Abstract
The spinocerebellar ataxia type 7 (SCA7) gene product, Ataxin-7 (ATXN7), localizes to the nucleus and has been shown to function as a component of the TATA-binding protein-free TAF-containing-SPT3-TAF9-GCN5-acetyltransferase transcription complex, although cytoplasmic localization of ATXN7 in affected neurons of human SCA7 patients has also been detected. Here, we define a physiological function for cytoplasmic ATXN7. Live imaging reveals that the intracellular distribution of ATXN7 dynamically changes and that ATXN7 distribution frequently shifts from the nucleus to the cytoplasm. Immunocytochemistry and immunoprecipitation demonstrate that cytoplasmic ATXN7 associates with microtubules (MTs), and expression of ATXN7 stabilizes MTs against nocodazole treatment, while ATXN7 knockdown enhances MT degradation. Interestingly, normal and mutant ATXN7 similarly associate with and equally stabilize MTs. Taken together, these findings provide a novel physiological function of ATXN7 in the regulation of cytoskeletal dynamics, and suggest that abnormal cytoskeletal regulation may contribute to SCA7 disease pathology.
INTRODUCTION
Spinocerebellar ataxia type 7 (SCA7) is one of nine polyglutamine (polyQ) diseases, a group of inherited neurodegenerative disorders including Huntington's disease, spinal and bulbar muscular atrophy/Kennedy's disease, spinocerebellar ataxia types 1, 2, 3, 6, 7 and 17, and dentato-rubro-pallido-luysian atrophy (1,2). Cerebellar ataxia and progressive retinal degeneration attribute to neuronal loss of Purkinje cells in the cerebellum and drop-out of cone-photoreceptor cells in the retina are characteristic symptoms of SCA7 (3). The polyQ tract sequence is located at the N-terminal region of the causative gene product, ATXN7 protein. Normal ATXN7 contains 4–35 cytosine-adenine-guanine repeats, whereas pathogenic variants show an expansion of 36–306 repeats (3).
ATXN7 is highly homologous to the yeast protein, Sgf73, which acts as a subunit of the SAGA chromatin remodelling and transcription complex (4,5). SAGA complex includes Spt, Ada and Gcn5 acetylase, and acts as a histone acetyltransferase complex like cAMP response element binding protein-binding protein (CBP) (6). In human and mammals, SAGA complex has multiple homologues: TATA-binding protein-free TAF complex (TFTC) (7,8), SPT3/TAF9/GCN5 acetyltransferase complex (STAGA) (9) and PCAF/GCN5 complex (10). All these complexes contain PCAF or GCN5 that is homologous to yeast Gcn5 (6). These co-activator complexes are recruited by specific transcription factors to enhancer/promoter regions of a target gene, and induce unwinding of DNA from nucleosomes via histone acetylation to upregulate transcription of the target gene (11–13).
Nuclear localization of ATXN7 is essential for transcription function. However, partial or transient cytoplasmic localization of ATXN7 was reported in vivo and in vitro. First, cytoplasmic localization of ATXN7 was observed in patient and control brains (14–16). Second, ATXN7 possesses a nuclear export signal sequence (NES) in addition to nuclear localization sequence (NLS), and it shuttles between nucleus and cytoplasm (17), like another SCA protein (18). Besides the simple explanation that the transcriptional function can be switched off by the nuclear export, these findings suggest that ATXN7 may perform certain cytoplasmic functions.
Here, we report that cytoplasmic ATXN7 has MT-associating and -stabilizing properties. Association with MTs was observed for both normal and mutant ATXN7, just as previous work found that both normal and mutant ATXN7 can regulate transcription (19). As altered ATXN7 function in the SCA7 brain is likely important for disease pathogenesis, our discovery of a novel cytoplasmic function for ATXN7 may provide another mechanism for SCA7 disease pathology.
RESULTS
ATXN7 distribution dynamically changes in nucleus and cytoplasm
It is essential to use cell lines expressing ATXN7 endogenously to address the physiological function of ATXN7. We thus selected HeLa cells, as HeLa cells are known to express ATXN7 endogenously (19). Localization of normal and mutant ATXN7 was analysed by expressing fusion proteins of DsRED and normal (10Q) or mutant (92Q) full-length ATXN7 in HeLa cells. As reported previously (17), live-imaging analysis with transfected HeLa cells revealed that ATXN7 protein exists both in the nucleus and in the cytoplasm, and its expression levels and nuclear to cytoplasmic ratio change dynamically during the cell cycle or morphological transformation (Supplementary Material, Fig. S1 and Movies). To our surprise, the intracytoplasmic distribution of ATXN7 also changed. We noticed that both normal (10Q) and mutant (92Q) ATXN7 transiently showed filamentous structures in the cytoplasm, which is reminiscent of the MT network (Supplementary Material, Fig. S1 and Movies).
Cytoplasmic distribution of endogenous ATXN7 during cell cycle
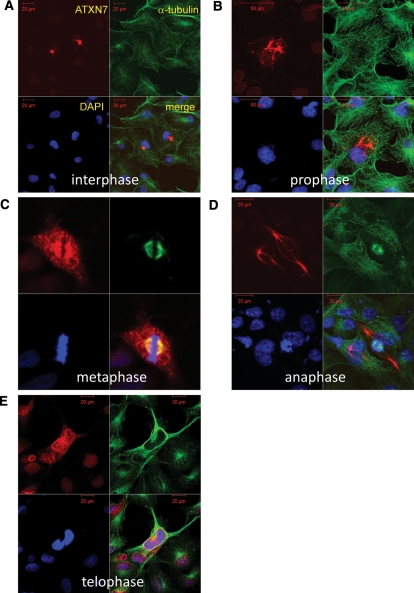
To further address ATXN7 cytoplasmic distribution and possible co-localization with MTs, we performed immunocytochemistry of non-transfected HeLa cells (Fig. 1). The specific antibody revealed various types of ATXN7 distribution that could be aligned in an order suggested by the information from live imaging. In a large proportion of cells, ATXN7 showed predominant nuclear distribution, and decreased cytoplasmic staining (Supplementary Material, Fig. S1). However, before going into M phase, HeLa cells at late G2 phase showed an increase in the concentration of cytoplasmic ATXN7 protein at the perinuclear region. The perinuclear foci did not merge initially with centrosomes in most cases (Fig. 1). However, when mitosis started, ATXN7 began to merge with α-tubulin, overlapped with the spindle, but distributed in a wider area (Fig. 1). When mitosis ended, ATXN7 separated from the spindle (Fig. 1), and redistributed to the nucleus (Fig. 1). However, even after mitosis, a small amount of ATXN7 remained in the cytoplasm at the perinuclear region and displayed fibrillar patterns in the cytoplasm (Fig. 1). During mitosis, ATXN7 did not stably overlap with actin (data not shown).
