Abstract
Polyalanine (poly-A) tracts exist in 494 annotated proteins; to date, expansions in these tracts have been associated with nine human diseases. The pathogenetic mechanism by which a poly-A tract results in these various human disorders remains uncertain. To understand the role of this mutation type, we investigated the change in functional properties of the transcription factor Arx when it has an expanded poly-A tract (ArxE), a mutation associated with infantile spasms and intellectual disabilities in humans. We found that although ArxE functions normally in the dorsal brain, its function in subpallial-derived populations of neurons is compromised. These contrasting functions are associated with the misregulation of Arx targets through the loss of the ability of ArxE to interact with the Arx cofactor Tle1. Our data demonstrate a novel mechanism for poly-A expansion diseases: the misregulation of a subset of target genes normally regulated by a transcription factor.
INTRODUCTION
Mutations in the Aristaless-related homeobox gene, ARX, result in a spectrum of phenotypes ranging from X-linked intellectual disability (XLID [MIM 300419]) to severe structural brain malformations (1). Individuals with ARX-associated XLID (with or without epilepsy) have structurally normal brains by neuroimaging; in contrast, brain malformations such as lissencephaly (LISX2 [MIM 300215]) are observed in individuals with more severe mutations, presumably as a result of aberrant radial cell migration. Although a relatively well-defined phenotype–genotype correlation exists for patients with ARX mutations, how different mutations in the same gene result in such divergent phenotypes is unknown. One hypothesis predicts that ARX-related XLID with or without seizures is the result of a mutation causing a selective cortical interneuron defect, while mutations that affect both cortical projection neurons and interneurons would give rise to the lissencephaly and associated neurologic phenotype.
The ARX sequence includes four poly-A tracts. Short expansions in the first or second tract have been found in patients with epilepsy and XLID but not gross brain anomalies (1). For example, the addition of only two alanines to the first poly-A tract [c.304_305ins(GCG)2; p.A103_A104insAA] is associated with XLID (2), while expansion in the same tract by seven alanines [c.333_334ins(GCG)7; p.A111_A112insAAAAAAA] results in X-linked Infantile Spasm Syndrome, infantile epileptic-dyskinetic encephalopathy or early and severe infantile epileptic encephalopathy with a burst-suppression pattern on EEG (ISSX1, EIEE1 [MIM 308350]) (1,3). Thus, in clear contrast to the polyglutamine tract expansion disorders (4), even small expansions in the first poly-A tract in ARX can lead to a severe developmental disorder.
Although a growing number of human syndromes have been associated with a poly-A tract expansion mutation, the effect of such expansions on protein function remains enigmatic (5,6). Although mutant protein aggregation has been implicated in the pathogenesis, such aggregation has mainly been observed upon protein overexpression and may not have physiologic relevance (5). In the case of Arx, poly-A expansion mouse models have not supported protein aggregation or the cell death phenotype that has been observed in vitro (7–9).
Arx is broadly expressed in the embryonic mouse forebrain (10,11). Within the dorsal telencephalon, it is expressed in the proliferative ventricular zone (VZ); in contrast, it is expressed more strongly in differentiating rather than proliferating neurons in the ventral telencephalon (12). In fact, ventral and dorsal expression of Arx appears to be driven by different enhancer regions (13). These differences suggest that the Arx protein may have distinct dorsal and ventral functions. Our previous studies indicate that partially eliminating Arx selectively from the mouse ventral forebrain results in a cortical interneuron deficit and causes an epilepsy remarkably similar to that in humans carrying an expansion of the first poly-A tract (14). In contrast, germline loss of Arx results in defects in both dorsal and ventral telencephalic progenitor cell development and a more severe brain phenotype (15,16). Recently, mice were engineered with an expansion in the first poly-A tract. Interestingly, these mice also have seizures and interneuron defects, akin to mice that lack Arx function selectively in the ventral forebrain (8,9). These findings raise the intriguing hypothesis that the mechanism by which an ARX/Arx poly-A mutation might cause disease is through the misregulation of only a subset of the ARX/Arx target genes. Herein we show differential gene regulation by ARX/Arx in the ventral versus dorsal telencephalon as a result of a poly-A tract expansion. These data provide a novel mechanism to understand the biologic basis for the phenotypic differences observed with different mutations in ARX/Arx in both humans and mice.
