Abstract
In follow-up of a recent genome-wide association study (GWAS) that identified a locus in chromosome 2p21 associated with risk for renal cell carcinoma (RCC), we conducted a fine mapping analysis of a 120 kb region that includes EPAS1. We genotyped 59 tagged common single-nucleotide polymorphisms (SNPs) in 2278 RCC and 3719 controls of European background and observed a novel signal for rs9679290 [P = 5.75 × 10−8, per-allele odds ratio (OR) = 1.27, 95% confidence interval (CI): 1.17–1.39]. Imputation of common SNPs surrounding rs9679290 using HapMap 3 and 1000 Genomes data yielded two additional signals, rs4953346 (P = 4.09 × 10−14) and rs12617313 (P = 7.48 × 10−12), both highly correlated with rs9679290 (r2 > 0.95), but interestingly not correlated with the two SNPs reported in the GWAS: rs11894252 and rs7579899 (r2 < 0.1 with rs9679290). Genotype analysis of rs12617313 confirmed an association with RCC risk (P = 1.72 × 10−9, per-allele OR = 1.28, 95% CI: 1.18–1.39) In conclusion, we report that chromosome 2p21 harbors a complex genetic architecture for common RCC risk variants.
INTRODUCTION
Kidney cancer accounts for nearly 4% of cancer incidence and 2% of cancer mortality in the United States, with over 58 000 new cases and 13 000 deaths estimated for 2010 (1). Renal cell carcinoma (RCC) represents 90% of renal malignancies in adults (2,3). Several epidemiological risk factors have been established for risk for sporadic RCC, namely hypertension, obesity and smoking (4,5). Genetic risk factors contribute to RCC risk as observed in pedigrees with von Hippel–Lindau (VHL) syndrome and other rare syndromes, such as hereditary papillary renal cell carcinoma, Birt–Hogg–Dube and hereditary leiomyomatosis and renal cell cancer (6–8). Also, the lifetime risk for developing RCC is doubled for those with a first-degree relative with RCC (9–11).
Genome-wide association studies (GWAS) have emerged as a tool to discover common variants with small effect sizes in cancer (12). Recently, a GWAS for RCC identified two single-nucleotide polymorphisms (SNPs) that mapped to chromosome 2p21, rs7579899 [P = 3.2 × 10−9, per-allele odds ratio (OR) = 1.15, 95% confidence interval (CI): 1.10–1.21] and rs11894252 (P = 4.3 × 10−9, per-allele OR = 1.14, 95% CI: 1.09–1.19) (pair-wise r2 = 1.00) (13). The interval includes the endothelial PAS domain protein 1 (EPAS1) gene, a key component of the VHL pathway, which has been implicated in renal carcinogenesis (14). It is also notable that germline mutations in EPAS1, the gene that encodes HIF2α, have been detected in familial erythrocytosis (15), and common genetic variants have been associated with erythrocyte abundance supporting a role of EPAS1 in adaptation to hypoxia and high altitude (16,17).
We conducted a fine-mapping analysis of the 120 kb region flanking the EPAS1 gene on 2p21. We genotyped 59 tagged SNPs in 2278 RCC cases and 3719 controls from two nested case–control studies [the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial and the Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC) study in Finland], a hospital-based Central and Eastern European RCC (CEERCC) case–control study and Caucasians from the United States Kidney Cancer (USKC) population-based case–control study (see Materials and Methods and Fig. 1).
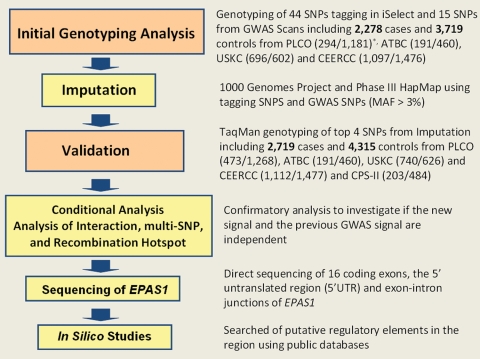
Figure 1.
Study design, sample sizes and stages of analyses. *The numbers in the parentheses next to each study in ‘Initial Genotyping Analysis’ and ‘Validation’ correspond to the number of cases and the number of controls, respectively.