Figure 1.
Ataxin-7 subcellular distribution changes dramatically during the cell cycle. Here, we see representative images in HeLa cells immunostained for ataxin-7 (red), α-tubulin (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). The subcellular localization of ataxin-7 (ATXN7) is presented for the different progressive stages of mitosis. (A) Most cells displayed the typical nuclear localization during interphase, though occasional cells showed perinuclear ATXN7 foci, reminiscent of centrosomes. The perinuclear foci of ATXN7 did not merge with α-tubulin in most cells. (B) Perinuclear ATXN7 foci become very prominent at prophase. (C) ATXN7 and α-tubulin co-localize at the spindle during mitosis, although ATXN7 is much more broadly distributed in comparison with α-tubulin. (D) ATXN7 localization shifts to the nucleus, and in particular to chromatin-containing regions during anaphase. (E) By telophase, ATXN7 appears mostly nuclear, though some ATXN7 staining persists in the cytoplasm after mitosis. Full video images are shown in Supplementary Material, Movies.
ATXN7 associates with MTs
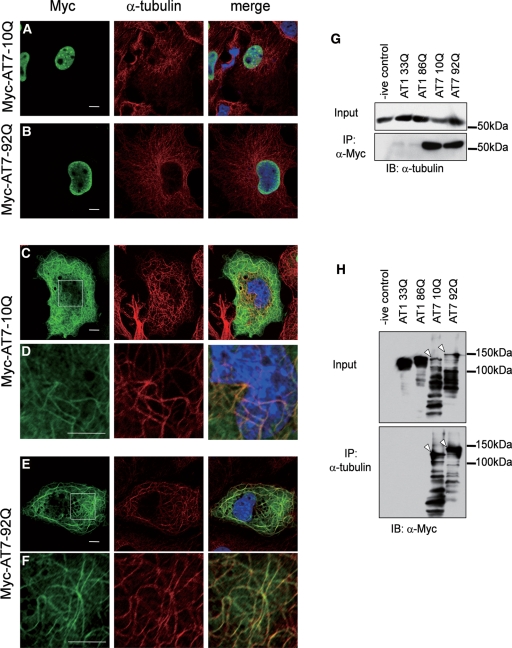
To directly test interaction between ATXN7 and MTs, myc-ATXN7-10Q or 92Q transfected cells were double-immunostained with anti-Myc and anti-α-tubulin antibodies. The transfected myc-ATXN7 enabled us to analyse the low level of cytoplasmic ATXN7 and revealed co-localization of the two proteins in a filamentous reticular structure after mitosis (Fig. 2A). Staining patterns of neighbouring cells clearly excluded detection of α-tubulin signals through the green filter. Interestingly, both normal and abnormal ATXN7 showed similar reticular patterns in the cytoplasm (Fig. 2A). As observed in live imaging, a fraction of transfected HeLa cells showed nuclear-restricted localization of ATXN7 (Fig. 2A).
Figure 2.
Both normal ATXN7 and mutant ATXN7 associate with MTs. (A–F) To analyse the cytoplasmic distribution of ATXN7 at higher resolution, HeLa cells were transfected with normal (10Q) or mutant (92Q) full-length ATXN7 with a Myc-tag, and double-labelled with anti-Myc (green) and anti-α-tubulin (red) antibodies. In addition to nuclear localization (A, B), normal ATXN7 (C, D) and mutant ATXN7 (E, F) showed a reticular pattern merged with α-tubulin in a part of cells. The white rectangles in panels (C) and (E) are shown enlarged in panels (D) and (F). Scale bars represent 10 μm. (G–H) Protein lysates from whole-cell extracts of transfected HeLa cells separated by SDS–PAGE, subjected to immunoprecipitation with anti-Myc (G) or anti-α-tubulin (H) antibodies, and then immunoblotted with the indicated antibodies. Lane 1 is empty vector control. Lanes 2 and 3 are cells transfected with normal (33Q) or mutant (86Q) ataxin1 vectors as negative controls. Lanes 4 and 5 are normal (10Q) or mutant (92Q) ATXN7-transfected cells. Note that both immunoprecipation of ataxin-7 or of tubulin demonstrate an interaction, and this is seen for both normal and mutant ataxin-7 (arrows, H).
To examine physical interaction between ATXN7 and α-tubulin, cell lysates from the transfected HeLa cells were subjected to immunoprecipitation with anti-Myc antibody. Ataxin-1 with Myc-tag (Myc-ATXN1) was also used to test the specificity of interaction between ATXN7 and α-tubulin. As expected, Myc-ATXN7—but not Myc-ATXN1—co-precipitated α-tubulin (Fig. 2B). Conversely, immunoprecipitation by anti-α-tubulin antibody co-precipitated Myc-ATXN7, but not Myc-ATXN1 (Fig. 2C). These results supported a specific association between ATXN7 and α-tubulin/MTs in HeLa cells. No remarkable difference was observed in the amount of co-precipitated α-tubulin between normal (10Q) and mutant (92Q) ATXN7. In order to exclude non-specific interaction of anti-Myc antibody to MTs, 1C2 antibody, which detects expanded polyQ tracts, was also used for immunocytochemistry. Filamentous structures derived from mutant ATXN7 were still observed (data not shown).