RESULTS
Arx with a poly-A tract expansion (ArxE) functions normally in radial cell migration
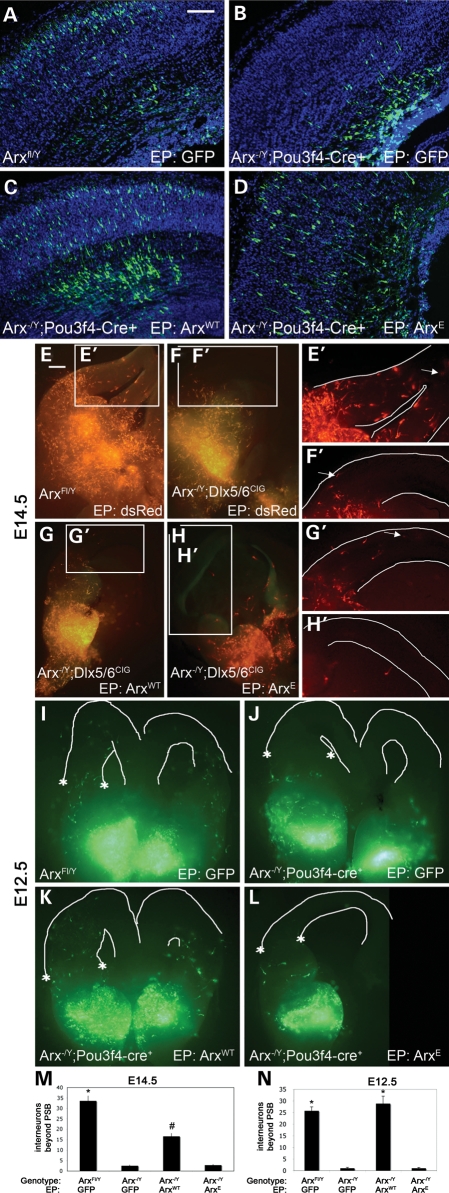
To understand how poly-A tract expansion affects development, we first examined the ability of Arx with an expansion of eight alanines in the first poly-A tract (ArxE) to function in radial and non-radial cell migration (NRCM). We had previously generated male mice with essentially no forebrain Arx expression (Arx-/Y;Pou3f4-cre) (17). The mutant embryos demonstrate microcephaly and NRCM defects, as seen with germline deletions (15,16). As found previously, we also observed a radial migration defect from the dorsal telencephalic VZ (Fig. 1A and B) (18). The defect was evident for the furthest migrating cells (Fig. 1 and Supplementary Material, Fig. S1, bins 9 and 10); although differences may be present closer to the VZ, the variability in the data precluded significance. Full-length Arx, delivered into the pallial VZ by in utero electroporation at E14.5, was able to fully rescue the radial migration defect (Fig. 1C). Interestingly, in utero delivery of ArxE on E14.5 was also able to rescue the migration defect (Fig. 1D), although the number of furthest-migrating neurons was reduced compared with rescue with Arx (Supplementary Material, Fig. S1A). This lack of complete rescue may be explained in part by ArxE degradation, as overexpressed ArxE forms aggregates in ∼30% of cells (7), and protein levels are reduced in the Arx(GCG)7/Y mutant mouse (8). Because Arx is down-regulated outside of the pallial VZ and is not expressed in neurons derived from this region once they begin to migrate (12), we assume the ArxE fully rescues all dorsal functions, in addition to migration; however, it remains possible that later functions, which we have not tested and are not yet known, cannot be rescued in the dorsal telencephalon.
Figure 1.
In utero electroporation (EP) of ArxWT, ArxE or control constructs into Arx-/Y;Pou3f4-cre+ (mutant) embryos compared with EP into wild-type littermates demonstrates that ArxE has a partial ability to rescue radial cell migration in the mutant mice; in contrast, slice electroporation of ArxE cannot rescue migration. (A) Wild-type embryos electroporated at E14.5 with a control vector expressing GFP were harvested at E18. Sections are counterstained with DAPI (blue). (B) An Arx-/Y;Pou3f4-cre+ brain electroporated with a control construct shows a defect in radial migration. (C and D) Arx-/Y; Pou3f4-cre+ embryos electroporated with ArxWT (C) or ArxE (D) show rescue of radial migration. Scale bar (upper right corner in A): 100 mm. (E) Normal NRCM is observed in E14.5 wild-type brains after EP of a control dsRed construct. White lines define the pial and ventricular surfaces. (F) In contrast, brains in which Arx is conditionally deleted in interneurons (Arx-/Y;Dlx5/6CIG) have loss of NRCM. Electroporation of ArxWT (G) but not ArxE (H) is able to partially rescue this migration defect. (E'–H') Detail of interneurons migrating in the pallium; the furthest-migrating interneuron is indicated with an arrow. Results were the same for Arx-/Y;Pou3f4-cre slice culture. (I) Control electroporation in wild-type E12.5 slice culture. (J) Loss of migration in E12.5 Arx-/Y;Pou3f4-cre slice culture. (K) NRCM rescue with ArxWT at E12.5. (L) Failure of rescue by ArxE at E12.5. A line drawn between the asterisks' define the location of the pallial–subpallial boundary (I–L, line not drawn so data are not obscured). (M) Counts in wild-type brains and both types of mutant brains electroporated with ArxWT were significantly different from all others (n > 10 for each genotype-rescue combination; *, #P < 0.05). A trend toward rescue was observed with electroporation of ArxWT but not ArxE into mutant brains. (N) Quantification of NRCM after E12.5 rescue shows ArxWT rescued NRCM at E12.5 (n > 7 for each genotype–rescue combination; *P < 0.05). No statistical differences were found between migration for Arx-/Y;Dlx5/6CIG and Arx-/Y;Pou3f4-Cre+ mutant mice; quantification includes both mutant lines. Each genotype/construct combination is represented by at least three brains from three different litters of each mutant line at each time point. PSB, pallial-subpallial boundary; error bars, SEM; scale bar (in A): 500 mm.
The ArxE mutation fails to support NRCM
We next tested the ability of ArxE to rescue the non-radial migration of GABAergic interneurons from the ventral telencephalon to the dorsal cortex. Integration of interneurons into cortical circuitry is crucial for modulating and controlling the activity of the brain and it has been proposed that the loss of or defects in these populations can lead to epilepsy (19). We have previously shown that the loss of Arx leads to the improper cortical distribution of interneurons in both Arx-/Y;Pou3f4-cre mice (17), and Arx-/Y;Dlx5/6-cre-IRES-GFP (Arx-/Y;Dlx5/6CIG) mice, in which Arx is selectively deleted in the ventral Dlx5/6+ expression domain (14,20). These defects are consistent with the NRCM defect observed in germline Arx-/Y mice (15,16). Electroporation of ArxE directly into the medial ganglionic eminence (MGE) of slice cultures from E14.5 Arx-/Y;Dlx5/6CIG brains could not rescue NRCM (Fig. 1H and H'). However, even ArxWT only partially rescued migration when electroporated into the MGE of an E14.5 Arx-/Y;Dlx5/6CIG slice culture (Fig. 1G and G'). We postulated that ArxWT may be required earlier in development in the ventral forebrain (11). Consistent with this hypothesis, ArxWT electroporated into E12.5 Arx-/Y;Dlx5/6CIG slice cultures completely rescued NRCM (Fig. 1K and M). In striking contrast, ArxE was not able to rescue NRCM (Fig. 1L–N), despite robust protein expression (Supplementary Material, Fig. S1B).
