1 M perchloric acid followed by centrifugation. The supernatants were centrifuged through a 0.45 µm filter (NANOSEP GHP MF; Pall Norden AB). Endogenous levels of noradrenaline, DA, DOPAC, HVA and 5-hydroxytryptamine (5-HT) were determined in the supernatants by comparison to standards as follows: dl-arterenol-hydrochloride for NA; 3-hydroxytyramine-hydrochloride for DA; 3,4-dihydroxyphenyl-acetic acid for DOPAC; 4-hydroxy-3-methoxyphenylacetic acid for HVA and 5-hydroxytryptamine creatinine sulfate for 5-HT. Separations were performed on a reverse-phase column (Reprosil-Pur, C18-AQ 150 mm×4 mm, 5 µm particle diameter; Dr. Maisch HPLC GmbH). The mobile phase consisted of a 0.05 m sodium phosphate/0.03 m citric acid buffer containing 0.1 mm EDTA, with various amounts of methanol and sodium-l-octane sulphonic acid, at a flow rate of 0.7 ml/min. Monoamines and metabolites were detected using an electrochemical detector system (Coulochem III with a High Sensitivity Analytical Cell, ESA). The detector cell was set at 50/350 mV and the resultant peak areas were measured using appropriate software (EZ Chrom Elite, ESA). Monoamine and metabolite levels were expressed as nanograms per gram wet weight of the tissue.
Slice preparation and voltammetric recordings
Mice of ∼12 months of age (n = 7 Ndufs4-DA, n = 4 Control 1, n = 5 Control 2) were used for slice preparations. Animals were sacrificed by cervical dislocation, and brains were rapidly removed and placed in a modified, high-sucrose-containing ice-cold artificial cerebrospinal fluid (aCSF). Coronal hemisections (280 μm) containing the striatum were cut using a vibratome (Leica VT1000S). Slices were incubated in standard oxygenated aCSF at 34–35°C for 20–30 min, then allowed to stabilize at room temperature for >30 min prior to initiating recordings. During recordings, slices were continuously superfused with aCSF at a rate of 2 ml/min, and maintained at 28–30°C.
FSCV was performed according to previously published protocols (26). Carbon fibers (7 μm diameter) were vacuum-aspirated into borosilicate pipette glass. Pipettes were pulled using a conventional patch-pipette puller, and the ends of the carbon fiber were cut to allow ∼25–30 μm exposed length protruding from the pipette tip. Pipettes were back filled with a 4 m potassium acetate/150 mm KCl solution and connected to a standard patch pipette holder/headstage assembly. A patch clamp amplifier (HEKA EVA-8) was used to deliver voltage and measure current from the headstage. Voltammetric scan and stimulation timing protocols were performed using PCI-based A/D boards (National Instruments) and custom software (courtesy of Dr. Mark Wightman, Univ. of North Carolina). Scans consisted of sweeps from −0.4 to 1.0 V and back to −0.4 V, at a rate of 400 V/s, and were obtained every 100 ms. A 5 s (50 scan) control period preceded each electrically evoked response, and was used to obtain a background current that was digitally subtracted from the current obtained during the peak of the response. Currents were converted to concentration by generation of linear in vitro calibration curves for each electrode using 1–4 μm DA. The mean current response for all electrodes (n = 17) was 35 ± 3 nA/μm DA, and did not significantly differ among groups. All signals used in analyses matched the expected voltammetric profile for DA.
Electrically evoked DA signals in brain slices
Under stereoscopic magnification, carbon fibers were lowered to a depth of ∼100 μm in dorsal striatum. A bipolar stimulating electrode was positioned ∼75–100 μm from the carbon fiber. Single, constant current pulses (0–250 μA, 1 ms duration) were delivered between voltammetric scans to elicit the DA release. DA uptake was assessed by fitting the decay portion of each signal to a single exponential function. The obtained tau values are independent of the signal amplitude, and have previously been demonstrated to be related to the efficiency of DAT-mediated uptake of DA (Vmax/Km), and are sensitive to DAT inhibitors.
In experiments designed to assess the effects of Ndufs4 deletion on ‘phasic’ DA release (27), two to three voltammetric signals were obtained at each recording site using a single pulse and 2, 5 and 10 pulses delivered at 25 Hz. After signals at each site were averaged, the difference between the peak DA signal obtained immediately after burst stimulation or single pulses was determined as DAnp − DA1p, where DAnp is the amplitude of the voltammetric signal for n pulses, and DA1p is the amplitude of the voltammetric signal obtained following a single electrical pulse. The data were fit to a linear regression model (y = mx + b; Prism 5.02; GraphPad, San Diego, CA, USA), where the slope m represents the relative change in DA concentration per pulse (26).
MPTP administration
During 2 consecutive days, male mice were given single subcutaneous injections of saline or MPTP (30 mg/kg; Sigma). Animals were sacrificed 7 days after the second injection.