Defining the ATXN7 domain necessary for MT interaction
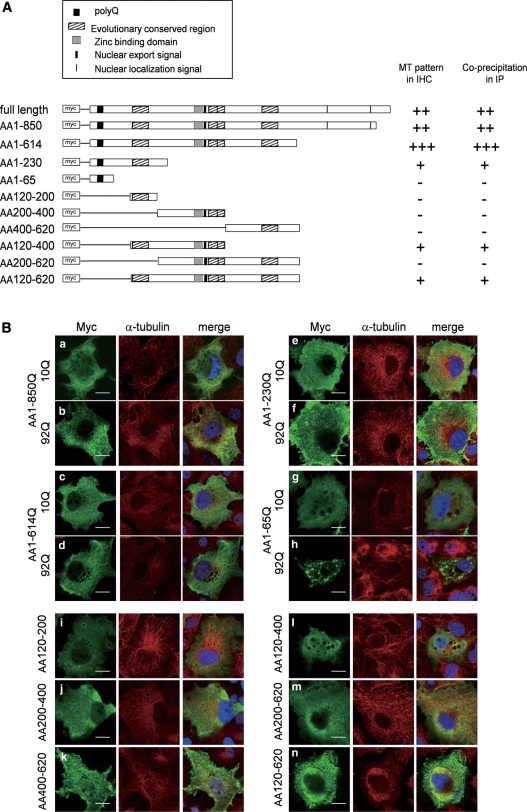
To determine the ATXN7 domain responsible for association with MTs, a series of deletion constructs was generated (Fig. 3A), and tested for their localization and binding to α-tubulin. In immunocytochemistry with anti-Myc and anti-α-tubulin antibodies, amino acids 1–850 and 1–614 showed the typical filamentous pattern (Fig. 3B). Amino acids 120–400 and 120–620 were also frequently distributed in the filamentous pattern. The filamentous pattern was sometimes seen with the amino acids 1–230 construct, but not found in other deletion constructs (Fig. 3B).
Figure 3.
ATXN7 domain required for co-localization with MTs. (A) Schematic presentation of full-length ATXN7 and the ATXN7 deletion constructs. PolyQ regions are indicated by a black box. Highly conserved regions, the zinc-binding domain, NLS and NES are also indicated. All constructs were Myc-tagged. The results of the co-localization studies (B) and immunoprecipitations (Fig. 4) are summarized here. (B) HeLa cells were transfected with the indicated ATXN7 deletion constructs, and were double-labelled with anti-Myc (green) and anti-α-tubulin (red) antibodies 2 days after transfection. Scale bars represent 20 μm.
Immunoprecipitation results supported interaction of amino acids 1–850 and 1–614 with α-tubulin, and amino acids 120–400 and 1–230 constructs also showed association with α-tubulin (Fig. 4A and B). Again, a weak association of amino acids 1–230 with α-tubulin was observed. These results consistently suggested that the amino acid residues from 120 to 400 mediate association of ATXN7 with α-tubulin, and amino acids 120–230 appear to be most important within this region. However, the strongest interaction was observed with the full-length ATXN7, suggesting that ATXN7 uses multiple regions for association with α-tubulin. A short fragment (amino acids 1–65) including polyQ tract sequence was prone to aggregate (Fig. 4B), whereas it did not interact with α-tubulin. The amino acids 1–65 fragment also showed a number of small dots in immunocytochemistry (Fig. 3B), while it did not form a single large inclusion body corresponding to the aggresome/centrosome.
Figure 4.
ATXN7 domains required for interaction with α-tubulin. (A, B) Protein lysates from whole-cell extracts of HeLa cells transfected with each deletion construct were separated by SDS–PAGE, subjected to immunoprecipitation with anti-Myc or anti-α-tubulin antibodies, and then immunoblotted with the indicated antibodies. Transfected ataxin-7 deletion constructs are indicated above each lane. Arrows in (B) indicate bands corresponding to the deletion construct protein product detected in the immunoprecipitation experiment, and the asterisk marks a non-specific band that does not correspond to the amino acids Δ400–620 construct.
Interactome databases including STRING at EMBL (URL: http://string.embl.de/) or published results from the Zoghbi group (20) do not show α-tubulin as an interacting partner of ATXN7, which might suggest that the interaction between ATXN7 and α-tubulin is indirect. On the other hand, although ATXN7 does not include previously known motif sequences, MTs’ binding motifs identified recently are rich in Glycine, Serine or Valine (21)—amino acids that are highly represented within the evolutionarily conserved domains of ATXN7. Furthermore, informatics analysis by MOTIF Search (URL: http://www.genome.jp/tools/motif/) identified a tau signature (22) at amino acids 429–437 (mouse ATXN7) and at amino acids 450–458 (human ATXN7), as well as a homologous sequence to Doublecortin, an MT-associated protein that stabilizes MTs (23,24) at amino acids 687–715 (mouse ATXN7). These observations support direct binding of ATXN7 to MTs. When we compared the binding affinity of α-tubulin to normal (10Q) and mutant (92Q) ATXN7, we did not observe any obvious differences (Fig. 4A and B) indicating that the interaction does not depend on the polyQ domain or its repeat length.
