Abstract
Bloom's syndrome (BS) is an autosomal recessive disorder that is invariably characterized by severe growth retardation and cancer predisposition. The Bloom's syndrome helicase (BLM), mutations of which lead to BS, localizes to promyelocytic leukemia protein bodies and to the nucleolus of the cell, the site of RNA polymerase I-mediated ribosomal RNA (rRNA) transcription. rRNA transcription is fundamental for ribosome biogenesis and therefore protein synthesis, cellular growth and proliferation; its inhibition limits cellular growth and proliferation as well as bodily growth. We report that nucleolar BLM facilitates RNA polymerase I-mediated rRNA transcription. Immunofluorescence studies demonstrate the dependance of BLM nucleolar localization upon ongoing RNA polymerase I-mediated rRNA transcription. In vivo protein co-immunoprecipitation demonstrates that BLM interacts with RPA194, a subunit of RNA polymerase I. 3H-uridine pulse-chase assays demonstrate that BLM expression is required for efficient rRNA transcription. In vitro helicase assays demonstrate that BLM unwinds GC-rich rDNA-like substrates that form in the nucleolus and normally inhibit progression of the RNA polymerase I transcription complex. These studies suggest that nucleolar BLM modulates rDNA structures in association with RNA polymerase I to facilitate RNA polymerase I-mediated rRNA transcription. Given the intricate relationship between rDNA metabolism and growth, our data may help in understanding the etiology of proportional dwarfism in BS.
INTRODUCTION
The ∼400 ribosomal DNA (rDNA) genes within human cells are distributed tandemly on the p-arms of the five acrocentric chromosomes—13, 14, 15, 21 and 22. These rDNA repeats, along with RNA polymerase I and numerous other proteins, are localized in interphase cells in a nuclear structure known as the nucleolus. The predominant function of nucleoli is the transcription of ribosomal RNA (rRNA) from rDNA, a process mediated by RNA polymerase I that occurs most prominently during S- and G2-phases of the cell cycle (1,2). Ribosomal RNA transcription is a major determinant of ribosome biogenesis, which in turn drives protein translation, cellular growth and proliferation (3). Animal models show that mutation of RNA polymerase I transcription factors inhibits rRNA transcription and impairs growth (4), while human syndromes caused by defects within the ribosome biogenesis pathway similarly display growth impairment (5).
The nucleolus contains three distinct sub-structural components, the fibrillar center (FC), dense fibrillar component (DFC) and the granular component (GC) [reviewed in (6)]. The FC and the DFC contain rDNA and RNA polymerase I; the DFC also contains factors required for rRNA processing (6,7). RNA polymerase I transcription most likely occurs at the FC–DFC interface, or entirely within the DFC (7). The GC is the outermost region of the nucleolus and contains factors necessary for ribosomal assembly (6). Proteomic analysis reveals a large number of putative RNA and DNA helicases, particularly those belonging to the DEAD-box family of RNA-dependent ATPases, that localize to all nucleolar regions and suggest a necessity for diverse helicases in ribosomal RNA synthesis, processing and assembly into ribosomes (8,9).
Bloom's syndrome (BS) is a rare autosomal recessive disorder characterized by a high predisposition to cancer and severe growth retardation (10). Cells from BS persons grow poorly in culture and have a decreased response to growth factors (11). Affected individuals invariably display intra-uterine growth retardation (IUGR) with a mean birth weight of ∼1.7 kg, and proportional dwarfism that persists throughout life with a mean adult height of 133 cm. The etiology of the BS growth defect remains unknown despite extensive clinical investigation (12).
Bloom's syndrome helicase (BLM), the protein absent in BS, belongs to the conserved recQ subfamily of ATP-dependent 3′-5′ DNA helicases (13,14). The BLM helicase localizes to PML bodies and nucleoli, most prominently during S-phase (15). The N-terminus of BLM is required for its accumulation to PML bodies, while nucleolar localization of BLM requires the C-terminal region that also directly binds rDNA repeats (16,17). Within rDNA sequences, BLM specifically associates with the 18S-coding region and Alu-repeat regions, upstream of the region where replication is initiated (17). Furthermore, clonally selected BS cells have less rDNA than BLM-proficient cells, suggesting the hypothesis that nucleolar BLM, by binding to rDNA, is necessary to maintain the stability of rDNA (16,17). The related recQ-like WRN helicase localizes to nucleoli in some human cell types and accelerates RNA polymerase I transcription (18,19). The Saccharomyces cerevisiae BLM ortholog Sgs1 facilitates rDNA replication and maintains the stability of rDNA repeats (20,21). Sgs1 is also essential for RNA polymerase I transcription in the absence of the Srs2 helicase, suggesting the possibility of a similar function for BLM in rDNA replication and transcription (22).
Here, we report that treatment of human cells with the RNA polymerase I inhibitor actinomycin D (AMD) results in redistribution of BLM from the nucleoli to the nucleoplasm and nucleolar periphery, consistent with an association of BLM with the RNA polymerase I transcription complex. In vivo protein co-immunoprecipitation demonstrates a physical interaction between BLM and the RNA polymerase I-specific subunit RPA194. 3H-uridine pulse-chase assays demonstrate a decreased production of the 45S rRNA transcript in BLM-deficient cells when compared with wild-type cells, indicating a slower rate of RNA polymerase I transcription in the absence of BLM. In vitro, BLM binds and unwinds GC-rich rDNA-like DNA20:DNA33 and RNA20:DNA33 duplexes predicted to form during rRNA transcription, but not DNA20:RNA33 or RNA20:RNA33 duplexes. We propose that BLM is part of an RNA polymerase I transcription complex in the nucleolus and modulates rDNA to remove secondary structures that, if left unresolved, stall RNA polymerase I transcription and increase recombination within rDNA repeats. These data may help in understanding the instability of rDNA repeats in BS cells (23), as well as the documented cellular (11) and whole body growth defect in BS (10,12).