FUNDING
This work was supported by the National Institutes of Health-Karolinska Institute Graduate Partnership Program for the Neurosciences (F.H.S.), The Swedish Research Council (grant numbers K2011-62X-21870-01-6, K2009-61X-03185-39-3), Swedish Brain Power, The Parkinson Foundation, The Karolinska Institutet and US Public Health Service, National Institute on Drug Abuse, Intramural Research Program.
ACKNOWLEDGEMENTS
We thank Eva Lindqvist, Karin Pernold and Karin Lundströmer for technical assistance and Dr Richard Palmiter for sharing Ndufs4 conditional knockout mice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Thomas B., Beal M.F. Parkinsons disease. Hum. Mol. Genet. 2007;16(Spec 2)):R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 2.Langston J.W., Ballard P., Tetrud J.W., Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 3.Burns R.S., Chiueh C.C., Markey S.P., Ebert M.H., Jacobowitz D.M., Kopin I.J. A primate model of parkinsonism: selective destruction of dopaminergic neurons in the pars compacta of the substantia nigra by N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Proc. Natl Acad. Sci. USA. 1983;80:4546–4550. doi: 10.1073/pnas.80.14.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopin I.J., Markey S.P. MPTP toxicity: implications for research in Parkinsons disease. Annu. Rev. Neurosci. 1988;11:81–96. doi: 10.1146/annurev.ne.11.030188.000501. [DOI] [PubMed] [Google Scholar]
- 5.Smeyne R.J., Jackson-Lewis V. The MPTP model of Parkinsons disease. Brain Res. Mol. Brain Res. 2005;134:57–66. doi: 10.1016/j.molbrainres.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Schapira A.H., Cooper J.M., Dexter D., Jenner P., Clark J.B., Marsden C.D. Mitochondrial complex I deficiency in Parkinsons disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 7.Bindoff L.A., Birch-Machin M., Cartlidge N.E., Parker W.D., Turnbull D.M. Mitochondrial function in Parkinson. Lancets disease. 1989;2:49. doi: 10.1016/s0140-6736(89)90291-2. [DOI] [PubMed] [Google Scholar]
- 8.Parker W.D., Boyson S.J., Parks J.K. Abnormalities of the electron transport chain in idiopathic Parkinsons disease. Ann. Neurol. 1989;26:719–723. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- 9.Betarbet R., Sherer T.B., MacKenzie G., Garcia-Osuna M., Panov A.V., Greenamyre J.T. Chronic systemic pesticide exposure reproduces features of Parkinsons disease. Nat. Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 10.Corral-Debrinski M., Horton T., Lott M.T., Shoffner J.M., Beal M.F., Wallace D.C. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 11.Soong N.W., Hinton D.R., Cortopassi G., Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat. Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 12.Bender A., Krishnan K.J., Morris C.M., Taylor G.A., Reeve A.K., Perry R.H., Jaros E., Hersheson J.S., Betts J., Klopstock T., et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 13.Bender A., Schwarzkopf R.-M., McMillan A., Krishnan K.J., Rieder G., Neumann M., Elstner M., Turnbull D.M., Klopstock T. Dopaminergic midbrain neurons are the prime target for mitochondrial DNA deletions. J. Neurol. 2008;255:1231–1235. doi: 10.1007/s00415-008-0892-9. [DOI] [PubMed] [Google Scholar]
- 14.Luoma P., Melberg A., Rinne J.O., Kaukonen J.A., Nupponen N.N., Chalmers R.M., Oldfors A., Rautakorpi I., Peltonen L., Majamaa K., et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- 15.Büeler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinsons disease. Exp. Neurol. 2009;218:235–246. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi W.-S., Palmiter R.D., Xia Z. Loss of mitochondrial complex I activity potentiates dopamine neuron death induced by microtubule dysfunction in a Parkinsons disease model. J. Cell. Biol. 2011;192:873–882. doi: 10.1083/jcb.201009132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruse S.E., Watt W.C., Marcinek D.J., Kapur R.P., Schenkman K.A., Palmiter R.D. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell. Metab. 2008;7:312–320. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi W.-S., Kruse S.E., Palmiter R.D., Xia Z. Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proc. Natl Acad. Sci. USA. 2008;105:15136–15141. doi: 10.1073/pnas.0807581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansson A., Hance N., Dufour E., Rantanen A., Hultenby K., Clayton D.A., Wibom R., Larsson N.-G. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc. Natl Acad. Sci. USA. 2004;101:3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvaruso M.A., Willems P., van den Brand M., Valsecchi F., Kruse S., Palmiter R., Smeitink J., Nijtmans L. Mitochondrial complex III stabilizes complex I in the absence of NUDFS4 to provide partial activity. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr446. doi:10.1093/hmg/ddr446. [DOI] [PubMed] [Google Scholar]