ATXN7 contributes to MT stabilization
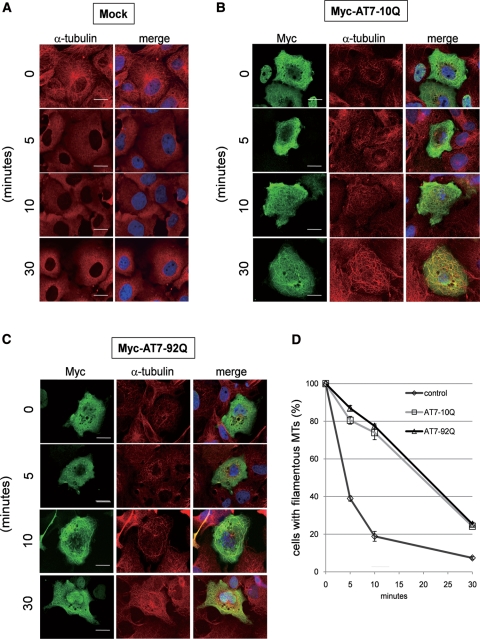
Normally, MTs radiate from their formation centre (25). However, as shown in Figure 2C–F, some of the ATXN7-transfected cells exhibited thicker, meandering MTs rather than the normal radiating form. This is known to represent an over-stabilized form of MTs (26), leading us to hypothesize that this result may arise from an MT-stabilizing effect of ATXN7. To test whether ATXN7 stabilizes MTs, we treated cells with nocodazole, a well-known inhibitor of MT polymerization (27) (Fig. 5A). Normal (10Q) or mutant (92Q) ATXN7-transfected cells were treated with 10 nm nocodazole for 5, 10 or 30 min (Fig. 5B and C). Even after 30 min treatment, the MT distribution was maintained filamentous by co-expression of ATXN7, while the filamentous pattern of MTs rapidly disappeared in untransfected cells (Fig. 5A). The stabilization effects were similar in cells expressing normal (10Q) and mutant (92Q) ATXN7 (Fig. 4D). We also tried to monitor MT stabilization by ATXN7 with live-cell imaging, whereas the signal of newly generated α-tubulin-EGFP was homogenous rather than filamentous in the cytoplasm and not suitable for the analysis.
Figure 5.
ATXN7 expression levels regulate MT stability. HeLa cells were transfected with ataxin-7 (AT7)-10Q/92Q-Myc or mock, and treated with 10 nm nocodazole for the indicated time. After treatment, cells were fixed and immunostained with anti-Myc (green) antibody, anti-α-tubulin (red) antibody and DAPI. Cells were analysed by confocal microscopy ((A) untransfected control, (B) normal (10Q) ATXN7-transfected, (C) mutant (92Q) ATXN7-transfected), and the percentage of filamentous MT-positive cells in ATXN7-transfected cells (n > 100) was scored at the indicated times after nocodazole treatment (D). Scale bars represent 20 μm.
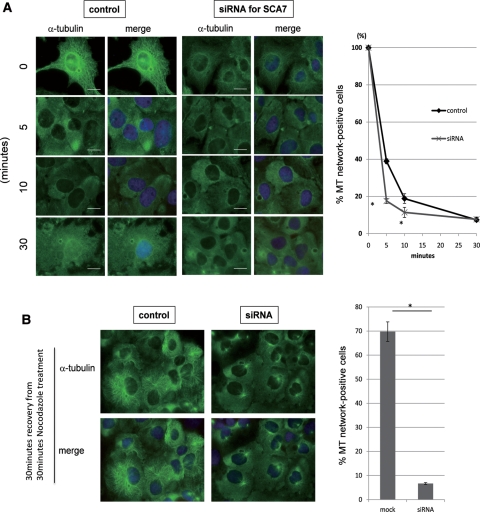
Furthermore, to test the effect of ATXN7 on MTs stabilization, we knocked down ATXN7 with a validated small-interfering RNA (siRNA; Fig. 6A). The filamentous MT network disappeared faster in siRNA-transfected cells in the presence of nocodazole (Fig. 6B). The recovery of the MT network after removal of nocodazole was also delayed. These findings support the conclusion that ATXN7 stabilizes MTs. Decreased expression of ATXN7 in siRNA-transfected cells was confirmed by western blot (Supplementary Material, Fig. S2).
Figure 6.
ATXN7 knockdown destabilizes MT networks. (A) Endogenous ATXN7 was knocked down by siRNA transfection and the MT network was examined by α-tubulin immunostaining. Ataxin-7 siRNA-transfected HeLa cells treated with nocodazole displayed a more rapid destruction of MT networks in the cytoplasm by nocodazole treatment at 5 and 10 min after treatment (P < 0.05, t-test). (B) Effect of ATXN7 knockdown on the recovery of MT network. HeLa cells subjected to ataxin-7 knockdown failed to recover their MT networks at 30 min after removal of nocodazole (P < 0.05, t-test).
DISCUSSION
In this study, we report dramatic co-localization of ATXN7 and MTs, biochemical interaction between ATXN7 and α-tubulin, and a functional role for ATXN7 in MT stabilization. Based upon live imaging of Myc-ATXN7 and immunocytochemistry of endogenous ATXN7, the extent of ATXN7 protein interaction with MTs dynamically changed in a cell cycle-dependent manner. However, after mitosis, ATXN7 persisted in the cytoplasm at a low level. The results from immunoprecipitation and co-localization analysis with deletion constructs suggested that a large region (amino acids 120–400) of ATXN7 participates in the MT interaction. The polyQ sequence in ATXN7 is therefore not essential for the interaction with MTs, and repeat length does not influence the interaction. Increasing the level of ATXN7 stabilized the MT network, while knockdown of ATXN7 destabilized the MT network. Taken together, these results reveal a new physiological function for ATXN7 in the cytosol.
A major concern for our observation is that over-expressed ATXN7 might be carried on the rail of MTs to aggresomes to form inclusion bodies as part of the cell's effort to sequester a misfolded, polyQ-rich protein (28). However, dynamic changes of ATXN7 in the nucleus and cytoplasm indicated that the association with MTs is neither aggregation nor protein degradation-related accumulation. The cytoplasmic distribution was also highly variable, assuming various patterns, suggesting that the association is not to promote protein degradation or sequestration. Moreover, immunocytochemistry of endogenous ATXN7 showed a similar reticular pattern, excluding the possibility that the cytoplasmic distribution of ATXN7 was an artefact of over-expression.