RESULTS
BLM re-localizes within the nucleus following inhibition of RNA polymerase I-mediated rRNA transcription
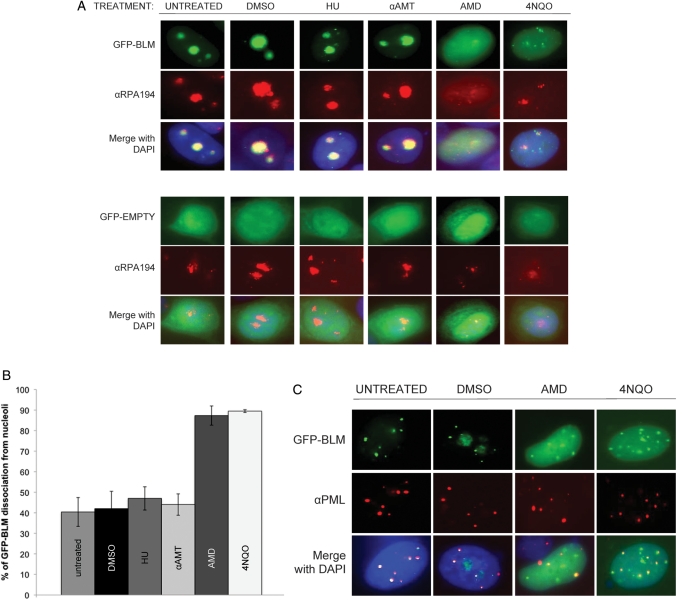
BLM localizes to the nucleolus (15), the site of RNA polymerase I-mediated rRNA transcription. To investigate the role of nucleolar BLM in rRNA transcription, we examined whether BLM localization is dependent upon RNA polymerase I-mediated rRNA transcription using the RNA polymerase I inhibitor AMD. AMD binds selectively to GC-rich DNA and strongly inhibits RNA polymerase I transcription (24) as rRNA genes have a high GC-content. RNA polymerase I and its associated factors distribute in a characteristic fashion following AMD treatment (25,26). A low concentration of AMD treatment changes the localization of RNA polymerase I from the center of the nucleoli to the nucleolar periphery (25). A high concentration of AMD results in the dispersal of RNA polymerase I throughout the nucleoplasm (26). In order to visualize the localization of BLM during AMD treatment, a GFP-tagged BLM construct was used in transfections of the breast cancer cell line MCF7. MCF7 cells were chosen to minimize artifacts of BLM over-expression as they express a relatively low level of endogenous BLM and an effective αBLM antibody for immunofluorescence is not available. pGFP–BLM-transfected MCF7 cells were treated with AMD and the sub-nuclear localization of GFP–BLM analyzed by immunofluorescence microscopy. Nucleolar localization of GFP–BLM was observed in untreated cells using BLM co-localization with the nucleolar protein nucleophosmin (NPM/B23) (Supplementary Material, Fig. S1A) and with the RNA polymerase I-specific subunit RPA194 (Fig. 1A); cells transfected with the pGFP control vector showed a diffuse nuclear staining of GFP (Fig. 1A). A short treatment with AMD results in a dramatic redistribution of NPM/B23, RPA194 and GFP–BLM from the nucleolus to the nucleoplasm (Fig. 1A and Supplementary Material, Fig. S1A). Exposure to AMD significantly decreased the nucleolar localization of GFP–BLM (Fig. 1B; 40% decrease); phase-contrast microscopy confirmed that nucleoli remained intact after AMD treatment (Supplementary Material, Fig. S1A). Such redistribution is consistent with that observed for RNA polymerase I after similar AMD treatments (26). We also tested the effect of the RNA polymerase II inhibitor α-amanitin (αAMT) on BLM localization. Our results indicated that GFP–BLM is retained in the nucleolus following treatment of cells with αAMT and demonstrate that although RNA polymerase II is inhibited at the AMD concentration used (26), the effect upon GFP–BLM nucleolar localization is specific to inhibition of RNA polymerase I and independent of RNA polymerase II.
Figure 1.
Nucleolar localization of BLM is dependent upon ongoing RNA polymerase I transcription. (A) The breast cancer cell line MCF7 was transiently transfected with pGFP–BLM and stained with αRPA194, a nucleolar protein, to demonstrate co-localization of GFP–BLM to nucleoli. MCF7 cells were treated with either actinomycin D (AMD), 4-nitroquinoline-1 oxide (4NQO), α-amanitin, hydroxyurea (HU), DMSO (negative control for AMD and 4NQO) or H2O (negative control for α-amanitin and HU), followed by staining with αRPA194 and visualization of transiently expressed GFP–BLM and RPA194. MCF7 cells were transfected with pGFP control vector and similarly treated to demonstrate a lack of an effect on GFP. (B) The results of scoring transiently transfected MCF7 cells for GFP–BLM localization following the indicated treatments are shown. Averages ± standard deviation were calculated for a minimum of 60 cells per treatment. (C) MCF7 cells were transiently transfected with pGFP–BLM, treated with AMD, 4NQO, DMSO (negative control for AMD and 4NQO) or H2O and stained with αPML to analyze the co-localization of GFP–BLM and PML when BLM dissociates from the nucleolus.
Finally, cells were treated with the DNA polymerase-stalling drug hydroxyurea (HU) to test whether the effect of AMD on BLM localization is a general response to nuclear stress. Nucleolar GFP–BLM was undisturbed (Fig. 1A and B). Similar results were obtained using the human embryonic kidney 293T cell line and the BS fibroblast cell line GM08505 (Supplementary Material, Fig. S1B). The recQ-like Werner syndrome helicase (WRN) functions in nucleolar rRNA transcription and localizes to nucleoli in a 4-nitroquinoline-1 oxide (4NQO)-sensitive manner (18). 4NQO induces DNA lesions usually corrected by nucleotide excision repair. We analyzed nucleolar localization of GFP–BLM in MCF7 cells following treatment with 4NQO and found, similarly to WRN, nucleolar dissociation of GFP–BLM (Fig. 1A), suggesting that these two related helicases may also function similarly in rRNA transcription (19). MCF7 cells transiently transfected with pGFP–BLM and treated with either AMD or 4NQO were stained with anti-PML to demonstrate that co-localization of PML and BLM is not disturbed following AMD or 4NQO treatment. These results show that the effect of AMD and 4NQO on GFP–BLM is specific to nucleolar BLM (Fig. 1C). These results indicate that localization of BLM within the nucleolus is associated with the ongoing RNA polymerase I-mediated rRNA transcription.
BLM interacts with the RNA polymerase I-specific subunit RPA194
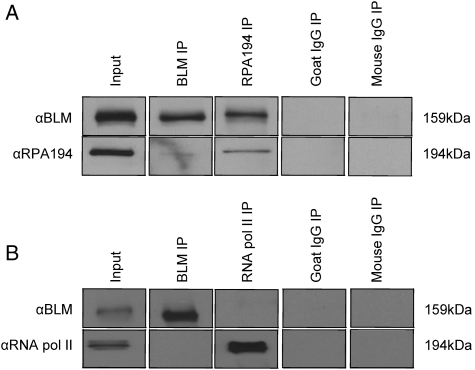
Further investigation of the role of BLM in RNA polymerase I-mediated rRNA transcription used protein co-immunoprecipitation experiments to demonstrate the interaction of BLM and at least one subunit of RNA polymerase I. RNA polymerase I is a nucleolar-specific polymerase solely dedicated to rRNA transcription. It is a multi-subunit enzyme, sharing some subunits with RNA polymerases II and III, although its largest subunit, RPA194, is not shared (27). αBLM and αRPA194 antibodies were used in co-immunoprecipitation experiments with nuclear lysates from MCF7 and 293T cells to demonstrate that BLM and RPA194 interact (Fig. 2 and Supplementary Material, Fig. S4). Co-immunoprecipitation of BLM and RPA194 was observed regardless of which antibody was used for immunoprecipitation (IP). An antibody specific to RNA polymerase II was unable to co-immunoprecipitate BLM and RNA polymerase II to control for the possibility that BLM non-specifically interacts with RNA polymerases (Fig. 2). The interaction of BLM and RNA polymerase I supports a function for BLM in the modulation of rDNA structures in association with RNA polymerase I to facilitate rRNA transcription.
Figure 2.