- 22.Ekstrand M.I., Terzioglu M., Galter D., Zhu S., Hofstetter C., Lindqvist E., Thams S., Bergstrand A., Hansson F.S., Trifunovic A., et al. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl Acad. Sci. USA. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abercrombie M. Estimation of nuclear population from microtome sections. Anat. Rec. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- 24.Baquet Z.C., Williams D., Brody J., Smeyne R.J. A comparison of model-based (2D) and design-based (3D) stereological methods for estimating cell number in the substantia nigra pars compacta (SNpc) of the C57BL/6J mouse. Neuroscience. 2009;161:1082–1090. doi: 10.1016/j.neuroscience.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birgner C., Nordenankar K., Lundblad M., Mendez J.A., Smith C., le Grevès M., Galter D., Olson L., Fredriksson A., Trudeau L.-E., et al. VGLUT2 in dopamine neurons is required for psychostimulant-induced behavioral activation. Proc. Natl Acad. Sci. USA. 2010;107:389–394. doi: 10.1073/pnas.0910986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Good C.H., Hoffman A.F., Hoffer B.J., Chefer V.I., Shippenberg T.S., Backman C.M., Larsson N.G., Olson L., Gellhaar S., Galter D., et al. Impaired nigrostriatal function precedes behavioral deficits in a genetic mitochondrial model of Parkinsons disease. FASEB J. 2011;25:1333–1344. doi: 10.1096/fj.10-173625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyland B.I., Reynolds J.N., Hay J., Perk C.G., Miller R. Firing modes of midbrain dopamine cells in the freely moving rat. Neuroscience. 2002;114:475–492. doi: 10.1016/s0306-4522(02)00267-1. [DOI] [PubMed] [Google Scholar]
- 28.Quintana A., Kruse S.E., Kapur R.P., Sanz E., Palmiter R.D. Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc. Natl Acad. Sci. USA. 2010;107:10996–11001. doi: 10.1073/pnas.1006214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keeney P.M., Xie J., Capaldi R.A., Bennett J.P. Parkinsons disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sossi V., de La Fuente-Fernández R., Holden J.E., Doudet D.J., McKenzie J., Stoessl A.J., Ruth T.J. Increase in dopamine turnover occurs early in Parkinsons disease: evidence from a new modeling approach to PET 18 F-fluorodopa data. J. Cereb. Blood Flow Metab. 2002;22:232–239. doi: 10.1097/00004647-200202000-00011. [DOI] [PubMed] [Google Scholar]
- 31.Galter D., Pernold K., Yoshitake T., Lindqvist E., Hoffer B., Kehr J., Larsson N.-G., Olson L. MitoPark mice mirror the slow progression of key symptoms and L-DOPA response in Parkinsons disease. Genes Brain Behav. 2009:1–9. doi: 10.1111/j.1601-183X.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zigmond M.J., Hastings T.G., Perez R.G. Increased dopamine turnover after partial loss of dopaminergic neurons: compensation or toxicity? Parkinsonism Relat. Disord. 2002;8:389–393. doi: 10.1016/s1353-8020(02)00019-6. [DOI] [PubMed] [Google Scholar]
- 33.Watabe M., Nakaki T. Mitochondrial complex I inhibitor rotenone inhibits and redistributes vesicular monoamine transporter 2 via nitration in human dopaminergic SH-SY5Y cells. Mol. Pharmacol. 2008;74:933–940. doi: 10.1124/mol.108.048546. [DOI] [PubMed] [Google Scholar]
- 34.Kaiya H., Namba M. Two types of dopaminergic nerve terminals in the rat neostriatum. An ultrastructural study. Neurosci. Lett. 1981;25:251–256. doi: 10.1016/0304-3940(81)90400-6. [DOI] [PubMed] [Google Scholar]
- 35.Sterky F.H., Lee S., Wibom R., Olson L., Larsson N.G. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc. Natl Acad. Sci. USA. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hastings T.G., Lewis D.A., Zigmond M.J. Role of oxidation in the neurotoxic effects of intrastriatal dopamine injections. Proc. Natl Acad. Sci. USA. 1996;93:1956–1961. doi: 10.1073/pnas.93.5.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caudle W.M., Richardson J.R., Wang M.Z., Taylor T.N., Guillot T.S., McCormack A.L., Colebrooke R.E., Di Monte D.A., Emson P.C., Miller G.W. Reduced vesicular storage of dopamine causes progressive nigrostriatal neurodegeneration. J. Neurosci. 2007;27:8138–8148. doi: 10.1523/JNEUROSCI.0319-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dagerlind A., Friberg K., Bean A.J., Hokfelt T. Sensitive mRNA detection using unfixed tissue: combined radioactive and non-radioactive in situ hybridization histochemistry. Histochemistry. 1992;98:39–49. doi: 10.1007/BF00716936. [DOI] [PubMed] [Google Scholar]
- 39.Wibom R., Hagenfeldt L., von Döbeln U. Measurement of ATP production and respiratory chain enzyme activities in mitochondria isolated from small muscle biopsy samples. Anal. Biochem. 2002;311:139–151. doi: 10.1016/s0003-2697(02)00424-4. [DOI] [PubMed] [Google Scholar]
- 40.Larsson N.G., Garman J.D., Oldfors A., Barsh G.S., Clayton D.A. A single mouse gene encodes the mitochondrial transcription factor A and a testis-specific nuclear HMG-box protein. Nat. Genet. 1996;13:296–302. doi: 10.1038/ng0796-296. [DOI] [PubMed] [Google Scholar]
- 41.Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]