Interactome studies of ATXN7 based on the yeast two-hybrid technique revealed that ATXN7 and the macular degeneration disease proteins EFEMP1 and FBLN5 share a molecular network sub-domain within the protein–protein interaction network (20). However, these interactome databases do not include α- or β-tubulin in the ATXN7 protein-interaction network. This is also the case with the String database at EMBL. As the formation of reticular patterns in the cytoplasm by ATXN7 depends upon cell cycle stage and is therefore transient, the interaction between ATXN7 and α-tubulin may not be detectable with a transcription-based interaction assay in yeast cells. Since the deletion analysis of ATXN7 did not narrow down the interacting region to a short motif, it is also possible that ATXN7 might utilize multiple co-factors for an indirect association with MTs. In addition to not predicting an interaction between ATXN7 and MTs, the ATXN7 interactome database lists several forms of actin as candidate interactors (20). Despite this prediction, our live imaging of HeLa cells expressing actin-GFP and ATXN7-DsRed did not yield any evidence for an actin–ATXN7 interaction. The actin distribution at the cell periphery did not merge with the ATXN7 meshwork (Supplementary Material, Fig. S3A), and immunoprecipitation did not support strong or stable interaction between ATXN7 and β-actin (Supplementary Material, Fig. S3B). Cytochalasin B treatment for 30 min disrupted actin network but did not affect ATXN7 meshwork in HeLa cells (Supplementary Material, Fig. S4).
Nevertheless, actin may still mediate interaction between MT and ATXN7 in neurons. Actin fibres and MTs form independent cytoskeletal networks of different diameters and different distribution in cells. The two networks engage in crosstalk during diverse cellular processes including cell motility, neuronal path finding, cellular wound healing, cell division and cortical flow (29). This dynamic, spatiotemporal crosstalk is especially critical for neurons in growth cone formation and neurite branching. Actually, our preliminary immunohistochemistry showed colocalization of ATXN7 and α-tubulin in Purkinje cell dendrites (Supplementary Material, Figs S5 and S6) and merging of ATXN7 and β-actin signals in neuropil of molecular layer (Supplementary Material, Fig. S7). However, the molecular basis of the interaction between actin fibres and MT networks is not fully understood, except for some cases including ShortStop (Shot), the Drosophila spectraplakin protein that mediates interaction between these two cytoskeletal networks (30). Moreover, the distribution of ATXN7 observed in HeLa cells was clearly distinct from that of actin fibres, arguing against interactions between actin and ATXN7 in the context of MTs. Collectively, the role of actin in ATXN7–MT association needs further investigation.
Stability of MTs is crucial for neurons to maintain the cell body architecture and consistent neurite structures. PolyQ-expanded ATXN7 has been shown to be present in the nucleus and there it interferes with the function of the STAGA HAT complex, leading to transcriptional dysregulation of photoreceptor genes in the retina (19,31,32). Although ATXN7 has a well-described function in the nucleus, it is clear that proteins can perform multiple functions in various cellular compartments. Indeed, the polyQ disease protein huntingtin has been shown to play a role in axonal transport and vesicle-mediated transport in endocytosis (33–35), while at the same time, upon proteolytic cleavage, an N-terminal fragment of the huntingtin protein has been shown to be a transcription factor in the TFIID and TFIIF co-activator complexes (36). Hence, a nuclear function for ATXN7 does not exclude a possible independent role for it in the cytosol. If ATXN7 is interacting with MTs in the cytosol, an important question is whether this interaction might be altered upon polyQ expansion in SCA7 disease. Our results indicate that glutamine tract length does not affect ATXN7 interaction with MTs or its ability to modulate the MT network. Nonetheless, polyQ-expanded ATXN7 in the context of neurons could engage in inappropriate or altered interactions with co-factors whose dysregulation might affect intracellular protein trafficking, organelle delivery or axonal transport pathways. It thus remains to be seen if mutant ATXN7 might reduce the accessibility of normal ATXN7 or some other factor to MTs, and might impair ATXN7 functions in stabilization of MTs and/or cross-talk between MT and actin networks. Another aspect of ATXN7 biology to consider is that ATXN7 is cleaved at amino acids 256 and 344 by caspase-7 (37). Our western blot analysis of Myc-ATXN7-10Q or -92Q expressing cells with a specific anti-Myc antibody detected multiple bands of ATXN7 (Fig. 2C), suggesting that ATXN7 is cleaved at multiple sites. Importantly, the short polyQ-containing fragments of mutant ATXN7 are prone to aggregate and to form cytoplasmic accumulations. Therefore, it is possible that such short polyQ-containing fragments aggregate and sequester full-length ATXN7. The nuclear aggregation of ATXN7 also occurs, as reported in human brains. Such aggregates in the nucleus may reduce ATXN7 in the cytoplasm and impair the cytoplasmic function of ATXN7, finally leading to instability of MTs (Supplementary Material, Figs S8 and S9). Although further studies are required to test this hypothesis, our results provide an entirely novel perspective on the physiological function of ATXN7 in the cytoplasm, and could contribute to an enhanced understanding of SCA7 disease pathology with implications for therapy development.
MATERIALS AND METHODS
Plasmid construction
The pCMV-myc-ataxin7-10Q and 92Q plasmids were generated as described previously (38). Human ATXN7-10Q and 92Q cDNA were amplified by PCR using 5′-TCATAAGCTTCGATGTCGGAGCGGGCCGCG-3′ (AT7-HindIII-F) 5′-TCATGTCGACTCAGGGACGTGCCTTTGGCT-3′ (ATXN7-SalI-R) primers from pCMV-myc-ATXN7-10Q and 92Q (34), respectively. The PCR products were digested by HindIII and cloned into HindIII and SmaI sites of pDsRed-Monomer-C1 (Clontech Laboratories, Inc.,) (hATXN7-10Q and 92Q/pDsRed-Monomer-C1).