BLM associates with the RNA polymerase I-specific subunit RPA194. (A) Co-immunoprecipitations were performed with nuclear extracts from 293T cells using either αBLM or αRPA194 antibodies for IP (51). Proteins were separated using 8% SDS–PAGE, blotted and analyzed with αBLM and αRPA194 antibodies; goat IgG is an isotype-matched negative control for αBLM, mouse IgG is an isotype-matched negative control for αRPA194. (B) Co-immunoprecipitations were performed as in (A) but αRPB1 (RNA polymerase II subunit) was used; mouse IgG is an isotype-matched negative control for anti-RNA polymerase II. In (B), we were unable to detect an interaction between BLM and RNA polymerase II, demonstrating the specificity of its interaction with RPA194 in (A).
BLM-deficient cells display a slower rate of RNA polymerase I-mediated rRNA transcription
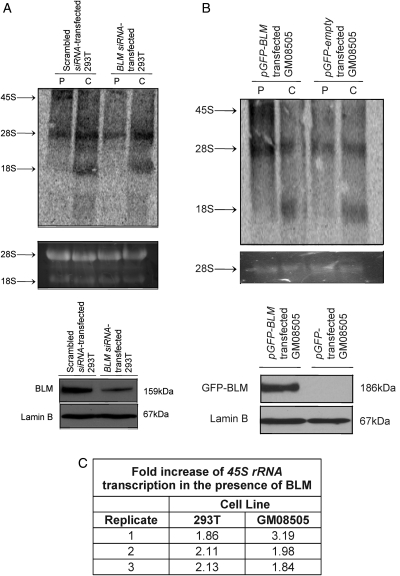
As nucleolar localization of GFP–BLM is associated with ongoing RNA polymerase I-mediated rRNA transcription, we asked whether BLM plays a role in rRNA expression. We used 293T cells transfected with an αBLM-directed siRNA to knockdown BLM expression or scrambled control siRNA in 3H-uridine pulse-chase assays to measure the rRNA transcription rate. Figure 3A shows that the abundance of radiolabeled 45S rRNA transcript diminishes following knockdown of BLM expression in 293T cells (Fig. 3C). αBLM-directed siRNA does not affect the level of the related recQ-like helicase WRN (Supplementary Material, Fig. S3). Additionally, we performed 3H-uridine pulse-chase assays using the BS fibroblast line GM08505 transiently transfected with pGFP–BLM or pGFP control vector. Figure 3B shows that the abundance of radiolabeled 45S rRNA transcript increases upon re-expression of BLM via GFP–BLM in GM08505 cells (Fig. 3C). In contrast, the helicase-dead GFP-BLM-D795A was unable to rescue the 45S rRNA transcript defect of GM08505 cells (Supplementary Material, Fig. S2G). Our results suggest that BLM facilitates RNA polymerase I-mediated rRNA transcription and emphasize the requirement for BLM helicase activity. To support these data, 293T cells were used in in vivo biotin-labeled nuclear run-on assays to measure the 45S rRNA transcription rate, as RNA polymerase I-mediated rRNA transcription initially produces the 45S rRNA transcript (24). GAPDH, an RNA polymerase II-transcribed gene, served as normalization for RNA polymerase I-mediated transcription (Supplementary Material, Fig. S2A). Supplementary Material, Figure S2C shows that siRNA-mediated knockdown of BLM expression (Supplementary Material, Fig. S2B) decreases the GAPDH-normalized 45S rRNA transcription rate. P-value (P= 0.03) was determined with the Wilcoxon signed-rank test as the experiment was conducted in a pair-wise manner (Supplementary Material, Fig. S2D). 293T cells treated with AMD or mock-treated with dimethylsulfoxide (DMSO) were used in in vivo biotin-labeled nuclear run-on assays to verify the inhibitory effect of AMD on RNA polymerase I-mediated transcription (Supplementary Material, Fig. S2E). Furthermore, 3H-uridine pulse-chase analyses in age- and sex-matched BS (GM03403 and GM16375) versus wild-type lymphoblastoid cells (GM01806 and GM11973) similarly show that BLM is required to maintain a normal rRNA transcription rate (Supplementary Material, Fig. S2F). Our findings demonstrate that BLM expression and helicase activity are required for efficient rRNA transcription.
Figure 3.
BLM deficiency slows RNA polymerase I-mediated 45S rRNA transcription rate. (A) 293T cells transfected with either an αBLM-directed siRNA or scrambled control siRNA were pulse-labeled with 3H-uridine for 30 min (P) and chased for 1 h (C) with cold uridine. Isolated RNAs were separated on a 1% MOPS-formaldehyde agarose gel, transferred to a nylon membrane and analyzed by autoradiography. 45S, 28S and 18S rRNA species are indicated next to autoradiograph (top panel). Ethidium bromide staining demonstrates equal loading of RNA (middle panel). Western blot demonstrates efficiency of BLM knockdown; lamin B serves as a protein loading control (bottom panel). (B) 3H-uridine pulse-chase analysis in BS fibroblasts (GM08505) transfected with either pGFP–BLM or pGFP; figure is labeled as in (A). (C) The pulse-chase analyses in 293T and GM08505 cells were analyzed using ImageQuant software to measure the 45S rRNA transcript abundance in the BLM-proficient cells (293T scrambled siRNA-transfected or GM08505 pGFP–BLM-transfected) compared with the BLM-deficient cells (293T αBLM-directed siRNA-transfected or GM08505 pGFP-transfected).
BLM binds and unwinds rDNA-like GC-rich DNA20:DNA33 and RNA20:DNA33 nucleic acid duplexes
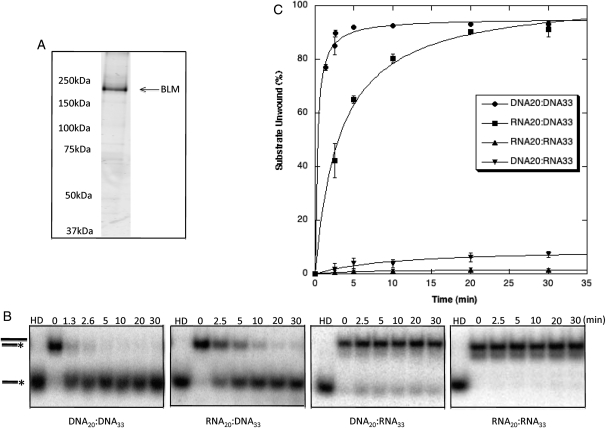
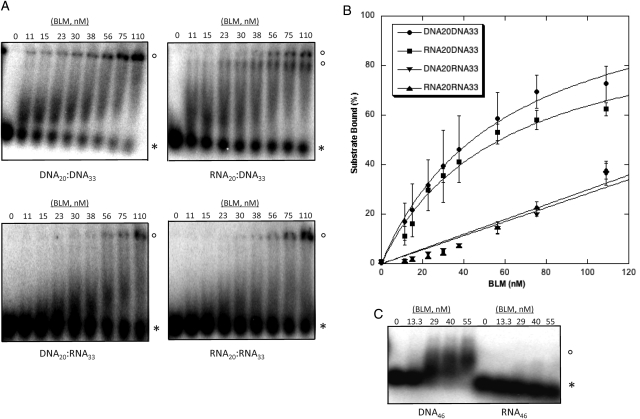
Altered localization of nucleolar BLM following AMD treatment, slower rates of RNA polymerase I-mediated rRNA transcription in the absence of BLM, and the association of BLM and RNA polymerase I suggest that BLM helicase functions may be required during rRNA transcription. As rRNA–rDNA duplexes can form in vivo (28) and inhibit movement of transcription complexes (29,30), we investigated the activity of BLM on RNA–DNA nucleic acid substrates. Recombinant BLM was expressed in yeast and fast protein liquid chromatography (FPLC)-purified (Fig. 4A) for in vitro helicase assays and electrophoretic mobility shift assays (EMSAs) using nucleic acid substrates with 20 bp of duplexed sequence and a 13-nucleotide 3′ single-stranded overhang. Previous in vitro studies have determined that BLM requires a 3′ overhang of at least eight nucleotides for unwinding of standard duplex DNA (31,32). Substrates in our experiments had either a 3′ overhang of DNA (DNA20:DNA33 and RNA20:DNA33) or RNA (DNA20:RNA33 and RNA20:RNA33). Substrates were incubated with BLM and products resolved using non-denaturing polyacrylamide gels. Figure 4 shows that BLM unwinds DNA overhang substrates (DNA20:DNA33 and RNA20:DNA33) following Michaelis–Menten kinetics with maximum unwinding: Umax, 95.26 ± 1.14% (T1/2= 0.28 min) and 103.84 ± 2.80% (T1/2= 3.24 min), respectively. BLM displays very low activity unwinding RNA overhang substrates, DNA20:RNA33 (Umax: 9.46 ± 1.57%, T1/2= 10.41 min) and RNA20:RNA33 (Umax: 1.77 ± 0.17%, T1/2= 6.16 min). Helicase assays, using duplexes containing a 26 nt 3′ overhang of either scrambled sequence, poly-T or poly-U sequences to control for secondary structure, show that BLM is unable to unwind duplexes with a 3′ RNA overhang regardless of its potential to form secondary structure (data not shown). These in vitro assays demonstrate the ability of BLM to unwind the RNA–DNA hybrid duplexes predicted to form during RNA polymerase I-mediated transcription, and extend the previously published unwinding results (31).
