Abstract
There is substantial evidence that impairment of peroxisome proliferator-activated receptor (PPAR)-γ-coactivator 1α (PGC-1α) levels and activity play an important role in Huntington's disease (HD) pathogenesis. We tested whether pharmacologic treatment with the pan-PPAR agonist bezafibrate would correct a deficiency of PGC-1α and exert beneficial effects in a transgenic mouse model of HD. We found that administration of bezafibrate in the diet restored levels of PGC-1α, PPARs and downstream genes to levels which occur in wild-type mice. There were significant improvements in phenotype and survival. In the striatum, astrogliosis and neuronal atrophy were attenuated and numbers of mitochondria were increased. Bezafibrate treatment prevented conversion of type I oxidative to type II glycolytic muscle fibers and increased the numbers of muscle mitochondria. Finally, bezafibrate rescued lipid accumulation and apparent vacuolization of brown adipose tissue in the HD mice. These findings provide strong evidence that treatment with bezafibrate exerts neuroprotective effects which may be beneficial in the treatment of HD.
INTRODUCTION
Huntington's disease (HD) is a dominantly inherited progressive neurodegenerative disease caused by an abnormal cytosine-adenine-guanine (CAG) repeat expansion in the huntingtin (htt) gene. The disease is characterized by progressive motor impairment, personality changes, psychiatric illness and gradual intellectual decline, leading to death 15–20 years after onset. Neuropathologic analysis shows a preferential and progressive loss of the medium spiny neurons (MSNs) in the striatum, as well as cortical atrophy, and degeneration of other brain regions later in the disease (1). There is as yet no cure for this disorder and no therapy to delay the onset of symptoms. The most extensively studied transgenic mouse model of HD are the R6/2 mice, which express exon-1 of the human htt gene and initially show behavioral and motor deficits at 6 weeks after birth. The mice subsequently develop clasping, weight loss, diabetes and reduced life span of 10–13 weeks (2).
Transcriptional dysregulation, protein aggregation, mitochondrial dysfunction and enhanced oxidative stress have been implicated in the disease pathogenesis. A critical role of peroxisome proliferator-activated receptor (PPAR)-γ-coactivator 1α (PGC-1α), a transcriptional master co-regulator of mitochondrial biogenesis, metabolism and antioxidant defenses, has been identified in HD. Interest in the role of PGC-1α in HD pathogenesis initially came from studies of PGC-1α knockout mice (PGC-1α KO), that display neurodegeneration in the striatum, which is also the brain region most affected in HD (3,4). PGC-1α plays a role in the suppression of oxidative stress and it also induces mitochondrial uncoupling proteins and antioxidant enzymes, including copper/zinc superoxide dismutase (SOD1), manganese SOD (SOD2) and glutathione peroxidase (Gpx-1) (5). Oxidative damage is a well-characterized feature which is documented in plasma of HD patients, HD postmortem brain tissue and in HD transgenic mice (6,7).
Using striata from human HD patients, striata from HD knock-in mice and the STHdhQ111 cell-based HD model, Cui et al. (8) showed marked reductions in mRNA expression of PGC-1α, and interference of mutant htt with the CREB/TAF4 complex was shown to be instrumental in this reduction. Down-regulation of PGC-1α significantly worsened the behavioral and neuropathologic abnormalities in a PGC-1α knock-out HD knock-in mouse model (PGC-1α KO/KI). Administration of a lentiviral vector expressing PGC-1α into the striatum of R6/2 mice was neuroprotective in that it increased the mean neuronal volume of MSNs (8). Caudate nucleus microarray expression data from human HD patients showed significant reductions in 24 out of 26 PGC-1α target genes (9). These authors also found reduced PGC-1α mRNA expression in striata of the N171-82Q transgenic mouse model of HD.
We subsequently carried out studies, which showed that the ability to upregulate PGC-1α in response to an energetic stress, produced by administration of the creatine analog, guanidinopropionic acid (GPA), was markedly impaired in HD transgenic mice (10,11). PGC-1α plays a critical role in mitochondrial biogenesis in muscle and in influencing whether muscle contains slow-twitch oxidative or fast-twitch glycolytic fibers (12). Impaired generation of ATP in muscle and a myopathy occurs in gene-positive individuals at risk for HD, HD patients and HD transgenic mice (13–15). We observed impaired PGC-1α activity in muscle of HD transgenic mice and in myoblasts and muscle biopsies from HD patients (10). We also showed a pathologic grade-dependent, significant reduction in numbers of mitochondria in striatal spiny neurons, which correlated with reductions in PGC-1α and the mitochondrial transcription factor a (Tfam) (16). Sequence variation in the PGC-1α gene modifies the age of onset of HD (17,18). Stimulation of extra-synaptic N-methyl-D-aspartate receptors, which is detrimental, impairs the PGC-1α cascade in HD mice (19). Impaired PGC-1α was shown to correlate with lipid accumulation in brown adipose tissue (BAT) of HD transgenic mice (20). These findings in concert strongly implicate reduced expression of PGC-1α in HD pathogenesis. If impaired PGC-1α transcriptional activity plays an important role in HD pathogenesis, then pharmacologic agents which increase its levels and activity should be beneficial.
Recently, the administration of the pan-PPAR agonist, bezafibrate, was shown to increase PGC-1α expression, mitochondrial DNA and ATP levels and also shown to increase life span and delay myopathy in a COX-10 subunit-deficient mouse model of mitochondrial myopathy (21). Bezafibrate enhances lipid metabolism and was shown to correct defects in oxidative phosphorylation in fibroblasts and/or myoblasts from patients with mitochondrial disorders (22,23). Bezafibrate is an effective cholesterol-lowering drug which is used to lower cholesterol and triglycerides and increase high-density lipoprotein. The PPARs are a subfamily of nuclear receptors, which are ligand-modulated transcription factors that regulate gene-expression programs of metabolic pathways. PPAR agonists increase oxidative phosphorylation capacity in mouse and human cells (23–25) and enhance mitochondrial biogenesis. Bezafibrate was shown to correct respiratory impairments in vitro (23), but has not been previously studied in HD. Since HD transgenic mice show reduced levels of PGC-1α and its downstream target genes, we examined whether bezafibrate treatment would increase PGC-1α expression and mitochondrial biogenesis and thereby result in improved phenotype, improved survival and reduced brain, muscle and BAT pathology in HD mice.