Polyglutamine-containing deletion constructs were generated by taking advantage of conveniently placed restriction sites in the multiple cloning sites of vector plasmid and restriction site inside ATXN7 sequence; XhoI for amino acids 1–850, KpnI for amino acids 1–614, PstI for amino acids 1–230 and ApaI for amino acids 1–65. For other constructs without polyQ, the target range of the original plasmid was amplified by PCR from pCMV-myc-ataxin7-10Q (34) and replaced into the myc-tagged original AT7 vector using EcoRI and XhoI sites. Primer sequences used for PCR amplification of deleted cDNA were 120F: tgccgaattcaaaaaccgcgaatg, 200F: tgccgaattcagtggaagcaaccg, 400F: tgccgaattccatccggactctca, 200R: tgggctcgagtgcactgcctcct, 400R: atgtctcgaggcgaatcaattcct and 620R: atgtctcgagagctcctaccgatt.
Cell culture and transfection
HeLa cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Sigma) with 5% fetal bovine serum (FBS; Invitrogen) under 5% CO2, at 37°C. Transfection was performed using Lipofectamine 2000 (Invitrogen). For transfection, all reagents were scaled down to one-half of the recommended volume to avoid their toxicity.
Immunocytochemistry
HeLa cells transfected with indicated plasmids were incubated for 36 h. After transfection, cells were fixed with 2% paraformaldehyde (PFA), permeabilized with 0.1% Triton X-100 and blocked with 5% non-fat dry milk. Cells were incubated overnight at 4°C with primary antibody, followed by incubation with secondary antibodies for 1 h at room temperature and with 4′,6-diamidino-2-phenylindole (DAPI) (1:3000 Vector Laboratories). Cells were observed under the confocal microscope (Zeiss LSM 510) after mounted by Fluoromount (Diagnostic biosystem). Primary and secondary antibodies were used as follows: Myc (Bethyl Laboratories, goat, 1:1000), α-tubulin (Sigma, mouse, 1:1000), Alexa Fluor488 (Invitrogen, goat, 1:1000) and Cy3 (Jackson, mouse, 1:1000).
Western blot
Anti-Myc (Sigma, 9E10) and anti-α-tubulin (Sigma) primary antibodies were used at 1:1000. Horseradishperoxidase-conjugated anti-mouse IgG (Amersham) were used at 1:3000. ECL (GE Healthcare) was used to detect bands following the commercial protocol.
Immunoprecipitation
HeLa cells plated in six-well plate were transfected by Lipofectamin 2000 (Invitrogen) and incubated for 36 h. The cells were lysed in 250 μl tris-NaCl-EDTA buffer (50 mm Tris–HCl, pH 7.8, 150 mm NaCl, 5 mm ethylene diaminetetraacetic acid, 0.25% Nonidet P-40 was added). After removing debris by centrifugation, cell lysates were incubated for 2 h at 4°C with 20 μl of protein G-agarose beads (GE Healthcare, 50% v/v) to remove non-specific binding proteins. After centrifugation, the supernatant was incubated with 1 μg of anti-Myc (Sigma, 9E10) or anti-α-tubulin antibody overnight at 4°C, followed by incubation with agarose beads for 2 h at 4°C. The beads were precipitated and washed five times with TNE buffer. Bound proteins were resolved by sodiumdodecyl sulphate–polyacrylamide gel electrophoresis, followed by western blotting with the indicated antibodies.
Nocodazole treatment and MT-stabilization assay
This assay was performed basically following the method by Hergovich et al. (39). In brief, HeLa cells plated on glass cover slips were transfected with myc-AT7-10Q/92Q and incubated for 36 h. Then, nocodazole (Sigma–Aldrich) was solved in dimethylsulphoxide and added to the culture medium (final concentration 10 nm). HeLa cells were cultured additionally for the indicated time at 37°C and stained as described above. Cells possessing MT network were counted three times for each condition.
siRNA knockdown of endogenous ATXN7
Pre-designated siRNA for human SCA7 was purchased from QIAGEN (catalogue number 1027416). From four different siRNAs, Hs-ATXN7-1 (SI00308273) was selected for our experiments. The sense sequences of Hs-ATXN7-1 and control siRNA (Qiagen) were 5′-CACCCGGTCTTTGACATGCAA-3′ and 5′-UUCUCCGAACGUGUCACGUdTdT-3′, respectively. The siRNAs were transfected into HeLa cells using Lipofectamine2000 (Invitrogen) according to the commercial protocol with minor modification. The transfected cells were incubated at 37°C for 48 h. The efficiency of gene knockdown was evaluated by western blot analysis. Nocodazole treatment and MT-stabilization assay have been done 48 h after transfection. After the stabilization assay, recovery of MTs was also examined by immunocytochemistry with anti-α-tubulin at 30 min after removal of nocodazole.
Immunohistochemistry of normal young mice
Fresh brains of C57BL/6 mice at four weeks were fixed in phosphate-buffered 4% PFA and embedded in paraffin. Sections (5 or 10 μm-thick) were dewaxed in xylene and rehydrated using a descending ethanol series. Sections were boiled in 0.01 m citrate buffer, pH 6.0, in a microwave oven three times and kept at room temperature for 30 min after final boil. Incubation in PBS with 1% bovine serum albumin (BSA) and 0.01% (v/v) Triton X-100 was performed for 30 min in order to block non-specific binding. Sections were incubated overnight at 4°C with primary antibody, followed by incubation with secondary antibodies for 1–2 h at room temperature and with DAPI (1:3000 Vector Laboratories). Finally, sections were mounted by Fluoromount (Diagnostic biosystem) and observed under the confocal microscope (Zeiss LSM 510). Primary and secondary antibodies were used as follows: AT7 (Thermo scientific, rabbit, 1:100), α-tubulin (Sigma, mouse, 1:200), β-actin (mouse, 1:200), Alexa Fluor488 (Invitrogen, mouse, 1:500) and Cy3 (Jackson, rabbit, 1:500).
Immunohistochemistry of ATXN7 transgenic mice
Generation of human ATXN7-92Q transgenic mouse was reported previously (40). The 33-week transgenic and littermate mice are perfused transcardially with 30 ml of 0.4 M phosphate buffer (PB) then 4% PFA in 0.4 m PB, pH 7.4. After perfused, brains were fixed in 4% PFA overnight, then transferred to 30% sucrose in 0.4 m PB for 48 h or until the brains became saturated in sucrose. The brain tissues were frozen and 30 µm sections were made by cryostat.