Figure 4.
BLM unwinds duplex substrates with a 3′ DNA overhang but not those with a 3′ RNA overhang. (A) The purity of BLM protein was determined using electrophoresis with 8% SDS–PAGE gels and staining with SYPRO Ruby Protein Gel Stain (Sigma). (B) Autoradiographs of representative gels illustrate unwinding activities. BLM (3.8 nm) was incubated with substrate for 0, 2.5, 5, 10, 20 or 30 min at 37°C as described in Materials and Methods. Products were resolved using 12% non-denaturing acrylamide gels. Unwinding is demonstrated by conversion of duplexed substrate to faster migrating single-stranded oligonucleotide. HD is heat-denatured substrate produced by heating to 95°C for 5 min. (C) Kinetics of BLM unwinding of RNA- and DNA-containing substrates. BLM unwinds DNA20:DNA33 and RNA20:DNA33 but does not appreciably unwind DNA20:RNA33 or RNA20:RNA33. Unwinding of each duplex substrate was calculated by comparing the amount of single-stranded substrate produced to the total amount of substrate in the reaction with correction for any un-annealed substrate in zero-time controls. Percent unwinding is graphed as a function of time.
We next performed EMSAs to investigate whether the hybrid duplex unwinding activity of BLM reflected different binding affinities for the substrates. EMSAs (Fig. 5) show that BLM binding to DNA20:DNA33 and RNA20:DNA33 follows Michaelis–Menten kinetics with Kd values of 60.46 ± 7.45 nm BLM and 58.92 ± 12.18 nm BLM, respectively. BLM binding to DNA20:RNA33 and RNA20:RNA33 is less favorable and does not follow Michaelis–Menten kinetics. The lower binding affinity of BLM for DNA20:RNA33 and RNA20:RNA33 may partially explain the reduced unwinding activity of BLM on these substrates. To address the possibility that the DNA20:RNA33 and RNA20:RNA33 substrates are not bound or unwound because of insufficient RNA overhang length, EMSAs were performed with single-stranded DNA46 and single-stranded RNA46. Figure 5C shows that while BLM binds DNA46, it does not efficiently bind RNA46. These data and the duplex-binding data suggest that BLM is unable to unwind DNA20:RNA33 and RNA20:RNA33 because of an inability to bind the single-stranded RNA overhang in these duplexes. Overall, these in vitro studies support a function for BLM at the interface of RNA and DNA metabolism in the nucleolus.
Figure 5.
BLM binds to DNA20:DNA33 and RNA20:DNA33, and less strongly to DNA20:RNA33 and RNA20:RNA33 duplexes. (A) Purified BLM (0, 11, 15, 23, 30, 38, 56, 75, 110 nm) was incubated with 32P-labeled substrates as described in Materials and methods. Reactions were separated using acrylamide gel electrophoresis and analyzed using ImageQuant software. Duplexes bound by BLM migrate more slowly (open circles) than unbound duplexes (asterisk). (B) Binding of each duplex was calculated by comparing the amount of bound complex to the total amount of duplex in the reaction. Percent binding is graphed as a function of BLM concentration. (C) Purified BLM (0, 13.3, 29, 40 and 55 nm) was incubated with 32P-labeled single-stranded DNA46 or RNA46 as described in Materials and Methods. Reactions were separated using 1% agarose gels that were dried and analyzed. Substrate bound by BLM migrates more slowly (open circles) than unbound substrate (asterisk).
DISCUSSION
Cellular and animal models demonstrate that defects in RNA polymerase I-mediated rRNA transcription or ribosome function negatively impact growth (4,33). In cell culture, deletion of the RNA polymerase I transcription factor Tif-1a leads to cell cycle arrest and apoptosis while the Tif-1a−/− mouse model, although an embryonic lethal, produces small embryos (4). Deletion of the non-essential Rpl29 ribosomal subunit slows protein synthesis and proliferation in murine fibroblasts and produces viable yet proportionally small mice (33). Similar to these models of perturbed ribosome biogenesis and function, cultured BS cells and persons with BS invariably display a growth defect (10–12), and although Blm−/− leads to embryonic lethality, before death and at all stages of embryogenesis, the embryos are very small (34). Importantly, BLM localizes to the nucleoli (15), associates with rDNA in vivo and is required to maintain rDNA integrity (17). As ribosome biogenesis from rRNA transcription to ribosome subunit assembly occurs predominantly in the nucleolus, we investigated a role for BLM in RNA polymerase I-mediated transcription.