RESULTS
Bezafibrate induces a battery of genes in the PGC-1α signaling pathway
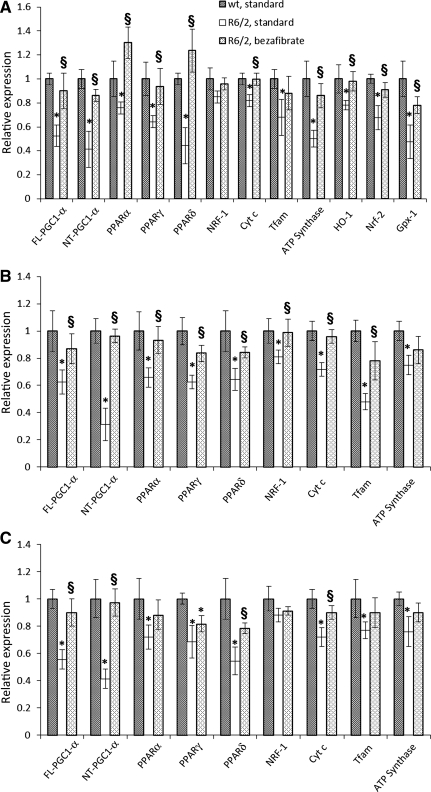
We utilized R6/2 mice with an N-terminal genomic fragment containing exon 1 with ∼130 CAG repeats (2). R6/2 mice and their wild-type littermates were raised on a diet containing 0.5% bezafibrate or standard chow, starting right after weaning. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis revealed that both the full-length (FL) and N-truncated (NT) isoforms of PGC-1α are downregulated in R6/2 mice brain, muscle and BAT compared with their wild-type littermates (Fig. 1A–C). Both FL- and NT-PGC-1α isoforms were significantly induced in the brain and peripheral tissues of R6/2 mice that were fed the bezafibrate diet and their levels were not significantly different from those of the wild-type controls. The ability of PGC-1α to activate a diverse set of metabolic programs in different tissues depends on its ability to form heteromeric complexes with a variety of transcription factors, including nuclear respiratory factor-1 (NRF-1) and PPARs. PPAR-α and PPAR-γ are co-activated with PGC-1α in a positive feedback loop and regulate glucose metabolism, fatty acid oxidation and mitochondrial biogenesis (24,26). The mRNA expression levels of the three isoforms of PPARs (α, γ and δ) and that of Cytochrome c (Cyt c), Tfam and ATP synthase were lower in the R6/2 brain, muscle and BAT when compared with their wild-type littermates; however, levels of NRF-1 were unchanged in brain and BAT. Bezafibrate administration restored the mRNA levels of PGC-1α and PPARs, downstream targets of PGC-1α, Cyt c and Tfam and ATP synthase in brain, muscle and BAT of R6/2 mice (Fig. 1).
Figure 1.
Bezafibrate restores the PGC-1α signaling pathway in R6/2 mice. (A) Relative expression of FL- and NT- isoforms of PGC-1α, PPARα, γ, δ, and the downstream target genes, NRF-1, Cyt c, Tfam, ATP synthase, as well as the oxidative stress response genes HO-1, Nrf-2 and Gpx-1 in brain of R6/2 mice on a standard diet or on the bezafibrate diet. The levels of each gene transcript were normalized to that of β-Actin and expressed as fold variation relative to the wild-type mice on a standard diet. The asterisks and symbols represent the significance levels calculated by unpaired, Student's two-tailed t-test: *P < 0.05 compared with the wild-type controls; §P < 0.05 compared with R6/2 controls (n = 5 and bars represent SEM). (B, C) Relative expression of FL-, NT-PGC-1α, PPARα, γ, δ, NRF-1, Cyt c, Tfam and ATP synthase in muscle (B) or BAT (C) of R6/2 mice on a standard diet or on the bezafibrate diet. β-Actin and wild-type mice on a standard diet were used as reference.
Concomitant with the stimulation of expression of genes involved in mitochondrial energy production, PGC-1α also induces genes responsible for countering reactive oxygen species (ROS) generated as by-products of oxidative metabolism (5,27). In R6/2 brains, we found that genes responsive to ROS, such as hemoxygenase-1 (HO-1), nuclear factor (erythroid-derived 2)-like 2 (Nrf-2) and Gpx-1 were significantly downregulated, and bezafibrate restored the levels of these genes to control levels in the R6/2 mice (Fig. 1A).
Improved phenotype and survival in R6/2 mice treated with bezafibrate
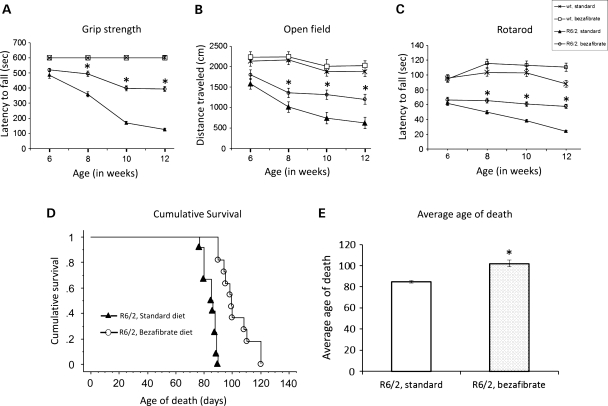
To assess muscle strength, we tested the R6/2 mice and their wild-type littermates on the grip strength test (Fig. 2A). As reported previously, R6/2 mice showed a robust deterioration of their grip strength as they aged, compared with their wild-type littermates with differences being significant at 8, 10 and 12 weeks of age (28). Bezafibrate-treated R6/2 mice showed an increase in grip strength (up to 2-fold) compared with the untreated R6/2 mice (Fig. 2A). To further assess motor function, general activity and exploration, we tested mice in the open field test. R6/2 transgenic mice were significantly hypoactive, as measured by the total distance covered, and in the bezafibrate-treated R6/2 group, a significant amelioration of the deficit was seen. The total distance covered was significantly greater in the treated group than in the R6/2 mice on a standard diet (Fig. 2B). Motor coordination was assessed by performance on an accelerated rotarod apparatus. Latency to fall was recorded for three trials per weekly assessment and scores were averaged (Fig. 2C). Consistent with previous literature, R6/2 mice showed progressive, robust deficits on rotarod, with a significantly reduced latency to fall starting at 6 weeks of age (28). Bezafibrate-treated mice remained on the rotarod longer than the untreated R6/2 mice, indicating better motor coordination in the treated mice (Fig. 2C).
Figure 2.
Bezafibrate improves the phenotype and extends survival in R6/2 mice. (A) Grip strength analysis of R6/2 mice and their wild-type littermates on bezafibrate diet. There is a rapidly progressive reduction of muscle strength that is improved in bezafibrate-treated R6/2 mice when compared with the R6/2 mice on a standard diet. *P < 0.001, when compared with the R6/2 controls (n = 8 for each genotype, bezafibrate or standard diet and bars represent SEM). (B) Measurement of exploratory activity in R6/2 mice at different ages. R6/2 mice are significantly hypoactive when compared with their wild-type littermates (*P < 0.001, n = 8). Bezafibrate significantly restores normal activity and exploration. (C) Assessment of motor coordination in R6/2 mice on bezafibrate diet. R6/2 mice showed progressive, robust deficits on rotarod, with a significantly reduced latency to fall starting at 6 weeks. Bezafibrate-treated mice remained on the rotarod longer than the untreated R6/2 mice. (D) Kaplan–Meier survival plot of R6/2 mice on the bezafibrate diet in comparison to R6/2 mice on a standard diet. No mice in wild-type groups (bezafibrate or standard diet) died in the observed time frame (n = 10 in each group). (E) Graph showing average age of death of R6/2 mice on the bezafibrate diet or a standard diet. *P < 0.05 (n = 10 in each group).
R6/2 mice normally die prematurely when compared with their wild-type littermates, between the age of 70 and 91 days (2). Symptomatic mice approaching the disease end-stage were examined twice daily (morning and late afternoon) to assess when they reached end-stage of the disease, as assessed by the righting reflex or failure to eat moistened chow placed beside mice over a 24h period (29,30). At this point, mice were euthanized by CO2 inhalation. This day was recorded as time of death of the mice. Figure 2D shows a Kaplan–Meier plot of the survival of R6/2 mice. In our hands, the R6/2 mice survived to 77–88 days of age, and the longest living mouse died at 90 days of age (mean = 84 ± 1.2 days). Bezafibrate-treated R6/2 mice lived 20% longer than the R6/2 mice on a standard diet (mean = 102 ± 3.2 days) (Fig. 2D and E).