Immunohistochemistry was performed on free-floating sections after blocking in 1% BSA and 0.1% Triton X-100 in 0.24% glycerin-containing PB. Sections were then incubated overnight at 4°C with primary antibody, followed by incubation with secondary antibodies for 1–2 h at room temperature and with DAPI (1:3000 Vector Laboratories). Finally, sections were mounted by Fluoromount (Diagnostic biosystem) and observed under the confocal microscope (Zeiss LSM 510). Primary and secondary antibodies were used as follows: AT7 (Thermo scientific, rabbit, 1:100), α-tubulin (Sigma, mouse, 1:200), Alexa Fluor488 (Invitrogen, mouse, 1:500) and Cy3 (Jackson, rabbit, 1:500).
Lentiviral vector: DsRED-ATXN7
Human ATXN7-10Q and 92Q cDNA were amplified by PCR using 5′-TCATAAGCTTCGATGTCGGAGCGGGCCGCG-3′ (AT7-HindIII-F) 5′-TCATGTCGACTCAGGGACGTGCCTTTGGCT-3′ (ATXN7-SalI-R) primers from pCMV-myc-ATXN7-10Q and 92Q (38), respectively. The PCR products were digested by HindIII and cloned into HindIII and SmaI sites of pDsRed-Monomer-C1 (Clontech Laboratories, Inc.) (hATXN7-10Q and 92Q/pDsRed-Monomer-C1). DeRed-hATXN7 10Q and 92Q cDNAs were amplified by PCR using 5′-CATGAATTCCACCATGGACAACACCGAGGACGTC-3′ (DsRed-EcoRI-F) and ATXN7-SalI-R primers from hATXN7-10Q and 92Q/pDsRed-Monomer-C1, respectively. The PCR products were digested with EcoRI and SalI and cloned into EcoRI and XhoI sites of pLVSIN-CMV-Pur (Takara Bio Inc.). pLVSIN-CMV-Pur was a self-inactivating-type lentiviral vector plasmid. These constructs were named DsRedm-hATXN7-10Q and 92Q/pLVSIN-CMV, respectively. The lentiviral-expression plasmids with helper plasmids, ViraPower Packaging Mix containing pPLP1, pPLP2 and pLP/VSVG (Technologies Japan Ltd.) were co-transfected into HEK293T cells by calcium-phosphate method. The media containing lentiviral vectors were collected at 40 h after transfection and concentrated by centrifugation at ×8000g at 4°C for 16 h. Pellets containing viral particles were further concentrated using Lenti-X Concentrator (Clontech Laboratories Inc.), divided into aliquots and kept at −80°C until use. The titres were measured using Lenti-X qRT-PCR Titration Kit (Clontech Laboratories Inc.). Each titre was 5 × 107 for Lenti-Dsred-AT7-10Q, and 2.4 × 107 for Lenti-Dsred-AT7-92Q.
Rat brain primary culture and infection of lentivirus
Briefly, cerebral cortexes from 18-day-old Wistar rat embryos (Japan SLC, Inc.,) were minced into fine pieces, rinsed with PBS and incubated with 0.05% trypsin at 37°C for 10–15 min and with DNase final concentration of 25 μg/ml and incubated for another 5 min at 37°C. The dissociated cells were washed twice with DMEM (Gibco) containing 50% FBS, 25 mm d-glucose, 4 mm l-glutamine and 25 µg/ml gentamycin. Cells were centrifuged at 100g for 1 min, resuspended in 5 ml 10% FBS/DMEM, gently triturated with blue tips and filtered through a nylon mesh (Falcon 2350; BD) in order to remove any debris. The isolated cells were centrifuged at 1000 rpm again for 5 min and collected as pellet. Finally, cells were resuspended in Neurobasal medium and plated 6 × 105cells/well in the six-well plate with the poly-l-lysine-coated glass slips (Sigma–Aldrich). Neurons were incubated in 5% CO2 at 37°C. Twenty-four hours after plating, viruses were added at multiplicity of infection of 7. Seventy-two hours after plating, arabinosylcytosine (Sigma–Aldrich) was added to the culture medium (4 μm) to prevent unnecessary growing of glial cells. Five days after plating, cells were washed several times using PBS and fixed using −20°C methanol for immunocytochemical analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement: None declared.
FUNDING
This work was supported by Strategic Research Program for Brain Sciences (SRPBS) and Grant-in-Aid for Scientific Research on Innovative Areas (Foundation of Synapse and Neurocircuit Pathology) from Ministry of Education, Culture, Sports, Science and Technology of Japan, CREST from Japan Science Technology Agency and Grant-in Aid from Research Committee for Ataxic Disease from Ministry of Health, Labour and Welfare of Japan to H.O., and by NIH grant R01 EY14061 to A.R.L.