AMD, a selective RNA polymerase I inhibitor, disrupts the nucleolar localization of RNA polymerase I and its related factors (25). We demonstrated that treatment of cells in culture re-localizes BLM from the nucleolus to the nucleoplasm and nucleolar periphery. This nucleolar redistribution pattern of BLM is consistent with an interaction of BLM with the RNA polymerase I transcription machinery and the interaction of BLM and rDNA (17). Using protein co-immunoprecipitation, we demonstrated that BLM physically interacts with the RNA polymerase I-specific subunit RPA194. The nucleolar re-localization of BLM in response to AMD as well and the physical association of BLM with RPA194, an RNA polymerase I subunit, suggests a role for BLM in rRNA transcription. Accordingly, the 45S rRNA transcription rate was measured in cells with either an innate or experimentally induced BLM deficiency, as 45S rRNA is the initial transcript produced by RNA polymerase I. Cells lacking BLM display a slower RNA polymerase I-mediated 45S rRNA transcription rate suggesting that BLM facilitates rRNA transcription. Published work demonstrates that BS lymphoblastoid cells in long-term culture (1 year or more) have ∼25% less rDNA than wild-type cells (17). Our observations of slowed rRNA transcription rates following short-term siRNA-mediated BLM depletion support a mechanism not explained by a decrease of template rDNA but rather one with the persistence of an impeding DNA structure. Our finding in BS cells that transient re-expression of BLM via GFP–BLM rescues the decreased rRNA transcription rate further supports this point. Similar data were reported in cells in the absence of the related recQ-like WRN helicase and suggest that BLM and WRN have the same or similar functions in RNA polymerase I-mediated transcription (19).
The formation of RNA–DNA hybrids during rRNA transcription and their ability to impede RNA polymerase I transcription complexes have been demonstrated in bacterial and yeast systems (29,30). When unwinding fully duplexed DNA, BLM requires a 3′ overhang of at least eight nucleotides and it is upon this strand that BLM translocates as it unwinds the duplex (14,31). BLM can unwind hybrid RNA–DNA substrates with a DNA overhang in vitro (31). Thus, we asked whether the unwinding ability of BLM using a hybrid duplex would proceed with a 3′ overhang of RNA. Our results show that BLM binds and unwinds DNA overhang substrates DNA20:DNA33 and RNA20:DNA33, but not RNA overhang substrates DNA20:RNA33 or RNA20:RNA33. The similarities between substrate unwinding and binding ability of BLM suggest that substrate binding may be the significant determinant of unwinding. This is consistent with previous studies of BLM using G4 DNA and Holliday substrates, in which G4 DNA is both better bound and unwound (35). Our results are also similar to those found for bacterial, archaeal and eukaryotic replicative helicases that unwind RNA–DNA hybrid duplexes only when binding to and translocating on the DNA strand (36). Our in vitro data support a role for BLM in rRNA transcription, rather than rRNA post-transcriptional processing or maturation, as helicases involved in these latter processes possess the ability to unwind RNA–RNA duplexes (37).
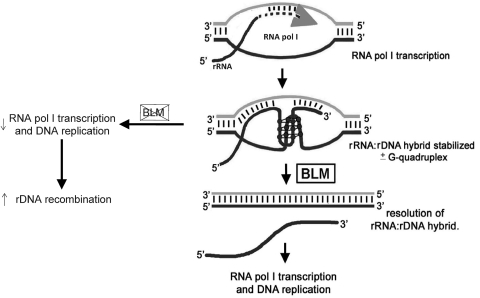
These experiments suggest a novel role for the BLM helicase in RNA polymerase I-mediated transcription. We propose a model by which nucleolar BLM maintains rDNA stability and promotes RNA polymerase I transcription (Fig. 6). The GC-rich nature of rDNA and rRNA transcripts may promote the formation of RNA–DNA hybrids, generating R-loops (38). R-loop formation within rDNA, and its inhibition of rRNA transcription, occurs in Escherichia coli and S. cerevisiae (29,30). Thus, the propensity of actively transcribed rDNA to form RNA–DNA hybrids is conserved. BLM readily unwinds RNA–DNA hybrids and R-loops (31); our data support these suggested functions of BLM while emphasizing a requirement for a 3′ DNA overhang. BLM may disrupt RNA–DNA hybrids and R-loops formed during RNA polymerase I transcription, as these structures otherwise inhibit progression of the transcription complex (29). Additionally, co-transcriptionally formed stable RNA–DNA hybrids, and thus R-loops, may induce genomic instability and recombination (39), a phenomenon termed transcription-associated recombination [reviewed in (28)]. Excessive variability in the number of Q-bright satellites in BS lymphocytes indicates a high frequency of recombination between rDNA gene clusters (23). Recent work using BS cells and various DNA repair-defective cells demonstrates that the recombination within the rDNA is relatively unique to cells lacking BLM (40). Therefore, this model may explain both the slower rate of RNA polymerase I transcription and the well-known high frequency of recombination and rDNA instability in BS cells (17,23).
Figure 6.
Model for the role of BLM in rRNA transcription. During RNA polymerase I-mediated 45S rRNA transcription, RNA–DNA hybrids may form between the nascent rRNA transcript and the template rDNA (28). We propose that BLM unwinds the RNA–DNA hybrid so that transcription and replication proceed unaffected. In the absence of BLM, the RNA–DNA hybrid may remain unresolved resulting in retardation of further rRNA transcription, stalling of transcription and replication forks, and the induction of recombination within the rDNA (37).
Severe proportional dwarfism was among the first characteristics documented in BS, and one for which a satisfactory explanation is still lacking (12). Our data suggest the hypothesis that deficient rRNA transcription may contribute to the growth deficiency typical of BS. All BS neonates exhibit IUGR, with typical birth weights of 1.5 and 1.8 kg for males and females, respectively, 2.6 and 3.2 kg SD below the mean considering the average gestational ages of 35 and 38 weeks, respectively (12). In the general population, ∼92% of babies born with IUGR eventually exhibit catch-up growth during childhood (41). This contrasts with the observation that none of the BS infants exhibit catch-up growth. Proportional dwarfism is invariably present in BS with a typical adult height and weight of ∼133 cm and 40 kg (12). These observations suggest that the etiology of the growth defect in BS is fundamentally different from that observed in many other common causes of growth stunting, such as growth hormone deficiency, thyroid hormone deficiency or malnutrition. Accordingly, these clinical parameters have been evaluated in BS persons, ranging from 9 months to 28 years of age. Growth hormone secretion, thyroid hormone levels and intestinal absorption are found within the normal range, excluding these as probable etiological factors in growth retardation (12).
In contrast, the growth characteristics of BS persons are similar to those observed in some rare disorders designated as ‘ribosomopathies’. Namely, Diamond–Blackfan anemia includes anemia as well as growth stunting, and is owing to mutations in various genes encoding ribosomal proteins which in turn decrease ribosome biogenesis (5). Approximately 20% of those affected by Diamond–Blackfan anemia exhibit IUGR, of which only 40% will exhibit catch-up growth during childhood, not quite analogous to that seen in BS yet distinctly different from that typical of IUGR in the general population (42). Short stature is also a prominent feature of Shwachman–Diamond syndrome, a disorder caused by mutations in the SBDS gene. The specific function of the SBDS gene product is unknown, but it associates with the 60S ribosomal subunit; its mutation leads to defective ribosome biogenesis (5,43). Cartilage hair hypoplasia, its variant metaphyseal dysplasia without hypotrichosis, and anauxetic dysplasia are congenital syndromes displaying significant growth retardation; all are owing to various mutations in the RMRP gene that encodes the untranslated RNA component of RNase MRP (44–46). RNase MRP is an endoribonuclease required for processing the precursor 45S rRNA into mature rRNA and, when deficient, leads to defective rRNA processing and impaired ribosome biogenesis (5). Mutation of the recQ-like helicase WRN is responsible for Werner syndrome, a disorder characterized by premature aging, an increased incidence of malignancy, as well as growth stunting and short stature. Similar to BLM, the WRN helicase localizes to the nucleoli and facilitates rRNA transcription (19). These disorders clearly suggest links between impaired ribosome biogenesis and growth deficiency.