Bezafibrate rescues neuropathologic features in R6/2 mice
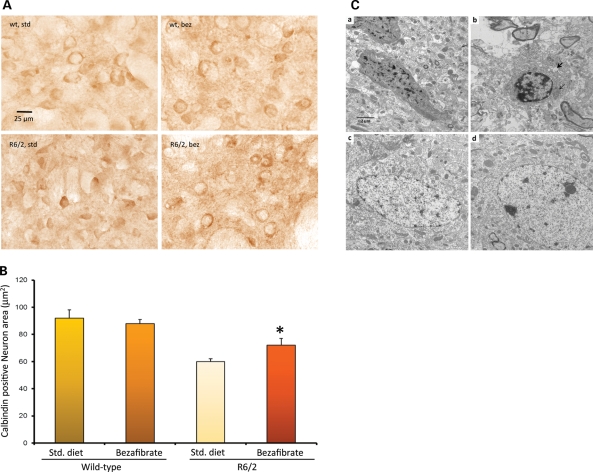
The neuropathologic features of HD are general atrophy of the brain, with losses of projection neurons in the deeper layers of the cortex and calbindin-immunoreactive MSNs in the caudate-putamen (1). We performed a stereological analysis of calbindin-immunoreactive medium spiny neuronal perikarya in the striatum of 12-week-old R6/2 mice. Consistent with other studies, we found a reduction of neuron size in R6/2 mice when compared with wild-type controls (Fig. 3A) (29). Induction of PGC-1α expression by bezafibrate treatment was accompanied by increases in the calbindin-positive neuron area in the R6/2 mice (Fig. 3A and B).
Figure 3.
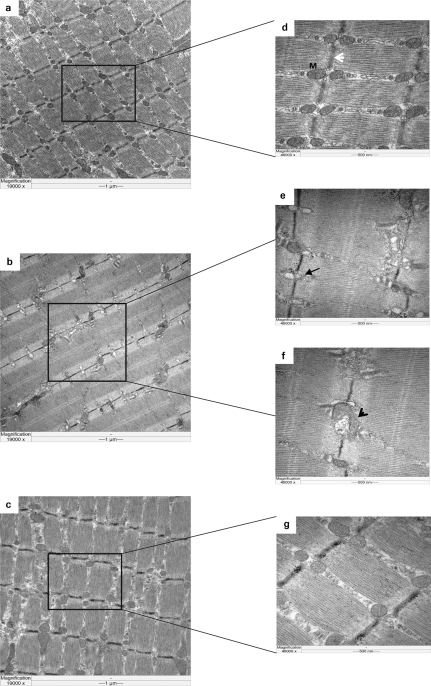
Bezafibrate prevents neurodegeneration and increases mitochondrial density. (A) Calbindin staining in the striatum of 12-week-old R6/2 mice and their wild-type littermates on bezafibrate or standard diet. (B) Stereological analysis of calbindin-immunoreactive medium spiny neuronal perikarya in the striatum. The decrease in neuron size is significantly ameliorated by bezafibrate treatment. *P < 0.05 (n = 6 in each group). (C) Electron micrographs showing degenerated neurons in the striatum of R6/2 mice (a, b) and its amelioration by bezafibrate (c, d). a, b: Apoptotic neurons with condensed cytoplasm and abnormal nuclear shape showing margination and condensation of chromatin. The presence of large cytoplasmic vacuoles (bold arrow) and lysosome-like dense bodies is also noted. Degenerated mitochondria (light arrow) and lot of empty spaces can also be seen. c, d: In striata from bezafibrate-treated R6/2 mice, the cytoplasm of the neuron is preserved and the axonal and dendritic profiles in the neuropil are relatively intact. Scale bars, 2 μm. Magnification 10 000×.
We further examined the striatum of R6/2 mice treated with bezafibrate and those on standard diet at the ultrastructural level. We observed several apoptotic neurons with condensed cytoplasm and abnormal nuclear shape showing margination and condensation of chromatin (Fig. 3Ca,b). Enlarged extracellular spaces, cytoplasmic vacuoles and lysosome-like dense bodies were also noted. Moreover, degenerated or degenerating mitochondria could also be seen. Ultrastructural abnormalities in the brains of HD mice have previously been noted, including the presence of dark neurons and abnormally shaped nuclei (31,32). In the bezafibrate-treated R6/2 mice, amelioration of these abnormalities was noted. In particular, in the striata from bezafibrate-treated R6/2 mice, the cytoplasm of the neurons is preserved, and the axonal and dendritic profiles in the neuropil are relatively intact (Fig. 3Cc,d). We also measured mitochondrial density. For this purpose, the intact mitochondria count per cell was noted and divided by the area of the cytoplasm, yielding the mitochondrial density (Table 1). Several neurons were counted per animal, and a group average is presented in Table 1. A significant increase in numbers of mitochondria in the striata of bezafibrate-treated R6/2 mice was seen, when compared with the R6/2 mice on standard chow.
Table 1.
Measurement of mitochondrial density in striatum region of the brain from R6/2 mice and their wild-type littermates on the bezafibrate diet or a standard diet
| Treatment/genotype | Mitochondrial density |
|---|---|
| wt, standard | 7.12 |
| wt, bezafibrate | 8.82 |
| R6/2, standard | 3.66* |
| R6/2, bezafibrate | 6.36§ |
Ten to fifteen neurons were counted per animal (n = 3).
*P < 0.05 when compared with wild-type controls.
§P < 0.05 when compared with R6/2 controls.
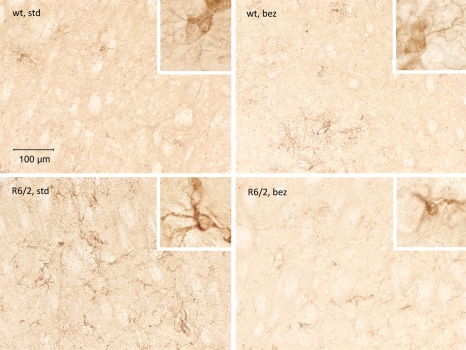
Glial fibrillary acidic protein (GFAP) immunostaining identifies reactive gliosis, an early marker of CNS damage in HD (33). We compared GFAP staining in the striatum of bezafibrate-treated and untreated wild-type and R6/2 mice (Fig. 4). Compared with age-matched wild-type mice striatum, the striatum of R6/2 mice showed a remarkable increase in GFAP immunoreactivity, indicated by intense labeling throughout astroglial cell bodies and their fibrous processes and the presence of hypertrophied astrocytes (inset). An amelioration of astrogliosis was observed in striata of R6/2 mice treated with bezafibrate (Fig. 4).
Figure 4.
Bezafibrate attenuates astrogliosis in R6/2 brains. Photomicrographs showing GFAP immunoreactivity in the striatum of wild-type and R6/2 mice with or without bezafibrate treatment. GFAP-labeled hypertrophied astrocytes (inset) are evident in the striatum of R6/2 mice. Astrogliosis in the HD striatum is reduced by bezafibrate treatment.