Arx and ArxE bind DNA target sequences with similar affinity
Our in vivo and slice culture data indicated that ArxE can function normally in the dorsal forebrain, but is defective in the ventral forebrain suggest that Arx performs different roles in these two areas. Arx is primarily a transcriptional repressor (17,21–23), which binds to the 5′ promoter region of Shox2, Ebf3 and Lmo1 and represses their expression (17). Although the mechanism of repression employed by Arx is incompletely understood, the source of activity appears to reside in the N-terminal octapeptide domain of the Arx protein. This domain is similar to the Engrailed homology repressor domain (eh-1) (24), and recruits Groucho/transducin-like enhancer of split co-factor proteins (TLE1–4) (22).
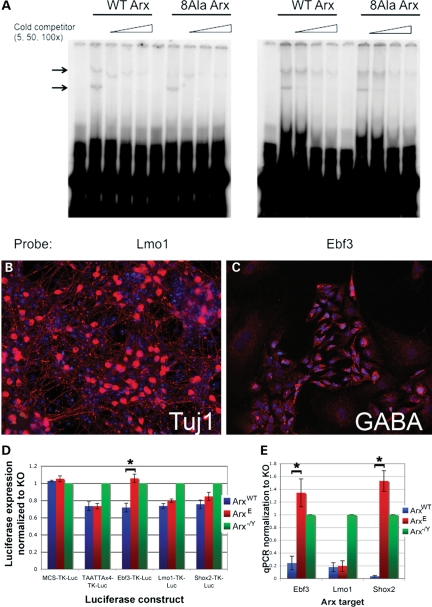
Given that other proteins with poly-A expansion mutations have lost the ability to bind to DNA (25,26), we first investigated whether ArxE retained the ability to bind to its known target sequence. Surprisingly, electrophoretic mobility shift assays (EMSAs) demonstrated that ArxE was able to bind DNA with similar affinity to wild-type Arx (Fig. 2A). Thus, the mechanism of action resulting from an expansion in the first alanine tract of Arx was not the loss of its ability to bind target sequences.
Figure 2.
ArxE has not lost the ability to bind to Arx targets, but has lost the ability to repress a subset of them. (A) EMSA comparing ArxE and ArxWT binding to Lmo1 and Ebf3-binding sites shows that ArxE has not lost the ability to bind to Arx targets. Arrows indicate specific bands for Lmo1 and Ebf3, as determined by the loss of the bands upon addition of cold competitor of increasing concentrations. The lower band is most likely monomer, and the upper band is dimer, based on the absence of a band showing degraded protein (data not shown). Both upper and lower bands from four sets of experiments were quantitated using ImageJ and used to calculate Kd values. The Kd Arx/Kd ArxE was calculated for Ebf3 upper band (2.18) and lower band (1.65) as well as the upper (0.89) and lower band for Lmo1 (according to the protocol of (35). These data indicate both Arx and ArxE bind both targets with similar affinity. (B–E) Measurement of Arx targets in differentiated ES cells shows loss of repression by ArxE for Ebf3 and Shox2 but not Lmo1. Three lines of ES cells were differentiated into neuronal populations: wild-type R1 (ArxWT), a line with Arx deleted (Arx-/Y), and a line engineered for Arx to have a poly-A expansion in the first alanine track (ArxE). (B) Tuj1 staining (DAPI counterstain) demonstrates extensive neuronal differentiation. (C) Many cells show GABAergic differentiation. (D) Luciferase assay shows that ArxE does not properly regulate Ebf3 expression. Luciferase data were normalized to Renilla expression. Data for the Arx-transfected data points are presented as percentage activation (±SEM) relative to empty vector-transfected cells (*P = 0.005). (E) Quantitative PCR data indicate the differential regulation of Arx targets in ES cells carrying the Arx poly-A expansion (±SEM, ArxWT versus ArxE for Ebf3 P = 0.004, for Shox2 P = 0.00009).
A subset of ventral Arx targets is misregulated by ArxE
We next examined whether the transcriptional repressive capacity of ArxE was retained using a luciferase reporter assay for defined target sequences (17). The assay was performed using ArxE, Arx-/Y or wild-type R1 embryonic stem (ES) cells differentiated into neurons, including a significant population of GABAergic neurons (27) (Fig. 2B and C, Supplementary Material, Fig. S2). Dissociated embryoid bodies were electroporated with luciferase reporter constructs as described in Materials and Methods. After differentiation, luciferase measurements demonstrated that ArxE is able to repress Lmo1, but not Ebf3 (Fig. 2D). Shox2 showed a trend toward loss of repression, although the difference was not significant (Supplementary Material, Fig. S2G). To verify the luciferase data, quantitative polymerase chain reaction (qPCR) was performed on reverse-transcribed RNA from the three ES cell lines. The results agreed with those from the luciferase experiments, and further demonstrated significant loss of Shox2 repression by ArxE (Fig. 2E, Supplementary Material, Fig. S2H). These data revealed that ArxE retains the ability to bind DNA but has lost repressive function for a subset of target genes.
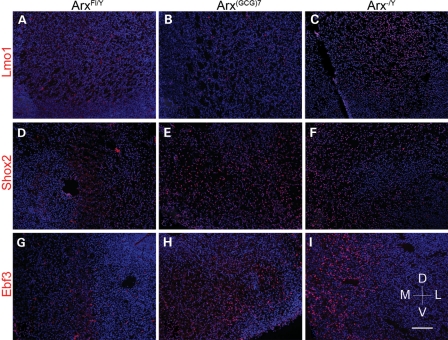
To confirm these in vitro data, we next tested the ability of ArxE to repress Arx targets in the brains of Arx (GCG)7/Y mutant mice with an expansion in the first poly-A tract (8) (Fig. 3). In accordance with the ES cell data, the targets Ebf3 and Shox2, but not Lmo1, are upregulated at E18 in the MGE of Arx (GCG)7/Y brain sections. Based on the ES cell and in vivo data, we conclude ArxE can only repress a subset of the normal Arx targets. Moreover, given that ArxE appears to bind DNA in the promoter regions with an affinity similar to Arx, the failure to repress at specific targets may depend on promoter context.