Many BS persons have deregulated insulin signaling, observed as either insulin resistance in younger children, or as insulin-resistant diabetes mellitus in young adults in their twenties (12). The presentation of insulin resistance is unique in BS, as it begins in early childhood and BS children are quite thin; this contrasts with the typical presentation of insulin-resistant diabetes mellitus in older overweight adults. Whereas postnatal growth hormone is a major mitogen, insulin is the predominant mitogen in utero. A familiar illustration is the macrosomia characteristic of neonates born to poorly controlled diabetic mothers whose fetuses are exposed to high insulin levels in utero (47). Importantly, growth hormone and insulin both signal through the insulin receptor substrate-1 (IRS-1) pathway (48). In addition to the classical roles of IRS-1 in the insulin-signaling pathway, IRS-1 localizes to the nucleoli, physically interacts with the RNA polymerase I transcription factor UBF and promotes rRNA transcription (49), suggesting that the growth-promoting effect of insulin is mediated in part through its effects on ribosome biogenesis. Furthermore, Irs1−/− mice are insulin-resistant and have prenatal and postnatal growth retardation (50) similar to that from impaired ribosome biogenesis. Thus, the absence of nucleolar BLM may limit the full potential of insulin- and growth hormone-stimulated rRNA transcription, also limiting growth in BS. Our data may help in understanding the relationship between the cellular and metabolic abnormalities as well as the growth deficiency observed in BS.
MATERIALS AND METHODS
Cell lines
MCF7 and 293T cells were obtained from ATCC and cultured in Dulbecco's Modified Eagle Medium (Invitrogen) containing 10% heat-inactivated fetal bovine serum (FBS; Hyclone). GM08505 cells were obtained from Coriell Cell Repository and cultured in Minimal Essential Medium (Invitrogen) containing 10% heat-inactivated FBS (Hyclone). GM01806, GM03403, GM11973 and GM16375 lymphoblastoid cells were obtained from Coriell Cell Repository and cultured in RPMI (Invitrogen) containing 15% heat-inactivated FBS (Hyclone). All cells were cultured at 37°C and 5% CO2.
Transfection, immunofluorescence and antibodies
pGFP–BLM was generated by cloning BLM cDNA into pEGFP-C1 (Clontech). MCF7 cells were transfected with the pGFP–BLM expression vector using Effectene Transfection Reagent (Qiagen) according to manufacturer's instructions. Forty-eight hours after pGFP–BLM transfection, cells were treated with either 5 µg/ml AMD or 30 µg/ml αAMT for 1 h at 37°C and then processed for immunofluorescence. MCF7 cells were treated with 4NQO according to published protocols (18). Forty-eight hours after pGFP–BLM transfection, cells were treated with 0.8 µg/ml 4NQO in DMSO (or mock-treated with DMSO) for 1 h at 37°C, then into fresh media for 2 h and processed for immunofluorescence. Finally, for HU treatment, 48 h after pGFP–BLM transfection, MCF7 cells were treated with 2 mm HU for 16 h, and then processed for immunofluorescence. Following drug treatments, cells were washed in phosphate-buffered saline (PBS), fixed with 10% formamide (Sigma), stained with anti-RPA194 (Santa Cruz, sc-48385), anti-nucleophosmin (NPM, Abcam FC82291) or anti-PML (Santa Cruz, sc-966), mounted on glass slides and subsequently scored for localization of GFP–BLM using a Zeiss Axiovert 200M microscope and Axiovision 4.5 software. Western blotting was performed according to standard procedures using anti-BLM (Bethyl Laboratories, A300-110A), anti-Lamin B (Santa Cruz Biotech, sc-6217), anti-RPA194 (Santa Cruz Biotech, sc-48385), anti-RNA polymerase II (Abcam, ab817) or anti-WRN (Abcam, ab66606). GM08505 cells were transfected with pGFP–BLM, pGFP-BLM-D795A or pGFP-empty using FuGENE HD Transfection Reagent (Roche) according to manufacturer's instructions. 293T cells were transfected with either BLM Silencer pre-designed siRNA (Ambion) (5′-GGAAGUUGUAUGCACUACCTT-3′) or Silencer negative control siRNA (Ambion) using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to manufacturer's instructions.
Protein co-immunoprecipitation
Protein co-immunoprecipitations used 293T nuclear lysates prepared according to published protocols (51). Antibodies used in co-immunoprecipitation included anti-BLM (Santa Cruz Biotech, sc-7790), anti-RPA194 (Santa Cruz Biotech, sc-48385) and anti-RNA polymerase II (Abcam, ab817). Protein–antibody complexes were captured with Dynabeads Protein G (Invitrogen, 100-04D), washed, eluted and separated using 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
Pulse-chase assays
Pulse-chase analysis was performed as previously described (52). Cells were pulse-labeled for 30 min in medium containing 2.5 µCi/ml 3H-uridine and chased in medium containing 0.5 mm uridine for the indicated amount of time. RNA was isolated using an RNeasy Midi Kit (Qiagen) or TRI-Reagent (MRC), separated by electrophoresis in a 1% formaldehyde–MOPS agarose gel, transferred to a nylon membrane and exposed to either Kodak BioMax MS film with BioMax TranScreen LE Intensifying Screen or placed in a Tritium Storage Phosphor Screen (Amersham Biosciences) for several days. Methylene blue or ethidium bromide staining demonstrated equal RNA loading. Band intensities were determined by densitometry analysis with ImageQuant software.
Biotin-labeled nuclear run-on assay
In vivo biotin-labeled nuclear run-on assays were performed as previously described (53) with minor modifications. After siRNA transfection, cells were collected, washed in PBS and lysed in lysis buffer [final 13.3 mm Tris, pH 7.5, 340 mm sucrose, 13.3 mm NaCl, 53 mm KCl, 2 mm ethylenediamine tetraaceticacid (EDTA), 0.5 mm spermidine, 0.13 mm spermine, 0.1% Triton X-100, 2 mm MgCl2] for 5 min on ice. Lysates were added to a sucrose cushion solution (final 13.3 mm Tris, pH 7.5, 1.2 M sucrose, 13.3 mm NaCl, 53 mm KCl, 2 mm EDTA, 0.5 mm ethylene glycol tetraacetic acid, 0.5 mm spermidine, 0.13 mm spermine, 2 mm MgCl2) and nuclei were collected by centrifugation at 2400g for 30 min at 4°C. Nuclei were re-suspended in freezing buffer (50 mm Tris, pH 8.3, 40% glycerol, 5 mm MgCl2, 0.1 mm EDTA) and isolated nuclei incubated in transcription buffer (2X: 200 mm KCl, 20 mm Tris, pH 8.0, 5 mm MgCl2, 4 mm dithiothreitol, 4 mm adenosine triphosphate, guanosine triphosphate, cytosine triphosphate, 200 mm sucrose, 20% glycerol) with 1 mm biotin-16-UTP (Roche) for 1 h at 30°C. Transcription was terminated by passage through a 25 G needle and DNase I (Roche) treatment according to manufacturer's instructions. Total RNA was isolated using Tri-Reagent (MRC). Eight micrograms of total RNA was bound to Dynabeads M-280 streptavidin (Invitrogen) for 1 h according to manufacturer's instructions. Beads were washed four times with 500 μl of 2X SSC, 15% formamide, 0.2% Tween-20 for 5 min with gentle rotation, then once in 1 ml of 2X SSC. RNA was eluted in H2O by heating to 95°C for 1 min, and used in cDNA synthesis using ThermoScript RT-PCR system (Invitrogen) according to manufacturer's instructions. cDNA was amplified using primers specific for GAPDH (forward primer: GACATCAAGAAGGTGGTGAAG, reverse primer: CCAGGAAATGAGCTTGACAAAG) and the 5′ external transcribed spacer of 45S rDNA (forward primer: GCCGGGTCCGAGCCGCGACGG, reverse primer: GCGGCGGGCGGGACGGCGAGG) using Taq DNA polymerase (Roche), separated by agarose gel electrophoresis and analyzed using ImageQuant software.