Evidence for amelioration of oxidative stress in R6/2 mice treated with bezafibrate
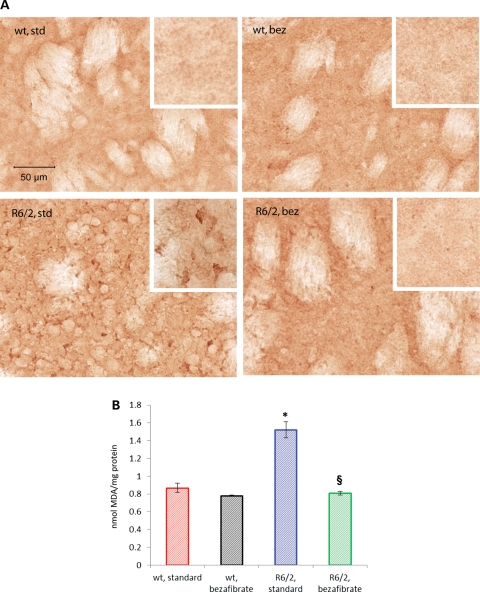
In HD, the generation of ROS and the resulting oxidative stress are implicated in the neurodegeneration and neuronal death (34; reviewed in 35). Postmortem human HD brain tissue shows increased levels of oxidative damage markers, including increased cytoplasmic lipofuscin, DNA strand breaks, oxidized DNA bases, protein nitration, carbonyls and lipid oxidative damage. Levels of malondialdehyde (MDA), a marker for oxidative damage to lipids, are elevated in human HD striatum and cortex when compared with age-matched controls (34). We observed increased immunoreactivity for MDA in R6/2 striatum, which was ameliorated by the bezafibrate diet (Fig. 5A). Consonant with the immunohistochemical data, high-performance liquid chromatography (HPLC) analysis also revealed elevated levels of MDA in R6/2 brains which were significantly reduced with bezafibrate treatment (Fig. 5B).
Figure 5.
Amelioration of oxidative stress in striatum of R6/2 mice by bezafibrate. (A) MDA immunostaining in the striatum of R6/2 mice and wild-type littermates on bezafibrate or standard diet. Insets show regions at a higher magnification. (B) Bar graph showing measurement of MDA levels by HPLC. *P < 0.001 when compared with wild-type controls. §P < 0.001 when compared with R6/2 controls (n = 6 in each group).
Bezafibrate prevents the fiber-type switching and structural abnormalities in muscle
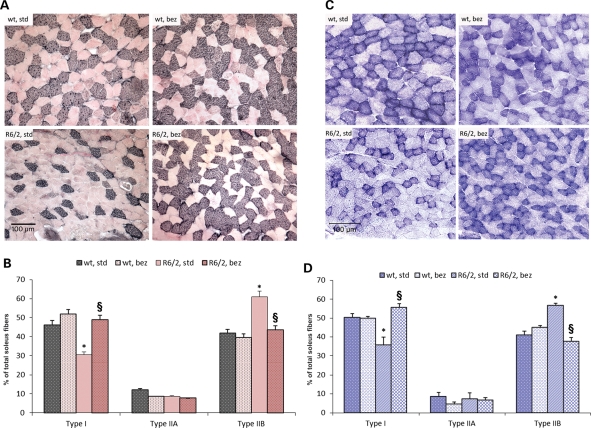
Muscle fibers can be classified as ‘slow-twitch’ fatigue-resistant fibers, which are dependent on PGC-1α and contain numerous mitochondria and use oxidative phosphorylation to generate ATP (type I and IIA fibers), and ‘fast-twitch fatiguable’ fibers (type IIX and IIB fibers), which have few mitochondria and which utilize glycolysis to generate ATP (12). PGC-1α levels are normally high in muscle enriched with type I fibers, such as the soleus muscle, and very low in type II fiber-rich muscles such as the extensor digitorum longus and the gastrocnemius (12). We examined the soleus muscle of R6/2 mice and their wild-type littermates for fiber typing using myosin heavy chain (MHC) immunostaining (Fig. 6A and B). For MHC staining, commercial antibodies to fast and slow isoforms of myosin were used, each with a different visualization system for specific identification of each fiber type on the same section. Results were visualized as dark immunoreactive black type I fibers; granular, pinkish gray type IIA; and light pink type IIB fibers (Fig. 6A). Quantitation of the MHC immunostaining revealed a significant decrease in type I fibers, and an increase in type II fibers in the soleus muscle of R6/2 mice, consistent with the reduced expression of PGC-1α. A reversal of this fiber-type switching was seen in R6/2 mice on the bezafibrate diet, with the type I and II fibers returning back to normal levels seen in wild-type mice (Fig. 6B).
Figure 6.
Fiber-type switching and abnormal ultrastructural abnormalities of muscle are reversed by bezafibrate. (A) MHC immunostaining in soleus muscle sections from wild-type and R6/2 mice with or without bezafibrate treatment. The type I fibers stain black, type IIA are granular, pinkish gray, whereas type IIB are light pink. (B) Quantitation of the MHC immunostaining. A significant reduction of type I fibers and a significant increase in type IIB fibers can be seen in the soleus muscle of R6/2 mice on a standard diet. A reversal of this fiber-type switching was seen in R6/2 mice on the bezafibrate diet, with the type I and II fibers returning back to normal levels seen in wild-type mice (n = 6 in each group). (C) Histochemical staining for SDH in soleus muscle sections from wild-type and R6/2 mice with or without bezafibrate treatment. (D) Quantitation of SDH histochemistry of soleus muscle. Decreased proportion of mitochondria-enriched oxidative type I fibers can be seen in soleus from R6/2 mice when compared with wild-type. An enrichment of type I fibers and a decrease in glycolytic type IIB fibers can be seen in soleus muscle from R6/2 mice on bezafibrate diet. These results confirm the findings seen with myosin immunostaining. *P < 0.05 when compared with wild-type controls. §P < 0.001 when compared with R6/2 controls (n = 6 in each group). (E) Transmission electron microscopic analysis of soleus muscle from wild-type and R6/2 mice on standard or bezafibrate diet. (a, d) A micrograph from the soleus muscle of a wild-type mouse. Note the arrangement of mitochondria (M) along the Z-line (white arrowhead). The micrographs (a–c) are taken at a lower magnification (19 000×) and those in d–g are taken at a higher magnification (48 000×). An altered morphology, number and alignment of mitochondria (black arrow) along the Z-lines can be seen in R6/2 mice under basal conditions (b, e, f). Structures resembling an autophagosome may also be noted (black arrowhead). Mitochondria are well organized and appear to be of normal shape and number in soleus muscle of R6/2 mice treated with bezafibrate (c, g).
We also performed succinate dehydrogenase (SDH) histochemistry. The intensity of SDH staining correlates with the mitochondrial activity and therefore with the fiber type (Fig. 6C and D). Typically, the type I fibers are enriched in mitochondria, whereas type IIB fibers have fewer mitochondria; therefore, type I fibers stain darker, whereas type IIB stain lightly for SDH. There was reduced SDH staining in the soleus of R6/2 mice on a standard diet, consistent with the reduced number of mitochondria. A significant increase in SDH staining was observed with bezafibrate treatment, which indicated an enrichment of mitochondria-abundant type I fibers (Fig. 6C and D). These results confirm the findings seen with myosin immunostaining described above.
We carried out further studies to determine the effects of bezafibrate on mitochondrial area and mitochondrial number using electron microscopy. In wild-type mice, mitochondria are uniform in size and align regularly along the Z-lines (Fig. 6E and Supplementary Material, Fig. S1), whereas in the R6/2 mice, the mitochondria are irregular in shape and poorly aligned. Similarly, PGC-1α-deficient mice show fewer and smaller mitochondria in soleus muscle (7). In the R6/2 mice treated with bezafibrate, mitochondria appeared to be of normal shape and their arrangement along the Z-lines was restored to normal (Fig. 6E).