Figure 3.
Aberrant expression of Ebf3 and Shox2, but not Lmo1, at E18 in Arx(GCG)7 mice. Wild-type and mutant E18 brain tissue were stained for Lmo1 (A–C), Shox2 (D–F) and Ebf3 (G–I) in red, with blue DAPI counterstain. The three proteins show no expression (equal to repression) in wild-type tissue (A, D, G), but are expressed (not repressed) in Arx-/Y tissue (C, F, I). In Arx(GCG)7/Y tissue, Lmo1 continues to be repressed (B), but Shox2 and Ebf3 are not repressed, as seen in the Arx-/Y tissue (E and H). All images are from the ventral subpallium, with the ventricle to the left, and dorsal at the top, as indicated by the compass. Staining was performed for n = 4 for each genotype/antibody combination. Scale bar (see I): 120 mm.
The misregulation of target genes required for NRCM is due to the context-dependent failure of ArxE to recruit the transcriptional co-repressor Tle1
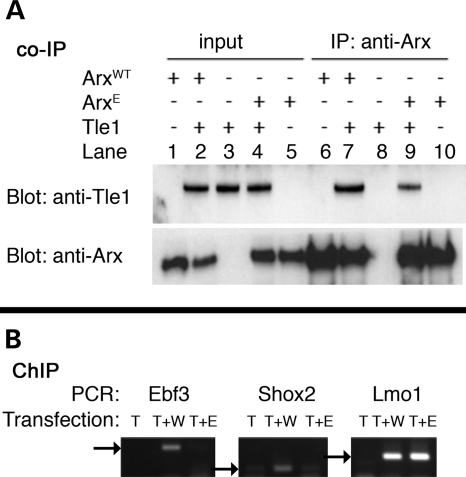
In order to understand how ArxE could have different actions at distinct promoters, we investigated its ability to bind its known co-repressor, Tle1. The Tle proteins are mammalian homologues of the Drosophila Groucho protein, and their main structural feature is the carboxy-terminal WD-repeat domain, which forms a β-propeller that mediates interactions with proteins, including transcription factors (28). Tle proteins function as corepressors, but do not bind DNA directly (24). Instead, they are recruited and bound by transcription factors. Transcription factors bind Tle proteins through one of two motifs in the WD-repeat domain: the Trp-Arg-Pro-Trp motif (or similar) or the Engrailed homology 1 motif, FxIxxIL. In the case of Arx, it binds to Tle proteins through its octapeptide (or engrailed) domain, SSYCIDSILG (22). We co-expressed ArxE and Tle1 in Neuro2a cells and then co-immunoprecipitated the proteins. We found that, although ArxE could pull down Tle1, it has reduced binding (Fig. 4A, lane 9). To further understand the role of promoter context interactions between Arx and Tle1, we co-expressed ArxWT or ArxE with Tle1, or Tle1 alone, and performed chromatin immunoprecipitation (ChIP) for Tle1, followed by PCR for the promoter regions of Ebf3, Lmo1 and Shox2 (Fig. 4B and Supplementary Material, Fig. S4). While the promoter regions of Ebf3, Lmo1 and Shox2 amplified by PCR when ArxWT was co-expressed with Tle1, only Lmo1 amplified when ArxE was co-expressed with Tle1 (although we verified amplification of the input, Supplementary Material, Fig. S3A). Altogether, our data support a novel model for the effect of the poly-A expansion in Arx, to prevent the recruitment or stabilization of Tle1 at a subset of Arx targets and therefore prevent the necessary repression of a subset of Arx targets.
Figure 4.
Loss of cooperative binding of Tle1 and ArxE. (A) Tle1 co-immunoprecipitates less efficiently with ArxE than with ArxWT. Neuro-2a cells normally do not express Tle1 (input, lanes 1 and 5). After co-expression of Tle1 and ArxWT, Tle1 can be pulled down with Arx (IP, lane 7). The amount of Tle1 pulled down by ArxE is reduced (IP, lane 9). (B) After co-transfection with Tle1 and ArxWT, ChIP with anti-V5 antibody to pull down Tle1 shows that the three Arx target sequences upstream of Ebf3, Shox2 and Lmo1 are amplified (arrows indicate band size). However, after co-transfection of Tle1 and ArxE, only Lmo1 is amplified. T, Tle1; T + W, Tle1 and ArxWT; T + E, Tle1 and ArxE (also see Supplementary Material, Fig. S4C for quantitation).
DISCUSSION
Short expansions in poly-A tracts are associated with a growing number of primarily developmental human disorders, including many involving the central nervous system (5). Protein folding anomalies resulting in protein aggregation and loss of DNA binding have been proposed to explain how these short expansions in a poly-A tract result in developmental disorders (5). In this study, we have identified a novel mechanism in which a poly-A tract expansion disrupts the function of a transcription factor by reducing its ability to recruit a cofactor required for normal transcriptional repression at a subset of target sites.