Protein purification
I. Hickson (University of Oxford, Oxford, UK) provided the pYES-BLM expression vector (pJK1). BLM was purified as previously described (54). Briefly, hexa-histidine (6X-His)-tagged BLM was over-expressed in S. cerevisiae. Yeast were lysed at 20 k psi using a French Press Cell Disrupter (Thermo) and lysates were separated by ultracentrifugation at 65 000g for 1 h at 4°C. The cleared lysate was purified by FPLC using Ni-NTA Superflow (Qiagen), followed by Q-Sepharose (Sigma). The purity of the resultant BLM was determined using 8% SDS–PAGE and staining of the gel with SYPRO Ruby Protein Gel Stain (Sigma) and analysis using ImageQuant software as previously described.
Helicase assays and EMSAs
Oligonucleotides were purchased from Invitrogen. Oligonucleotide sequences (5′–3′ orientation) are as follows: DNA20: CGCTAGCAATATTCTGCAGC, DNA33: GCTGCAGAATATTGCTAGCGGGAATTCGGCGCG, RNA20: CGCUAGCAAUAUUCUGCAGC, RNA33: GCUGCAGAAUAUUGCUAGCGGGAAUUCGGCGCG, DNA46: GCGCGGAAGCTTGGCTGCAGAATATTGCTAGCGGGAATTCGGCGCG, RNA46: GCGCGGAAGCUUGGCUGCAGAAUAUUGCUAGCGGGAAUUCGGCGCG RNA20, DNA20, DNA46 and RNA46 were 32P end-labeled using polynucleotide kinase (NEB) according to manufacturer's instructions. Substrates were duplexed by heating to 95°C for 5 min and slowly cooling. Helicase assays were performed as previously described (55), with the exception that reactions were performed with 20 μl final volume using 2fmol of substrate per reaction. Helicase products were separated on 12% non-denaturing polyacrylamide gels. For EMSA, reactions were set up identically to those in helicase assays but with ATP omitted. Binding products were separated using 4% polyacrylamide, 5% glycerol, 1X TBE gels electrophoresed at 4°C. EMSA reactions using single-stranded substrate were resolved on 1% agarose TBE gels electrophoresed at room temperature. Helicase and binding assays were analyzed using ImageQuant software.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (CA117898 to J.G.); The Bloom's Syndrome Foundation to (J.G.); The Ohio State University Pelotonia Fellowship Program to (P.G.); and The Ohio State University Medical Scientist Training Program (MSTP) to (P.G.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs Debbie Parris and George Thomas for experimental suggestions and helpful discussions, and Cathy Ebert and members of the Groden laboratory for helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Schwarzacher H.G., Wachtler F. The nucleolus. Anat. Embryol. 1993;188:515–536. doi: 10.1007/BF00187008. [DOI] [PubMed] [Google Scholar]
- 2.Ayrault O., Andrique L., Fauvin D., Eymin B., Gazzeri S., Seite P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene. 2006;25:7577–7586. doi: 10.1038/sj.onc.1209743. [DOI] [PubMed] [Google Scholar]
- 3.Birch J.L., Zomerdijk J. Structure and function of ribosomal RNA gene chromatin. Biochem. Soc. Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Grone H.J., Schutz G., Grummt I. Genetic inactivation of the transcription factor TIF-1A leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 5.Narla A., Ebert B.L. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang S. Building an efficient factory: where is pre-rRNA synthesized in the nucleolus? J. Cell Biol. 2002;157:739–741. doi: 10.1083/jcb.200204159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koberna K., Malinsky J., Pliss A., Masata M., Vecerova J., Fialova M., Bednar J., Raska I. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of ‘Christmas trees’ in situ. J. Cell Biol. 2002;157:743–748. doi: 10.1083/jcb.200202007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland H.G., Mumford G.K., Newton K., Ford L.V., Farrall R., Dellaire G., Caceres J.F., Bickmore W.A. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 2001;10:1995–2011. doi: 10.1093/hmg/10.18.1995. [DOI] [PubMed] [Google Scholar]
- 9.Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 10.German J. Blooms syndrome. I. Genetical and clinical observations in the first twenty-seven patients. Am. J. Hum. Genet. 1969;21:196–227. [PMC free article] [PubMed] [Google Scholar]
- 11.Lechner J.F., Kaighn M.E., Jetten A.M., Groden J., German J. Blooms syndrome cells have an abnormal serum growth response. Exp. Cell Res. 1983;145:381–388. doi: 10.1016/0014-4827(83)90016-2. [DOI] [PubMed] [Google Scholar]
- 12.Diaz A., Vogiatzi M.G., Sanz M.M., German J. Evaluation of short stature, carbohydrate metabolism and other endocrinopathies in Blooms syndrome. Horm. Res. 2006;66:111–117. doi: 10.1159/000093826. [DOI] [PubMed] [Google Scholar]
- 13.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Blooms syndrome gene product is homologous to recQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 14.Karow J.K., Chakraverty R.K., Hickson I.D. The Blooms syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 15.Yankiwski V., Marciniak R.A., Guarente L., Neff N.F. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl Acad. Sci. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yankiwski V., Noonan J.P., Neff N.F. The C-terminal domain of the Bloom syndrome DNA helicase is essential for genomic stability. BMC Cell Biol. 2001;2:11. doi: 10.1186/1471-2121-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schawalder J., Paric E., Neff N.F. Telomere and ribosomal DNA repeats are chromosomal targets of the Bloom syndrome DNA helicase. BMC Cell Biol. 2003;4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray M.D., Wang L., Youssoufian H., Martin G.M., Oshima J. Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp. Cell Res. 1998;242:487–494. doi: 10.1006/excr.1998.4124. [DOI] [PubMed] [Google Scholar]
- 19.Shiratori M., Suzuki T., Itoh C., Goto M., Furuichi Y., Matsumoto T. WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene. 2002;21:2447–2454. doi: 10.1038/sj.onc.1205334. [DOI] [PubMed] [Google Scholar]
- 20.Versini G., Comet I., Wu M., Hoopes L., Schwob E., Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heo S.J., Tatebayashi K., Ohsugi I., Shimamoto A., Furuichi Y., Ikeda H. Blooms syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells. 1999;4:619–625. doi: 10.1046/j.1365-2443.1999.00288.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee S.K., Johnson R.E., Yu S.L., Prakash L., Prakash S. Requirement of yeast Sgs1 and Srs2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 23.Therman E., Otto P.G., Shahidi N.T. Mitotic recombination and segregation of satellites in Blooms syndrome. Chromosoma. 1981;82:627–636. doi: 10.1007/BF00285772. [DOI] [PubMed] [Google Scholar]