Bezafibrate attenuated apparent vacuolization in the BAT of R6/2 mice
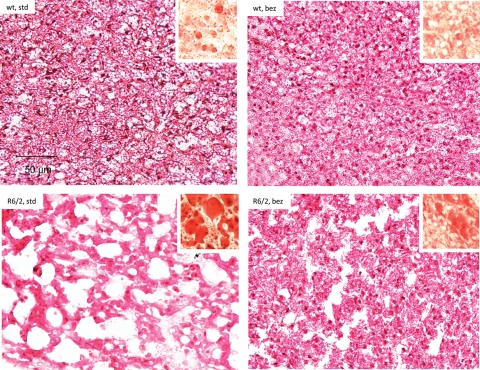
Using other mouse models of HD (N171-82Q and NLS-N171-82Q HD mice), we and others previously reported that HD mice have an impaired response to cold temperature, i.e. defective adaptive thermogenesis (9,11). In rodents, BAT is the principal tissue that mediates the adaptive thermogenesis and is distinguished from white fat by its high degree of vascularization and mitochondrial density (36). PGC-1α is expressed in BAT and is a key mediator of adaptive thermogenesis by activating uncoupling protein 1 (37). As seen with the other HD mice models, the H&E staining of BAT from the R6/2 mice showed marked reductions in cell density and nuclei numbers and increased apparent vacuolization when compared with wild-type mice (Fig. 7). The white-fat-like appearance of BAT was due to accumulation of neutral lipids as revealed by Oil red O staining (Fig. 7, inset). Bezafibrate reduced the apparent vacuolization in the BAT of the R6/2 transgenic mice when compared with R6/2 mice fed a standard diet, and reduced Oil red O staining was also observed, which was the cause of the vacuolated appearance (Fig. 7).
Figure 7.
Bezafibrate reduces apparent vacuolization of BAT in R6/2 mice. BATs of wild-type and R6/2 mice stained with hematoxylin–eosin showing increased apparent vacuolization in the R6/2 mice. Oil red O staining (red staining, inset) revealed abundant accumulation of larger lipid droplets in the R6/2 mice when compared with wild-type mice. Bezafibrate reduces the accumulation of lipids and apparent vacuolization in the R6/2 mice.
DISCUSSION
A number of bioenergetic and metabolic impairments are known to occur in HD patients: (1) increased lactate production in cerebral cortex and basal ganglia; (2) reduced phosphocreatine-to-inorganic phosphate ratio in resting muscle, the extent of which correlates with CAG repeat expansion length, and which is exacerbated after exercise; (3) abnormal mitochondrial membrane depolarization in lymphoblasts; (4) impaired complex II, III and IV activity of the mitochondrial oxidative phosphorylation pathway and reduced aconitase activity in the basal ganglia; (5) abnormal ultrastructure of mitochondria in cortical biopsies obtained from patients with both juvenile and adult-onset HD; and (6) pathologic grade-dependent reductions in numbers of mitochondria (16; reviewed in 38). We showed that the phenotypic and neuropathologic features of HD can be modeled in rodents and primates, with the mitochondrial toxin 3-nitropropionic acid (39,40).
Impaired expression and/or function of PGC-1α, the master co-regulator of mitochondrial biogenesis, has been implicated in the pathogenesis of several neurodegenerative disorders, including Parkinson's disease, Alzheimer's disease, Friedreich's ataxia and HD. A link of the transcriptional co-activator PGC-1α to HD pathogenesis was first suggested by observations in PGC-1α-deficient mice (3,4). PGC-1α KO exhibit impaired mitochondrial function, a hyperkinetic movement disorder and striatal degeneration, all features also observed in HD (3,4). Furthermore, impaired PGC-1α function and levels occur in striatal cell lines, transgenic mouse models of HD and in postmortem brain tissue from HD patients (8,9). Recent studies showed that expression of mutant htt in primary oligodendrocytes results in decreased expression of PGC-1α, and decreased expression of myelin basic protein (MBP) and deficient myelination were found in the R6/2 mouse model of HD (41). A decrease in MBP and deficient postnatal myelination occurs in the striatum of PGC-1α KO (41). In accordance with earlier studies, we show here that the PGC-1α signaling pathway is downregulated in the brain, muscle and BAT of R6/2 HD mice. We also found spongiform lesions in R6/2 striatum, along with the presence of astrogliosis, which is similar to observations made in PGC-1α-deficient mice (3,4). The neuropathologic observation of spongiform degeneration is of interest, because similar lesions occur in MnSOD null mice (42), and PGC-1α plays an important role in controlling expression of MnSOD (5).
In the present study, we found that the impairment of PGC-1α pathway can be reversed in brain, muscle and BAT from R6/2 HD mice, using the PPAR-panagonist, bezafibrate, which was previously shown to be effective in increasing life span and delaying the onset of symptoms in a mouse model of mitochondrial myopathy (21). Recently, in an early-onset partial COX-deficiency model (Surf1-KO mice), it was shown that bezafibrate induced expression of PPARα and PPARβ/δ, but also caused weight loss and hepatomegaly (43). It increased genes involved in fatty acid oxidation but did not increase mtDNA content and mitochondrial respiratory chain activities in Surf1−/− mice (43). In a more recent report, in a late-onset mitochondrial myopathy mice, bezafibrate delayed the accumulation of COX-negative fibers and mtDNA deletions (44). However, bezafibrate did not induce mitochondrial biogenesis in this model and produced marked hepatomegaly. The authors suggested that the adverse effects of hepatomegaly may mask beneficial effects of bezafibrate in mice. We also observed slight increase in liver weight, which is a known effect of PPAR stimulation in mice, but not in humans. Moreover, we found that R6/2 mice on standard diet have abnormal liver morphology, which is ameliorated by bezafibrate (Supplementary Material, Fig. S2). We, however, did observe an increase in PGC-1α, NRFs, Tfam and numbers of mitochondria. This shows that strain differences affect the efficacy of bezafibrate in producing increases in PGC-1α and downstream genes, because in both our work discussed here and that of Wenz et al. (21,25), there were increases in PGC-1α.
We found widespread beneficial effects of bezafibrate on phenotype, survival and histopathologic features in brain, muscle and BAT of R6/2 mice. The increase in survival observed with the bezafibrate diet (20%) is comparable to the highest range of percent increases in survival seen in other therapeutic trials in mouse models of HD (45). We showed that administration of creatine or triterpenoid compounds to N171-82Q mice increased survival by 19 or 21.9%, respectively, and that administration of coenzyme Q10 with remacemide or administration of mithramycin increased survival by 31.8 or 29.1% in R6/2 mice, respectively (46–49). Bezafibrate, therefore, improves the phenotype and survival of R6/2 mice in a comparable range to the best therapeutic interventions so far tested. Recently, administration of a PPARγ agonist, thiazolidinedione, was shown to produce beneficial effects on weight loss, mhtt aggregates and global ubiquitination profiles in R6/2 mice (50). Earlier, it was shown in STHdhQ111 cells that PPARγ activation by rosiglitazone prevents the mitochondrial dysfunction and oxidative stress that occurred when mutant striatal cells were challenged with pathological increases in calcium (51).
We found that the improved phenotype, increased survival and the induction of the PGC-1α signaling pathway were accompanied by reduced neuropathologic features and a significant increase in mitochondrial density in striatum of R6/2 mice treated with bezafibrate. PGC-1α plays a critical role in mitochondrial biogenesis, and in studies of cortical, midbrain and cerebellar granule neurons, both PGC-1α and PGC-1β control mitochondrial density (52). Overexpression of PGC-1β or PGC-1α, or activation of the latter by SIRT1, protects neurons from mutant htt-induced loss of mitochondria and cell death (52). The SIRT1 activator, resveratrol, increases the activity of PGC-1α and improves mitochondrial activity as a consequence of its deacetylation of PGC-1α, which increases its effects on liver, fat and muscle metabolism (53). We recently showed that resveratrol treatment of the N171-82Q transgenic mice increased PGC-1α and reduced the apparent vacuolization in BAT and reduced glucose levels, but there were no beneficial effects in the striatum due to poor brain penetration (54).