Supplementary Material
REFERENCES
- 1.Zoghbi H.Y., Orr H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Taylor J.P., Hardy J., Fischbeck K.H. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 3.Garden G.A., La Spada A.R. Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration. Cerebellum. 2008;7:138–149. doi: 10.1007/s12311-008-0027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timmers H.T., Tora L. SAGA unveiled. Trends Biochem. Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 5.McMahon S.J., Pray-Grant M.G., Schieltz D., Yates J.R., Grant P.A. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc. Natl Acad. Sci. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee K.K., Workman J.L. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 7.Wieczorek E., Brand M., Jacq X., Tora L. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 8.Brand M., Yamamoto K., Staub A., Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- 9.Martinez E., Kundu T.K., Fu J., Roeder R.G. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 10.Ogryzko V.V., Kotani T., Zhang X., Schiltz R.L., Howard T., Yang X.J., Howard B.H., Qin J., Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 11.Nagy Z., Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 12.Tse C., Sera T., Wolffe A.P., Hansen J.C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 14.Cancel G., Duyckaerts C., Holmberg M., Zander C., Yvert G., Lebre A.S., Ruberg M., Faucheux B., Agid Y., Hirsch E., Brice A. Distribution of Ataxin7 in normal human brain and retina. Brain. 2000;123:2519–2530. doi: 10.1093/brain/123.12.2519. [DOI] [PubMed] [Google Scholar]
- 15.Lindenberg K.S., Yvert G., Müller K., Landwehrmeyer G.B. Expression analysis of Ataxin-7 mRNA and protein in human brain: evidence for a widespread distribution and focal protein accumulation. Brain Pathol. 2000;10:385–394. doi: 10.1111/j.1750-3639.2000.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ströma A.L., Jonassona J., Harta P., Brännströmb T., Forsgrenc L., Holmberg M. Cloning and expression analysis of the murine homolog of the spinocerebellar ataxia type 7 (SCA7) gene. Gene. 2002;285:91–99. doi: 10.1016/s0378-1119(02)00399-2. [DOI] [PubMed] [Google Scholar]
- 17.Taylor J., Grote S.K., Xia J., Vandelft M., Graczyk J., Ellerby E.M., La Spada A.R., Truant R. Ataxin-7 can export from the nucleus via a conserved exportin-dependent signal. J. Biol. Chem. 2006;281:2730–2739. doi: 10.1074/jbc.M506751200. [DOI] [PubMed] [Google Scholar]
- 18.Irwin S., Vandelft M., Pinchev D., Howell J.L., Graczyk J., Orr H.T., Truant R. RNA association and nucleocytoplasmic shuttling by ataxin-1. J. Cell Sci. 2005;118:233–242. doi: 10.1242/jcs.01611. [DOI] [PubMed] [Google Scholar]
- 19.Helmlinger D., Hardy S., Sasorith S., Klein F., Robert F., Weber C., Miguet L., Potier N., Van-Dorsselaer A., Wurtz J.M., et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 20.Kahle J.J., Gulbahce N., Shaw C.A., Lim J., Hill D.E., Barabási A.L., Zoghbi H.Y. Comparison of an expanded ataxia interactome with patient medical records reveals a relationship between macular degeneration and ataxia. Hum. Mol. Genet. 2011;20:510–527. doi: 10.1093/hmg/ddq496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao B., Mao C. Identification of microtubule-binding domains on microtubule-associated proteins by major coat phage display technique. Biomacromolecules. 2009;10:555–564. doi: 10.1021/bm801224q. [DOI] [PubMed] [Google Scholar]
- 22.Weingarten M.D., Lockwood A.H., Hwo S.Y., Kirschner M.W. A protein factor essential for microtubule assembly. Proc. Natl Acad. Sci. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gleeson J.G., Lin P.T., Flanagan L.A., Walsh C.A. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 24.Horesh D., Sapir T., Francis F., Wolf S.G., Caspi M., Elbaum M., Chelly J., Reiner O. Doublecortin, a stabilizer of microtubules. Hum. Mol. Genet. 1999;8:1599–1610. doi: 10.1093/hmg/8.9.1599. [DOI] [PubMed] [Google Scholar]
- 25.Piperno G., LeDizet M., Chang X.J. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Biol. Chem. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiff P.B., Horwitz S.B. Taxol stabilizes microtubules in mouse fibroblast cells. Proc. Natl Acad. Sci. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoebeke J., Nijena G.V., Brabanderb M.D. Interaction of noncodazole (R 17934), a new anti-tumoral drug, with rat brain tubulin. Biochem. Biophys. Res. Commun. 1976;69:319–324. doi: 10.1016/0006-291x(76)90524-6. [DOI] [PubMed] [Google Scholar]
- 28.Muchowski P.J., Ning K., D'Souza-Schorey C., Fields S. Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. Proc. Natl Acad. Sci. 2002;99:727–732. doi: 10.1073/pnas.022628699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez O.C., Schaefer A.W., Mandato C.A., Forscher P., Bement W.M., Waterman-Storer C.M. Conserved microtubule–actin interactions in cell movement and morphogenesis. Nat. Cell. Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 30.Applewhite D.A., Grode K.D., Keller D., Zadeh A.D., Slep K.C., Rogers S.L. The spectraplakin short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Mol. Biol. Cell. 2010;21:1714–1724. doi: 10.1091/mbc.E10-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmlinger D., Hardy S., Abou-Sleymane G., Eberlin A., Bowman A.B., Gansmüller A., Picaud S., Zoghbi H.Y., Trottier Y., Tora L., et al. Glutamine-expanded Ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helmlinger D., Tora L., Devys D. Transcriptional alterations and chromatin remodeling in polyglutamine diseases. Trends Genet. 2006;22:562–570. doi: 10.1016/j.tig.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Gauthier L.R., Charrin B.C., Borrell-Pages M., Dompierre J.P., Rangone H., Cordelieres F.P., De Mey J., MacDonald M.E., Lessmann V., Humbert S., Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 34.Morfini G.A., You Y.M., Pollema S.L., Kaminska A., Liu K., Yoshioka K., Bjorkblom B., Coffey E.T., Bagnato C., Han D., et al. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat. Neurosci. 2009;12:864–871. doi: 10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velier J., Kim M., Schwarz C., Kim T.W., Sapp E., Chase K., Aronin N., DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp. Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- 36.Zhai Z., Jeong H., Cui L., Krainc D., Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 37.Young J.E., Gouw L., Propp S., Sopher B.L., Taylor J., Lin A., Hermal E., Logvinova A., Chen S.F., Chen S., Bredesen D.E., Truant R., Ptacek L.J., La Spada A.R., Ellerby L.M. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J. Biol. Chem. 2007;282:30150–30160. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- 38.Chen S., Peng G.H., Wang X., Smith A.C., Grote S.G., Sopher B.L., La Spada A.R. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum. Mol. Genet. 2004;13:53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- 39.Hergovich A., Lisztwan J., Barry R., Ballschmieter P., Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell. Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- 40.La Spada A.R., Fu Y.-H., Sopher B.L., Libby R.T., Wang X., Li L.Y., Einum D.D., Huang J., Possin D.E., Smith A.C., et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induced cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.