The epilepsy and intellectual disability phenotypes observed in patients with an expansion of the first poly-A tract in ARX are consistent with this mutation selectively affecting interneurons (19). The lack of a structural defect (lissencephaly) in patients with this type of mutation also suggests that radial migration is preserved. Two Arx poly-A tract expansion mouse models recently generated exhibit a partial interneuron phenotype (8,9). Both models survive postnatally and exhibit epilepsy, anxiety, learning and memory deficits. One of these models showed a mild loss of parvalbumin-containing interneurons in the cortex at 1 month of age; in contrast, interneurons were greatly reduced in the septum and striatum (8). In the second model, learning and anxiety abnormalities were observed along with EEG findings of seizures. In this model, reductions in Arx- and calbindin-expressing cortical interneurons along with a reduction in striatal interneurons were identified (9). These data, along with our data showing a selective interneuron defect in the absence of a radial migration defect, are consistent with the neurologic defects found in patients having no structural defect.
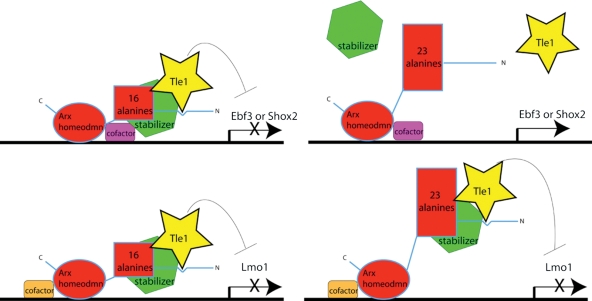
Our data are also consistent with the observed Arx poly-A tract mutation inheritance pattern. Although the in vitro data indicate that poly-A tract expansion results in nuclear inclusion formation (7), this mechanism would be expected to have a dominant-negative phenotype, but such a pattern of inheritance is not observed (1). The lack of inclusions in the mouse models further suggests that this is not the mechanism. Immunohistochemistry for Arx in the brains of Arx (GCG)7/Y mutant mice showed staining throughout the nucleus, with no evidence of nuclear or cytoplasmic inclusions (data not shown). In addition, TUNEL staining showed no difference at either E11.5 or E18.5 between the brains of Arx (GCG)7/Y mutant mice and those of wild-type mice (data not shown). However, pathology due to protein misfolding leading to protein dysfunction or to toxic levels of insoluble protein without visible inclusions cannot be ruled out. We have also precluded a DNA-binding mechanism, as has been proposed for poly-A tract expansion mutations in several other proteins (5). Our results support the idea that poly-A expansion leads to a change in conformation that prevents the recruitment of Tle1 to certain promoter regions (Fig. 5). We postulate that additional cofactors will be required in the transcriptional complex associated with ARX. As with other transcription factors, different cofactors will participate in transcriptional regulation at different sites. As proposed in our model, some cofactors will require physical interactions for stabilization of Tle1 for transcriptional repression (e.g. Ebf3 or Shox2) and others will not (e.g. Lmo1). Further studies will delineate the complete subset of Arx targets misregulated when the first poly-A is expanded, and investigate the specific functions of these targets in interneurons.
Figure 5.
Model of pathological mechanism resulting from expansion of first poly-A tract in Arx. The expansion results in a context-dependent loss of Tle recruitment to promoter sites. The context at the Ebf3 and Shox2 promoters does not allow Tle recruitment with ArxE (top right); however, the binding and activity of Tle are not perturbed in the case of Lmo1 (bottom right).
Of considerable interest is the finding that biological function is retained in the dorsal forebrain, but is defective in the ventral forebrain. The deficit caused by the Arx poly-A expansion mutation in the ventral forebrain is in part due to a failure to repress a subset of normal Arx targets resulting from the faulty recruitment of Tle1 to select target sites. Thus, the poly-A expansion leads to a partial loss of function. This conclusion is consistent with the ISSX-like phenotype seen in mice that have Arx deleted in only a subset of interneurons, suggesting that in humans, this disease may not reflect a complete loss of function of ARX in interneurons (14) (data not shown). In contrast, most Arx-/Y; Dlx5/6CIG mice die at or before birth; one interpretation of this finding is that interneuron expression and normal function of Arx is necessary for survival. Given Arx and Dlx5/6 have little overlap in expression outside the central nervous system, this conclusion would seem warranted; however, this does not exclude other central nervous system functions for Arx in Dlx5/6 expressing cells. In addition, while these data demonstrate a context-dependent failure of Arx to function in the ventral forebrain, the rescue of dorsal forebrain function suggests that an expansion in the first poly-A tract does not affect the role of Arx in cortical VZ cells. Alternatively, Tle1 may not be required for repression in the dorsal forebrain, or Arx may be primarily activating in this region.
In contrast to polyglutamine tracts, which can tolerate a broad number of residues with expansions in the hundreds often required to produce disease, poly-A tracts never consist of >20 alanines. Furthermore, they do not undergo the dynamic expansion that leads to the significant generation-to-generation variation in repeat length encoding glutamines (6). When a tract is expanded beyond its usual length, even if the resulting expanded tract has fewer than 20 alanines, disease results, as seen for nine diseases caused by poly-A tract expansions (5). Our data suggest a mechanism of disease for the dramatic effect seen in disorders caused by these mutations.
In conclusion, poly-A tract expansions are a relatively newly identified class of mutation that results in human disorders, many of which affect the nervous system. Although several possible pathogenetic mechanisms for a poly-A tract expansion have been proposed, limited in vivo data exist to support these other models. Herein, we present a novel mechanism involving a partial loss of function attributable to a context-dependent ability of Arx to recruit a co-repressor to the transcriptional regulatory complex. These findings are consistent with the phenotypes observed in humans and possibly explain how other poly-A tract expansions may function, although direct testing of these other genes will be required.
MATERIALS AND METHODS
In utero electroporation
All animal experiments were approved by the Children's Hospital of Philadelphia animal care and use committee. In utero electroporations were performed as described previously using GFP or DsRed expression vectors co-transfected at a ratio 5:1 with the Arx expression construct (pCAG-Arx-IRES-GFP or pCDNA3.1-Arx -V5-His) (29). Three and a half days after electroporation, the dams were sacrificed and the embryonic brains were harvested, fixed and processed for cryosectioning. Slides were imaged on a Leica DM6000B microscope.