In disease-free neurons, the generation of ROS is a normal by-product of cellular respiration, mediated by mitochondria. Accumulation of ROS in neurons and subsequent oxidative stress is attenuated by free radical scavengers, such as glutathione and SOD1, preventing subsequent damage (6,55). There is evidence for oxidative damage in HD (reviewed in 35). Markers of oxidative damage, including heme oxygenase (an inducible isoform that occurs in response to oxidative stress), 3-nitrotyrosine (a marker for peroxynitrite-mediated protein nitration) and MDA (a marker for oxidative damage to lipids), are elevated in human HD striatum and cortex when compared with age-matched control brain specimens (56). The extent and intensity of these markers mirror the dorsoventral pattern of progressive neuronal loss in the neostriatum, with increased immunoreactive expression in the dorsal striatum when compared with the less severely affected ventral striatum. Consistent with the immunohistochemical data, analysis of biochemical assays in HD patients show significant increases in MDA and 4-hydroxynonenal brain levels, almost 8-fold greater than in control subjects (57).
PGC-1α plays a role in the suppression of oxidative stress and induces antioxidant enzymes, including copper/zinc SOD1, SOD2 and Gpx-1 (5). In concert with the increase in PGC-1α expression, we observed that the oxidative stress response genes such as HO-1, Nrf-2 and Gpx-1 also increase in the brains of R6/2 mice treated with bezafibrate. The levels of MDA measured by HPLC and MDA immunoreactivity in striatum were significantly reduced in R6/2 mice by bezafibrate treatment when compared with the control R6/2 mice. These observations provide strong evidence for amelioration of oxidative stress in R6/2 mice by upregulation of PGC-1α using bezafibrate. We recently showed that administration of triterpenoids, which activate the Nrf2/ARE transcriptional pathways, are neuroprotective in the N171-82Q transgenic mouse model of HD (49).
We showed that PGC-1α is reduced in muscle from HD transgenic mice and in muscle biopsies and myoblasts from HD patients (10). There was an impaired response to GPA treatment in the muscle and brains of NLS-N171-82Q HD mice. In wild-type mice, GPA treatment activated AMPK, which increased PGC-1α, NRF1 and Tfam, and this was accompanied by an increase in COX II/18s rRNA, consistent with mitochondrial biogenesis, increased mtDNA and increased numbers of mitochondria. This pathway, which leads to an increase in mitochondria in response to an energetic stress, was blocked in the NLS-N171-82Q HD mice (10,11). Bezafibrate treatment in R6/2 mice rescued the PGC-1α signaling pathway and restored the levels of downstream target genes involved in mitochondrial function, e.g. Cyt c, Tfam and ATP synthase. Bezafibrate also reversed the fiber-type switching back to normal and restored the normal morphology of muscle, shape, numbers and arrangement of mitochondria along the Z-lines in the soleus muscle of R6/2 mice.
PGC-1α is rapidly induced in response to cold exposure and regulates adaptive thermogenesis via uncoupling proteins (UCP-1), resulting in heat production in BAT. Significant hypothermia occurs in the N171-82Q, NLS-N171-82Q and R6/2 mouse models of HD (9). Following cold exposure, increases in UCP-1 expression are impaired in HD mice (9,11), and ATP/ADP ratios and mitochondrial numbers are decreased, similar to the findings in PGC-1α KO mice (3,4). We observed an apparent vacuolization in the BAT of the R6/2 mice, caused by accumulation of neutral lipids, which was reduced by bezafibrate treatment.
The important role of PGC-1α in the regulation of mitochondrial function, together with the association of mitochondrial dysfunction with HD pathogenesis, implies that activation of PGC-1α may be useful in the treatment of HD. In the present work, we show that stimulation of PPAR-PGC-1α axis by bezafibrate produces widespread beneficial effects in brain and peripheral tissues of R6/2 model of HD. Bezafibrate is an attractive agent for clinical studies as it has been used in humans for more than 25 years and it is well-tolerated with few side effects. It is therefore particularly attractive for clinical trials in neurodegenerative diseases such as HD. In other work, we found that bezafibrate exerts beneficial effects in BACHD mice (58), an FL mhtt transgenic mouse model of HD (Johri and Beal, unpublished data). Our study showing beneficial effects of bezafibrate in the R6/2 mouse model of HD provides strong evidence that bezafibrate may proved to be an effective neuroprotective agent for treatment of HD.
MATERIALS AND METHODS
Reagents
Bezafibrate, MDA (98% purity) and other chemicals were purchased from Sigma (St Louis, MO, USA). R6/2 mice were from Jackson Laboratory, Bar Harbor, ME, USA. Anti-calbindin was from Chemicon, Temecula, CA, USA; anti-MDA-modified protein was a gift from Dr Craig Thomas and anti-GFAP was from Dako, Denmark. The sequences of all the primers used in this study have been published elsewhere and/or are available on request (10,11,59).
Animal treatment
All experiments were conducted within National Institutes of Health Guidelines for Animal Research and were approved by the Weill Cornell Medical College Animal Care and Use Committee. The animals were kept on a 12-h light/dark cycle, with food and water available ad libitum. Mice were fed standard diet containing 0.5% bezafibrate or standard diet (Purina-Mills, Richmond, IN, USA), starting right after weaning up to 3 months of age. This dose was based on the beneficial effects previously reported in a mouse model of mitochondrial myopathy, in which it produced marked beneficial effects, without significant toxicity (21).
Real-time PCR
Total RNA was isolated from liquid nitrogen snap-frozen tissues using Trizol reagent, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). Genomic DNA was removed using RNase-free DNase (Ambion, Life Technologies, CA, USA) in RNA pellets resuspended in DEPC-treated water (Ambion, Life Technologies). Total RNA purity and integrity was confirmed by ND-1000 NanoDrop (NanoDrop Technologies, Thermo Fisher Scientific, DE, USA). Equal amounts of RNA were reverse-transcribed using the cDNA Synthesis Kit (Invitrogen, Life Technologies). Real-time RT-PCR was performed using the ABI prism 7900 HT sequence detection system (Applied Biosystems, Foster City, CA, USA). Expression of the gene β-Actin served as a control to normalize values. Relative expression was calculated using the ΔΔCt method.
Phenotype testing
Experimenters were blind to the genotype during all testing, at least until the appearance of a robust phenotype in the mutants. We utilized a phenotype testing battery consisting of rotarod, grip strength and open field. On the rotarod (Economex, Columbus Instruments, Columbus, OH, USA), mice were tested over three consecutive days, in three 5min trials, with an accelerating speed (from 0 to 40 rpm in 5 min) separated by a 30-min inter-trial interval. The latency to fall from the rod was recorded. Exploratory behavior was recorded in the open field (45 cm × 45 cm; height: 20 cm), for 10 min per day using a video tracking system (Ethovision 3.0, Noldus Technology, Attleborough, MA, USA) and averaged over 3 days. For the grip strength test, mice were held by the tail and placed on a wire-grill apparatus so that they grabbed the handle with both front paws and then gently pulled back until they released it. Each session consisted of five trials.
Immunohistochemistry
Mice intended for neuropathologic analysis were deeply anesthetized by intraperitoneal injection of sodium pentobarbital and perfused with 0.9% sodium chloride followed by 4% paraformaldehyde. Post-fixation, staining and processing of brain, muscle, BAT and liver samples were performed as described previously (10,11,49).
Transmission electron microscopy
Transmission electron microscopy was performed using previously published methods (10), except that for striatum the post-fixation was performed in 1% OsO4 in 0.1 m buffer instead of 1% OsO4–1.5%K-ferricyanide (soleus) for 60 min at room temperature.