- 24.Drygin D., Rice W.G., Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 25.Jordan P., Mannervik M., Tora L., Carmo-Fonseca M. In vivo evidence that TATA-binding protein/ SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell. Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svarcova O., Strejcek F., Petrovicova I., Avery B., Pedersen H.G., Lucas-Hahn A., Niemann H., Laurincik J., Maddox-Hyttel P. The role of RNA polymerase I transcription and embryonic genome activation in nucleolar development in bovine preimplantation embryos. Mol. Reprod. Dev. 2008;75:1095–1103. doi: 10.1002/mrd.20865. [DOI] [PubMed] [Google Scholar]
- 27.Seither P., Coy J.F., Pouska A., Grummt I. Molecular cloning and characterization of the cDNA encoding the largest subunit of mouse RNA polymerase I. Mol. Gen. Genet. 1997;255:180–186. doi: 10.1007/s004380050487. [DOI] [PubMed] [Google Scholar]
- 28.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hraiky C., Raymond M.A., Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J. Biol. Chem. 2000;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 30.Hage A.E., French S.L., Beyer A.L., Tollervey D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popuri V., Bachrati C.Z., Muzzolini L., Mosedale G., Costantini S., Giacomini E., Hickson I.D., Vindigni A. The human recQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 32.Mohaghegh P., Karow J.K., Brosh R.M., Bohr V.A., Hickson I.D. The Blooms and Werners syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirn-Safran C.B., Oristian D.S., Focht R.J., Parker S.G., Vivian J.L., Carson D.D. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev. Dyn. 2007;236:447–460. doi: 10.1002/dvdy.21046. [DOI] [PubMed] [Google Scholar]
- 34.Chester N., Kuo F., Kozak C., O'Hara C.D., Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Blooms syndrome gene. Genes Dev. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber M.D., Lee D.C., Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin J.H., Kelman Z. The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates. J. Biol. Chem. 2006;281:26914–26921. doi: 10.1074/jbc.M605518200. [DOI] [PubMed] [Google Scholar]
- 37.Kressler D., Hurt E., Babler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Salazar M., Thompson B.D., Kerwin S.M., Hurley L.H. Thermally induced DNA.RNA hybrid to G-quadruplex transitions: possible implications for telomere synthesis by telomerase. Biochemistry. 1996;35:16110–16115. doi: 10.1021/bi961442j. [DOI] [PubMed] [Google Scholar]
- 39.Huertas P., Aguilera A. Cotranscriptionally formed stable DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Killen M.W., Stults D.M., Adachi N., Hanakahi L., Pierce A.J. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 2009;18:3417–3428. doi: 10.1093/hmg/ddp282. [DOI] [PubMed] [Google Scholar]
- 41.Albertsson-Wikland K., Wennergren G., Wennergren M., Vilbergsson G., Rosberg S. Longitudinal follow-up of growth in children born small for gestational age. Acta Paediatr. 1993;82:438–443. doi: 10.1111/j.1651-2227.1993.tb12718.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen S., Warszawski J., Bader-Meunier B., Tchernia G., Da Costa L., Marie I., Dommergues J.P. Diamond–Blackfan anemia and growth status: the French registry. J. Pediatr. 2005;147:669–673. doi: 10.1016/j.jpeds.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 43.Makitie O., Ellis L., Durie P.R., Morrison J.A., Sochett E.B., Rommens J.M., Cole W.G. Skeletal phenotype in patients with Shwachman–Diamond syndrome and mutations in SBDS. Clin. Genet. 2004;65:101–112. doi: 10.1111/j.0009-9163.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- 44.Ridanpaa M., van Eenennaarn H., Pelin K., Chadwick R., Johnson C., Yuan B., vanVenrooij W., Pruijn G., Salmela R., Rockas S., et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 45.Bonafe L., Schmitt K., Eich G., Giedion A., Superti-Furga A. RMRP gene sequence analysis confirms a cartilage-hair hypoplasia variant with only skeletal manifestations and reveals a high density of single-nucleotide polymorphisms. Clin. Genet. 2002;61:146–151. doi: 10.1034/j.1399-0004.2002.610210.x. [DOI] [PubMed] [Google Scholar]
- 46.Thiel C.T., Horn D., Zabel B., Ekici A.B., Salinas K., Gebhart E., Ruschendorf F., Sticht H., Spranger J., Muller D., et al. Severely incapacitating mutations in patients with extreme short stature indentify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am. J. Hum. Genet. 2005;77:795–806. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riskin A., Garcia-Prats J.A. Infant of a diabetic mother. 2011 In Basow, D.S., (ed). Up To Date, Waltham, MA. [Google Scholar]
- 48.Myers M.G., Jr., Grammer T.C., Wang L.M., Sun X.J., Pierce J.H., Blenis J., White M.F. Insulin receptor substrate-1 mediates phosphatidylinositol 3′-kinase and p70S6K signaling during insulin, insulin-like growth factor-1, and interleukin-4 stimulation. J. Biol. Chem. 1994;269:28783–28789. [PubMed] [Google Scholar]
- 49.Tu X., Batta P., Innocent N., Prisco M., Casaburi I., Belletti B., Baserga R. Nuclear translocation of insulin receptor substrate-1 by oncogenes and Igf-1. Effect on ribosomal RNA synthesis. J. Biol. Chem. 2002;277:44357–44365. doi: 10.1074/jbc.M208001200. [DOI] [PubMed] [Google Scholar]
- 50.Tamemoto H., Kadowaki T, Tobe K., Yagi T., Sakura H., Hayakawa T., Terauchi Y., Ueki K., Kaburagi Y., Satoh S., et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 51.Jiang K., Pereira E., Maxfield M., Russell B., Goudelock D.M., Sanchez Y. Regulation of Chk1 includes chromatin association and 14–3–3 binding following phosphorylation on Ser-345. J. Biol. Chem. 2003;278:25207–25227. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- 52.Schlosser I., Holzel M., Murnseer M., Burtcher H., Weidle U.H., Eick D. A role for c-MYC in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patrone G., Puppo F., Cusano R., Scaranari M., Ceccherini I., Puliti A., Ravazzola R. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. Biotechniques. 2000;29:1016–1017. doi: 10.2144/00295st02. 1012–1014. [DOI] [PubMed] [Google Scholar]
- 54.Russell B., Bhattacharyya S., Keirsey J., Sandy A., Grierson P., Perchiniak E., Kavecansky J., Acharya S., Groden J. Chromosome breakage is regulated by the interaction of the BLM helicase and topoisomerase IIα. Cancer Res. 2011;71:561–571. doi: 10.1158/0008-5472.CAN-10-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lillard-Wetherell K., Machwe A., Langland G.T., Combs K.A., Behbehani G.K., Schonberg S.A., German J., Turchi J.J., Orren D.K., Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.