Slice electroporation and culture
Slice cultures of embryonic mouse forebrain were prepared as described previously (30). After a 30 min incubation in 1:1 Dulbecco's modified Eagle's medium (DMEM):F12 (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (Invitrogen), 1 mm penicillin/streptomycin and 6.5 mg/ml glucose at 37°C in 5% CO2 in a standard sterile incubator, sections were injected with plasmid DNA and electroporated using a needle electrode and a block petridish electrode (Protech International Inc) to deliver three 5 ms electric pulses of 20 V. After a further 30 min incubation period, slices were transferred to DMEM plus N2 supplement (1:50, Invitrogen), 1 mm penicillin/streptomycin and 6.5 mg/ml glucose for 2 days and then imaged on a Leica dissecting microscope.
Electrophoretic mobility shift assay
EMSAs were performed as previously described with slight modifications (17). One microgram of a GST-Arx construct was incubated with 0.1 pmol of labeled probe in total 20 µl of binding buffer [20 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 6.5), 50 mm KCl, 1 mm MgCl2, 1 mm dithiothreitol, 5% glycerol, 1 µg poly(dI-dC) (Roche) with 0.5% Triton X-100]. The mixtures were loaded and run on a 4% 40:1 polyacrylamide:bis gel for Lmo1, and a 4% 80:1 polyacrylamide:bis gel for Ebf3. Cloning of the sumoylated Arx was accomplished by PCR and purification of SUMO and Arx. The two products were combined for 10 cycles to create a fusion vector, which was subcloned into pTYB2. The forward primer for SUMO included a streptavidin peptide-binding nucleotide sequence and an NdeI cut site; the reverse primer for Arx included an XhoI cut site for subcloning into pTYB2. The primer sequences are as follows: SUMO forward primer: 5′-ggaattccatatggcaagctggagccacccgcagttcgaaaagggtgca ATGTCGGACTCAGAAGTCAATCAAGAAG-3′; SUMO reverse primer: 5′-CCTCTTCCTGGTACTGATTGCTCAT ACCACCAATCTGTTCTCTGTGAGCC-3′; Arx forward primer: 5′-GGCTCACAGAGAACAGATTGGTGGT ATGAGCAATCAGTACCAGGAAGAGG-3′; Arx reverse primer: 5′-ccgctcgag GCACACCTCCTTCCCCGTG-3′. Protein was purified from E. coli using the IMPACT kit (NEB), according to the manufacturer's instructions, and concentrated with the Vivaspin 20 (Sartorius Stedim Biotech), following the manufacturer's instructions with the change of the buffer to 50 mm Tris–HCl (pH 6.5) containing 150 mm NaCl.
ES cell differentiation
ES cell lines were differentiated as previously described (27). The Arx-/Y line was created by cre-GFP electroporation into the ES cells used to make the ArxFl/Fl mice for our conditional mutant (17). The expansion of the poly-A tract in Arx was engineered as described (7).
Immunohistochemistry
Cells grown on cover slips were fixed for 5 min in 4% paraformaldehyde (with glutaraldehyde for α-GABA antibody), and washed in phosphate buffered saline (PBS). Cells were blocked in PBS with 10% normal goat serum, 0.5% Triton X-100 and the primary antibody diluted in block was placed on the cells for 1 h at room temperature, followed by secondary antibody. Cryosection slides were baked for 10 min at 55°C, and washed with PBS before citric acid antigen retrieval (Vector Laboratories). After blocking in 1% Triton and 10% serum (of the appropriate species) for 1 h at room temperature, primary antibody was applied overnight at 4°C. After washing, the appropriate biotin-conjugated secondary antibody was applied at 1:500 for 3 h, followed by the Alexa fluor Streptavidin-594 (Invitrogen) at 1:500 for 1 h. Finally, 4,6′-diamidino-2-phenylindole was applied at 1:1000 for 10 min before washing and cover slipping the slides. Primary antibodies used for these studies included α-Tuj1 (Covance MMS0435P, 1:500), α-GABA (Sigma A2052, 1:200), α-Arx (8) (1:1000), α-GAD67 (Millipore MAB5406, 1:200), α-Shox2 (Santa Cruz, sc-21898, 1:200), α-Lmo1 (Novus, NB100-57557, 1:200), α-Ebf3 (Abnova, H00253738-M05, 1:200) and α-glial fibrillary acidic protein (kindly provided by Dr V. Lee, University of Pennsylvania, School of Medicine).
Luciferase assay
During the ES cell differentiation protocol, embryoid bodies were dissociated with Accutase (Millipore), washed with PBS and resuspended in Nucleofector solution (Lonza). Amaxa electroporation of 2 μg of a firefly luciferase construct (17) and 200 ng of Renilla luciferase was performed following the manufacturer's protocol with a Nucleofector I device using program A-24. Cells were subsequently plated in triplicate at a density of 2 × 105 cells per cm2 onto poly-d-lysine- (Sigma) and laminin- (BD Biosciences) coated black-walled 96-well plates (Corning). On day 12 of differentiation, the luciferase assay was performed using the Dual-Glo Luciferase kit (Promega) according to the manufacturer's protocol. The experiment was performed three times.
Quantitative real-time PCR
RNA extraction and quantitative PCR were performed as previously described (17). Total RNA was extracted from differentiated ES cells using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Approximately 500 ng of total RNA was reverse transcribed with SuperScript II reverse transcriptase with random primers (Invitrogen), and 10 ng RNA equivalent of cDNA was used for real-time PCR. Real-time PCR was carried out using Taqman Gene Expression Assays (Applied Biosystems, Foster City, CA, USA). Assay IDs are available upon request. Each sample was run in triplicate, with multiplex probes for Actb, on a Stratagene MX3005P real-time PCR machine (La Jolla, CA, USA). Relative levels of mRNA expression were calculated according to the CT method (31), normalized by comparison to Actb mRNA expression. The experiment was performed three times.