High-performance liquid chromatography
The HPLC determination of MDA was carried out by a method modified from a previous report (60). The HPLC system consisted of a Waters 717plus autosampler, 515 isocratic pump and 470 scanning fluorescence detector (Waters, Milford, MA, USA). Pump flow rate was 1.0 ml/min with mobile phase comprised of acetonitrile buffer (40:60, v/v). The buffer was 50 mm potassium monobasic phosphate (anhydrous) with an adjusted pH of 6.8 using 5 m potassium hydroxide. The fluorescence detector was set at an excitation wavelength of 515 nm and emission wavelength of 553 nm. The column was an ESA 150 × 3 mm C18 column with particle size 3 μm (ESA, Inc., Chelmsford, MA, USA) placed in a column warmer set to 30°C. Peak heights were integrated by an ESA 501 chromatography data system (ESA, Inc.).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institutes of Health grant (P01AG14930) and by the Huntington Disease Society of America coalition for the cure to M.F.B.
Supplementary Material
References
- 1.Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. doi:10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. doi:10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 3.Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. doi:10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Leone T.C., Lehman J.J., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., et al. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. doi:10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. doi:10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Browne S.E., Beal M.F. Oxidative damage in Huntington's disease pathogenesis. Antioxid. Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. doi:10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 7.Hersch S.M., Gevorkian S., Marder K., Moskowitz C., Feigin A., Cox M., Como P., Zimmerman C., Lin M., Zhang L., et al. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2′dG. Neurology. 2006;66:250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. doi:10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- 8.Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. doi:10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D., et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. doi:10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Chaturvedi R.K., Adhihetty P., Shukla S., Hennessy T., Calingasan N., Yang L., Starkov A., Kiaei M., Cannella M., Sassone J., et al. Impaired PGC-1α function in muscle in Huntington's disease. Hum. Mol. Genet. 2009;18:3048–3065. doi: 10.1093/hmg/ddp243. doi:10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaturvedi R.K., Calingasan N.Y., Yang L., Hennessey T., Johri A., Beal M.F. Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington's disease following chronic energy deprivation. Hum. Mol. Genet. 2010;19:3190–3205. doi: 10.1093/hmg/ddq229. doi:10.1093/hmg/ddq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. doi:10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 13.Gizatullina Z.Z., Lindenberg K.S., Harjes P., Chen Y., Kosinski C.M., Landwehrmeyer B.G., Ludolph A.C., Striggow F., Zierz S., Gellerich F.N. Low stability of Huntington muscle mitochondria against Ca2+ in R6/2 mice. Ann. Neurol. 2006;59:407–411. doi: 10.1002/ana.20754. doi:10.1002/ana.20754. [DOI] [PubMed] [Google Scholar]
- 14.Kosinski C.M., Schlangen C., Gellerich F.N., Gizatullina Z., Deschauer M., Schiefer J., Young A.B., Landwehrmeyer G.B., Toyka K.V., Sellhaus B., et al. Myopathy as a first symptom of Huntington's disease in a Marathon runner. Mov. Disord. 2007;22:1637–1640. doi: 10.1002/mds.21550. doi:10.1002/mds.21550. [DOI] [PubMed] [Google Scholar]
- 15.Turner C., Cooper J.M., Schapira A.H. Clinical correlates of mitochondrial function in Huntington's disease muscle. Mov. Disord. 2007;22:1715–1721. doi: 10.1002/mds.21540. doi:10.1002/mds.21540. [DOI] [PubMed] [Google Scholar]
- 16.Kim J., Moody J.P., Edgerly C.K., Bordiuk O.L., Cormier K., Smith K., Beal M.F., Ferrante R.J. Mitochondrial loss, dysfunction and altered dynamics in Huntington's disease. Hum. Mol. Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. doi:10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weydt P., Soyal S.M., Gellera C., Didonato S., Weidinger C., Oberkofler H., Landwehrmeyer G.B., Patsch W. The gene coding for PGC-1α modifies age at onset in Huntington's disease. Mol. Neurodegener. 2009;4:3. doi: 10.1186/1750-1326-4-3. doi:10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taherzadeh-Fard E., Saft C., Andrich J., Wieczorek S., Arning L. PGC-1alpha as modifier of onset age in Huntington disease. Mol. Neurodegener. 2009;4:10. doi: 10.1186/1750-1326-4-10. doi:10.1186/1750-1326-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto S., Pouladi M.A., Talantova M., Yao D., Xia P., Ehrnhoefer D.E., Zaidi R., Clemente A., Kaul M., Graham R.K., et al. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat. Med. 2009;15:1407–1413. doi: 10.1038/nm.2056. doi:10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan J., Hickey M.A., Zhang P., Chesselet M.F., Reue K. Adipose tissue dysfunction tracks disease progression in two Huntington's disease mouse models. Hum. Mol. Genet. 2009;18:1006–1016. doi: 10.1093/hmg/ddn428. doi:10.1093/hmg/ddn428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenz T., Diaz F., Spiegelman B.M., Moraes C.T. Activation of the PPAR/PGC-1α pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. doi:10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Tenenbaum A., Motro M., Fisman E.Z. Dual and pan-peroxisome proliferator-activated receptors (PPAR) co-agonism: the bezafibrate lessons. Cardiovasc. Diabetol. 2005;4:14. doi: 10.1186/1475-2840-4-14. doi:10.1186/1475-2840-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastin J., Aubey F., Rotig A., Munnich A., Djouadi F. Activation of peroxisome proliferator-activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients cells lacking its components. J. Clin. Endocrinol. Metab. 2008;93:1433–1441. doi: 10.1210/jc.2007-1701. doi:10.1210/jc.2007-1701. [DOI] [PubMed] [Google Scholar]
- 24.Hondares E., Mora O., Yubero P., Rodriguez de la Concepcion M., Iglesias R., Giralt M., Villarroya F. Thiazolidinediones and rexinoids induce peroxisome proliferator-activated receptor-coactivator (PGC)-1α gene transcription: an autoregulatory loop controls PGC-1α expression in adipocytes via peroxisome proliferator-activated receptor-γ coactivation. Endocrinology. 2006;147:2829–2838. doi: 10.1210/en.2006-0070. doi:10.1210/en.2006-0070. [DOI] [PubMed] [Google Scholar]
- 25.Wenz T., Wang X., Marini M., Moraes C.T. A metabolic shift induced by a PPAR panagonist markedly reduces the effects of pathogenic mitochondrial tRNA mutations. J. Cell. Mol. Med. 2010;15:2317–2325. doi: 10.1111/j.1582-4934.2010.01223.x. doi:10.1111/j.1582-4934.2010.01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huss J.M., Kelly D.P. Nuclear receptor signaling and cardiac energetics. Circ. Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. doi:10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 27.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. doi:10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Menalled L., El-Khodor B.F., Patry M., Suarez-Farinas M., Orenstein S.J., Zahasky B., Leahy C., Wheeler V., Yang X.W., MacDonald M., et al. Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models. Neurobiol. Dis. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. doi:10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrante R.J., Andreassen O.A., Dedeoglu A., Ferrante K.L., Jenkins B.G., Hersch S.M., Beal M.F. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J. Neurosci. 2002;22:1592–1599. doi: 10.1523/JNEUROSCI.22-05-01592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubin C., Knoblaugh S., Ladiges W. Phenotypic characterization of genetically engineered mice. In: Hau J., Van Hoosier G.L. Jr, editors. Handbook of Laboratory Animal Science: Essential Principles and Practices. 2nd edn. Vol. 1. Boca Raton, FL: CRC Press; 2003. pp. 218–220. [Google Scholar]
- 31.Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., Bates G.P. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. doi:10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 32.Yu Z.X., Li S.H., Evans J., Pillarisetti A., Li H., Li X.J. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J. Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedreen J.C., Folstein S.E. Early loss of neostriatal striosome neurons in Huntington's disease. J. Neuropathol. Exp. Neurol. 1995;54:105–120. doi: 10.1097/00005072-199501000-00013. doi:10.1097/00005072-199501000-00013. [DOI] [PubMed] [Google Scholar]
- 34.Browne S.E., Bowling A.C., MacGarvey U., Baik M.J., Berger S.C., Muqit M.M., Bird E.D., Beal M.F. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann. Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. doi:10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 35.Stack E.C., Matson W.R., Ferrante R.J. Evidence of oxidant damage in Huntington's disease: translational strategies using antioxidants. Ann. NY Acad. Sci. 2008;1147:79–92. doi: 10.1196/annals.1427.008. doi:10.1196/annals.1427.008. [DOI] [PubMed] [Google Scholar]
- 36.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. doi:10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 37.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. doi:10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 38.Browne S.E., Beal M.F. The energetics of Huntington's disease. Neurochem. Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. doi:10.1023/B:NERE.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 39.Beal M.F., Brouillet E., Jenkins B.G., Ferrante R.J., Kowall N.W., Miller J.M., Storey E., Srivastava R., Rosen B.R., Hyman B.T. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J. Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brouillet E., Hantraye P., Ferrante R.J., Dolan R., Leroy-Willig A., Kowall N.W., Beal M.F. Chronic mitochondrial energy impairment produces selective striatal degeneration and abnormal choreiform movements in primates. Proc. Natl Acad. Sci. USA. 1995;92:7105–7109. doi: 10.1073/pnas.92.15.7105. doi:10.1073/pnas.92.15.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang Z., Valenza M., Cui L., Leoni V., Jeong H.K., Brilli E., Zhang J., Peng Q., Duan W., Reeves S.A., et al. Peroxisome-proliferator-activated receptor gamma coactivator 1α contributes to dysmyelination in experimental models of Huntington's disease. J. Neurosci. 2011;31:9544–9553. doi: 10.1523/JNEUROSCI.1291-11.2011. doi:10.1523/JNEUROSCI.1291-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinerfeld D., Traini M.D., Weinberger R.P., Cochran B., Doctrow S.R., Harry J., Melov S. Endogenous mitochondrial oxidative stress: neurodegeneration, proteomic analysis, specific respiratory chain defects, and efficacious antioxidant therapy in superoxide dismutase 2 null mice. J. Neurochem. 2004;88:657–667. doi: 10.1046/j.1471-4159.2003.02195.x. doi:10.1046/j.1471-4159.2003.02195.x. [DOI] [PubMed] [Google Scholar]
- 43.Viscomi C., Bottani E., Civiletto G., Cerutti R., Moggio M., Fagiolari G., Schon E.A., Lamperti C., Zeviani M. In vivo correction of COX deficiency by activation of the AMPK/PGC-1α axis. Cell Metab. 2011;14:80–90. doi: 10.1016/j.cmet.2011.04.011. doi:10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yatsuga S., Suomalainen A. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr482. doi:10.1093/hmg/ddr482. [DOI] [PubMed] [Google Scholar]
- 45.Hersch S.M., Ferrante R.J. Translating therapies for Huntington's disease from genetic animal models to clinical trials. NeuroRx. 2004;1:298–306. doi: 10.1602/neurorx.1.3.298. doi:10.1602/neurorx.1.3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andreassen O.A., Dedeoglu A., Ferrante R.J., Jenkins B.G., Ferrante K.L., Thomas M., Friedlich A., Browne S.E., Schilling G., Borchelt D.R., et al. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington's disease. Neurobiol. Dis. 2001;8:479–491. doi: 10.1006/nbdi.2001.0406. doi:10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- 47.Beal M.F., Ferrante R.J. Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat. Rev. Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. doi:10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 48.Ferrante R.J., Ryu H., Kubilus J.K., D'Mello S., Sugars K.L., Lee J., Lu P., Smith K., Browne S., Beal M.F., et al. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington's disease. J. Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. doi:10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stack C., Ho D., Wille E., Calingasan N.Y., Williams C., Liby K., Sporn M., Dumont M., Beal M.F. Triterpenoids CDDO-ethyl amide and CDDO-trifluoroethyl amide improve the behavioral phenotype and brain pathology in a transgenic mouse model of Huntington's disease. Free Radic. Biol. Med. 2010;49:147–158. doi: 10.1016/j.freeradbiomed.2010.03.017. doi:10.1016/j.freeradbiomed.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiang M.C., Chen C.M., Lee M.R., Chen H.W., Chen H.M., Wu Y.S., Hung C.H., Kang J.J., Chang C.P., Chang C., et al. Modulation of energy deficiency in Huntington's disease via activation of the peroxisome proliferator-activated receptor gamma. Hum. Mol. Genet. 2010;19:4043–4058. doi: 10.1093/hmg/ddq322. doi:10.1093/hmg/ddq322. [DOI] [PubMed] [Google Scholar]
- 51.Quintanilla R.A., Jin Y.N., Fuenzalida K., Bronfman M., Johnson G.V. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J. Biol. Chem. 2008;283:25628–25637. doi: 10.1074/jbc.M804291200. doi:10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wareski P., Vaarmann A., Choubey V., Safiulina D., Liiv J., Kuum M., Kaasik A. PGC-1α and PGC-1β regulate mitochondrial density in neurons. J. Biol. Chem. 2009;284:21379–21385. doi: 10.1074/jbc.M109.018911. doi:10.1074/jbc.M109.018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. doi:10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Ho D.J., Calingasan N.Y., Wille E., Dumont M., Beal M.F. Resveratrol protects against peripheral deficits in a mouse model of Huntington's disease. Exp. Neurol. 2010;225:74–84. doi: 10.1016/j.expneurol.2010.05.006. doi:10.1016/j.expneurol.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Beal M.F. Mitochondria, NO and neurodegeneration. Biochem. Soc. Symp. 1999;66:43–54. doi: 10.1042/bss0660043. [DOI] [PubMed] [Google Scholar]
- 56.Browne S.E., Ferrante R.J., Beal M.F. Oxidative stress in Huntington's disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. doi:10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoy N., Mackay G.M., Forrest C.M., Christofides J., Egerton M., Stone T.W., Darlington L.G. Tryptophan metabolism and oxidative stress in patients with Huntington's disease. J. Neurochem. 2005;93:611–623. doi: 10.1111/j.1471-4159.2005.03070.x. doi:10.1111/j.1471-4159.2005.03070.x. [DOI] [PubMed] [Google Scholar]
- 58.Gray M., Shirasaki D.I., Cepeda C., Andre V.M., Wilburn B., Lu X.H., Tao J., Yamazaki I., Li S.H., Sun Y.E., et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. doi:10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johri A., Starkov A.A., Chandra A., Hennessey T., Sharma A., Orobello S., Squitieri F., Yang L., Beal M.F. Truncated peroxisome proliferator-activated receptor-γ coactivator 1α splice variant is severely altered in Huntington's disease. Neurodegener. Dis. 2011;8:496–503. doi: 10.1159/000327910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agarwal R., Chase S.D. Rapid, fluorimetric-liquid chromatographic determination of malondialdehyde in biological samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002;775:121–126. doi: 10.1016/s1570-0232(02)00273-8. doi:10.1016/S1570-0232(02)00273-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.