Co-IP
pcDNA 3.1-Tle1-V5 was TOPO cloned following PCR for Tle1 after reverse transcription of RNA extracted from the brain of a BL/6 mouse. AMAXA nucleofection (Lonza) was used to electroporate pCMV-ArxWT-IRES-GFP or pCMV-ArxE-IRES-GFP, with or without pcDNA3.1-Tle1-V5 into Neuro-2a cells. After 48 h, protein was harvested at 4°C for 10 min using lysis buffer [20 mm Tris, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100, Complete Mini Protease Inhibitor Tablet (Roche), Phosphatase Inhibitor Cocktail II (Sigma), pH 7.4]. The concentration of Triton X-100 in the lysate was diluted to 0.5%, and the lysate was pre-cleared by incubation with recombinant Protein G agarose beads (Invitrogen). After overnight incubation at 4°C with 1 μg of α-TLE antibody (Santa Cruz, sc-13373), protein G beads were added to lysate for 1 h at room temperature. The beads were washed six times with lysis buffer (0.1% Triton X-100). Twenty microliters of NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen) and 2 μl of NuPAGE antioxidant (Invitrogen) were added to the beads, which were heated at 80°C for 10 min before loading into a NuPAGE 10% Bis–Tris gel (Invitrogen). After running, the protein was transferred to Immobilon transfer membrane (Millipore), and immunoblotted with α-Arx (C-14) (Santa Cruz, sc-48843) and α-TLE.
Chromatin immunoprecipitation (ChIP)
ChIP was performed following instructions for the EZ ChIP Immunoprecipitation Kit (Millipore) after FuGENE HD co-transfection of Neuro2a cells with pcDNA3.1-Tle1-V5 and either pCMV-Arxwt-IRES-GFP, pCMV-ArxE-IRES-GFP or pCMV-IRES-GFP (empty vector) with 3 μg of α-V5 antibody (Invitrogen) per reaction. Non-qPCR was subsequently performed using primers as previously published (17,32).
Statistical analysis
Analyses were performed using R, SAS and Excel. Hypothesis tests were two-sided and used a type I error rate of 0.05. We adjusted for multiple comparisons using either a Bonferroni correction (in utero electroporation, luciferase and qPCR results) or a post hoc Tukey adjustment.
In utero electroporation
We fit a log linear model using the negative binomial distribution to account for possible overdispersion (see Supplementary Material, Fig. S1) (33,34). We tested the global hypothesis, whether the distribution of cells among treatment/genotype groups differed across bins using a likelihood ratio test comparing the full model to a reduced model containing only an effect of bins. Conditional on the primary test showing significance, the Wald test was used to determine where differences occurred.
Slice electroporation
A one-way analysis of variance, with a post hoc Tukey adjustment, was performed to determine whether frequencies of interneurons that migrated beyond the pallial–subpallial notch differed among types of mutants.
Luciferase and qPCR
T-tests were used to compare levels between specific groups.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health grants [NS46616 to J.A.G., HD26979 to J.A.G. (core director), NS64666 to M.P.N. and HD07516 (T32 to J. Raper) supported M.P.N.].
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ilya M. Nasrallah for the Arx constructs, Carl Fulp for the luciferase constructs, Rachel Mascareno for care of ES cells and Golden lab members for helpful discussion of the work.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Friocourt G., Parnavelas J.G. Mutations in ARX result in several defects involving GABAergic neurons. Front. Cell Neurosci. 2010;4:4. doi: 10.3389/fncel.2010.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienvenu T., Poirier K., Friocourt G., Bahi N., Beaumont D., Fauchereau F., Ben Jeema L., Zemni R., Vinet M.C., Francis F., et al. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum. Mol. Genet. 2002;11:981–991. doi: 10.1093/hmg/11.8.981. [DOI] [PubMed] [Google Scholar]
- 3.Guerrini R., Moro F., Kato M., Barkovich A.J., Shiihara T., McShane M.A., Hurst J., Loi M., Tohyama J., Norci V., et al. Expansion of the first PolyA tract of ARX causes infantile spasms and status dystonicus. Neurology. 2007;69:427–433. doi: 10.1212/01.wnl.0000266594.16202.c1. [DOI] [PubMed] [Google Scholar]
- 4.Bauer P.O., Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J. Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 5.Messaed C., Rouleau G.A. Molecular mechanisms underlying polyalanine diseases. Neurobiol. Dis. 2009;34:397–405. doi: 10.1016/j.nbd.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Albrecht A., Mundlos S. The other trinucleotide repeat: polyalanine expansion disorders. Curr. Opin. Genet. Dev. 2005;15:285–293. doi: 10.1016/j.gde.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Nasrallah I.M., Minarcik J.C., Golden J.A. A polyalanine tract expansion in Arx forms intranuclear inclusions and results in increased cell death. J. Cell Biol. 2004;167:411–416. doi: 10.1083/jcb.200408091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitamura K., Itou Y., Yanazawa M., Ohsawa M., Suzuki-Migishima R., Umeki Y., Hohjoh H., Yanagawa Y., Shinba T., Itoh M., et al. Three human ARX mutations cause the lissencephaly-like and mental retardation with epilepsy-like pleiotropic phenotypes in mice. Hum. Mol. Genet. 2009;18:3708–3724. doi: 10.1093/hmg/ddp318. [DOI] [PubMed] [Google Scholar]
- 9.Price M.G., Yoo J.W., Burgess D.L., Deng F., Hrachovy R.A., Frost J.D., Jr, Noebels J.L. A triplet repeat expansion genetic mouse model of infantile spasms syndrome, Arx(GCG)10 + 7, with interneuronopathy, spasms in infancy, persistent seizures, and adult cognitive and behavioral impairment. J. Neurosci. 2009;29:8752–8763. doi: 10.1523/JNEUROSCI.0915-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miura H., Yanazawa M., Kato K., Kitamura K. Expression of a novel aristaless related homeobox gene ‘Arx’ in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 1997;65:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 11.Cobos I., Broccoli V., Rubenstein J.L. The vertebrate ortholog of Aristaless is regulated by Dlx genes in the developing forebrain. J. Comp. Neurol. 2005;483:292–303. doi: 10.1002/cne.20405. [DOI] [PubMed] [Google Scholar]
- 12.Colombo E., Galli R., Cossu G., Gecz J., Broccoli V. Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev. Dyn. 2004;231:631–639. doi: 10.1002/dvdy.20164. [DOI] [PubMed] [Google Scholar]
- 13.Colasante G., Collombat P., Raimondi V., Bonanomi D., Ferrai C., Maira M., Yoshikawa K., Mansouri A., Valtorta F., Rubenstein J.L., et al. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J. Neurosci. 2008;28:10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh E., Fulp C., Gomez E., Nasrallah I., Minarcik J., Sudi J., Christian S.L., Mancini G., Labosky P., Dobyns W., et al. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain. 2009;132:1563–1576. doi: 10.1093/brain/awp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura K., Yanazawa M., Sugiyama N., Miura H., Iizuka-Kogo A., Kusaka M., Omichi K., Suzuki R., Kato-Fukui Y., Kamiirisa K., et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 16.Colombo E., Collombat P., Colasante G., Bianchi M., Long J., Mansouri A., Rubenstein J.L., Broccoli V. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J. Neurosci. 2007;27:4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulp C.T., Cho G., Marsh E.D., Nasrallah I.M., Labosky P.A., Golden J.A. Identification of Arx transcriptional targets in the developing basal forebrain. Hum. Mol. Genet. 2008;17:3740–3760. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friocourt G., Kanatani S., Tabata H., Yozu M., Takahashi T., Antypa M., Raguenes O., Chelly J., Ferec C., Nakajima K., et al. Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J. Neurosci. 2008;28:5794–5805. doi: 10.1523/JNEUROSCI.1067-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato M., Dobyns W.B. X-linked lissencephaly with abnormal genitalia as a tangential migration disorder causing intractable epilepsy: proposal for a new term. “interneuronopathy”. J. Child. Neurol. 2005;20:392–397. doi: 10.1177/08830738050200042001. [DOI] [PubMed] [Google Scholar]
- 20.Stenman J., Toresson H., Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J. Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colasante G., Sessa A., Crispi S., Calogero R., Mansouri A., Collombat P., Broccoli V. Arx acts as a regional key selector gene in the ventral telencephalon mainly through its transcriptional repression activity. Dev. Biol. 2009;334:59–71. doi: 10.1016/j.ydbio.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie O., Ponte I., Mangelsdorf M., Finnis M., Colasante G., Shoubridge C., Stifani S., Gecz J., Broccoli V. Aristaless-related homeobox gene, the gene responsible for West syndrome and related disorders, is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience. 2007;146:236–247. doi: 10.1016/j.neuroscience.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 23.Seufert D.W., Prescott N.L., El-Hodiri H.M. Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev. Dyn. 2005;232:313–324. doi: 10.1002/dvdy.20234. [DOI] [PubMed] [Google Scholar]
- 24.Jennings B.H., Pickles L.M., Wainwright S.M., Roe S.M., Pearl L.H., Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Brown L.Y., Brown S.A. Alanine tracts: the expanding story of human illness and trinucleotide repeats. Trends Genet. 2004;20:51–58. doi: 10.1016/j.tig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Trochet D., Hong S.J., Lim J.K., Brunet J.F., Munnich A., Kim K.S., Lyonnet S., Goridis C., Amiel J. Molecular consequences of PHOX2B missense, frameshift and alanine expansion mutations leading to autonomic dysfunction. Hum. Mol. Genet. 2005;14:3697–3708. doi: 10.1093/hmg/ddi401. [DOI] [PubMed] [Google Scholar]
- 27.Maroof A.M., Brown K., Shi S.H., Studer L., Anderson S.A. Prospective isolation of cortical interneuron precursors from mouse embryonic stem cells. J. Neurosci. 2010;30:4667–4675. doi: 10.1523/JNEUROSCI.4255-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buscarlet M., Perin A., Laing A., Brickman J.M., Stifani S. Inhibition of cortical neuron differentiation by Groucho/TLE1 requires interaction with WRPW, but not Eh1, repressor peptides. J. Biol. Chem. 2008;283:24881–24888. doi: 10.1074/jbc.M800722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabata H., Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 30.Nasrallah I.M., McManus M.F., Pancoast M.M., Wynshaw-Boris A., Golden J.A. Analysis of non-radial interneuron migration dynamics and its disruption in Lis1± mice. J. Comp. Neurol. 2006;496:847–858. doi: 10.1002/cne.20966. [DOI] [PubMed] [Google Scholar]
- 31.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Buscarlet M., Hermann R., Lo R., Tang Y., Joachim K., Stifani S. Cofactor-activated phosphorylation is required for inhibition of cortical neuron differentiation by Groucho/TLE1. PLoS ONE. 2009;4:e8107. doi: 10.1371/journal.pone.0008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venables W.N., Ripley B.D., Hazel M. Hussong Fund. Modern Applied Statistics with S. New York: Springer; 2002. [Google Scholar]
- 34.Team R.D.C. R Foundation for Statistical Computing. Austria: Vienna; 2008. [Google Scholar]
- 35.Jung Y., Mikata Y., Lippard S.J. Kinetic studies of the TATA-binding protein interaction with cisplatin-modified DNA. J. Biol. Chem. 2001;276:43589–43596. doi: 10.1074/jbc.M108299200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.