RESULTS
On the basis of the initial genotyping analysis, a new SNP rs9679290 was identified to be associated with RCC risk approaching the threshold for genome-wide significance (P = 5.75 × 10−8, per-allele OR = 1.27, 95% CI: 1.17–1.39) (Fig. 2A), whereas, in this subset of the GWAS data set, the signals for the two highly correlated SNPs (rs11894252 and rs7579899) identified in the previous GWAS were not as strong (P = 1.35 × 10−3, per-allele OR = 1.17, 95% CI: 1.06–1.28 and P = 2.13 × 10−3, per-allele OR = 1.16, 95% CI: 1.05–1.28, respectively) (Table 1). Notably, rs9679290 is not correlated with either rs11894252 or rs7579899 (r2 < 0.1) (Supplementary Material, Table S1).
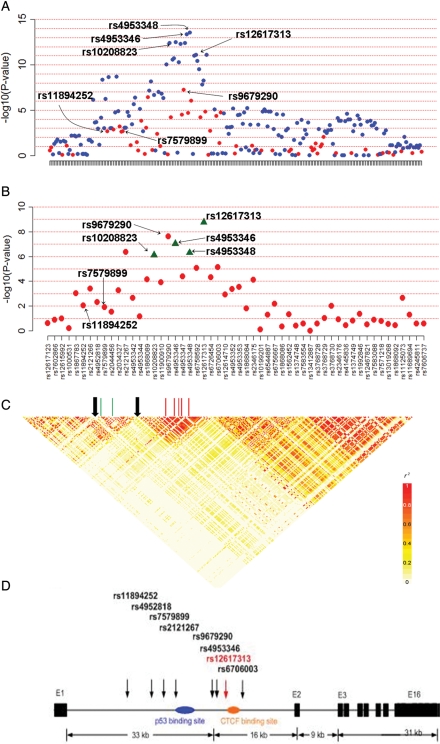
Figure 2.
Association results and LD plots for the 2p21 region. (A) The P-values (–log 10 scale) of association tests from the SNPs of the initial genotyping analysis (red dots) and from the analysis using imputed SNPs (blue dots). (B) The P-values of the four genotyped validated SNPs (green triangles) and SNPs in the initial genotyping analysis (red dots). (C) The LD structure (pairwise r2) and the location of the recombination hotspots (the black arrows) in the region. (D) EPAS1 gene structure, location of the SNPs genotyped and transcription factor binding sites in the region.
Table 1.
SNPs selected for three stage fine mapping of 2p21 region
| SNP ID | Genotype controls |
Combined (2278/3719) | PLCO (294/1181) | ATBC (191/460) | USKC (696/602) | CEERCC (1097/1476) | CPSII (203/484) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | ||
| Initial genotyping | rs9679290 | 476/980/498 | 952/1616/635 | 1.27 (1.17–1.39) | 5.75 × 10−8 | 1.45 (1.20–1.76) | 1.32 × 10−4 | 1.46 (1.10–1.93) | 8.09 × 10−3 | 1.21 (1.02–1.43) | 2.74 × 10−2 | 1.19 (1.04–1.37) | 1.12 × 10−2 | ||
| rs2121267 | 665/954/336 | 917/1614/675 | 1.25 (1.15–1.37) | 3.73 × 10−7 | 1.45 (1.19–1.75) | 2.10 × 10−4 | 1.32 (0.98–1.75) | 6.41 × 10−2 | 1.18 (0.99–1.39) | 6.23 × 10−2 | 1.22 (1.05–1.39) | 5.54 × 10−3 | |||
| rs6758592 | 259/560/303 | 310/560/219 | 1.26 (1.15–1.39) | 8.60 × 10−7 | 1.59 (1.17–2.17) | 2.63 × 10−3 | 1.48 (1.10–2.01) | 9.60 × 10−3 | 1.21 (1.02–1.44) | 2.48 × 10−2 | 1.20 (1.05–1.38) | 7.42 × 10−3 | |||
| rs6706003 | 468/1004/490 | 905/1640/671 | 1.21 (1.11–1.32) | 1.48 × 10−5 | 1.39 (1.14–1.68) | 8.45 × 10−4 | 1.30 (0.98–1.72) | 6.53 × 10−2 | 1.14 (0.97–1.35) | 1.19 × 10−1 | 1.18 (1.03–1.35) | 1.92 × 10−2 | |||
| rs2346175 | 81/250/143 | 420/791/386 | 1.31 (1.16–1.48) | 1.74 × 10−5 | 1.45 (1.20–1.76) | 1.11 × 10−4 | 1.35 (1.02–1.79) | 3.66 × 10−2 | 1.16 (0.96–1.42) | 1.29 × 10−1 | |||||
| rs11894252 | 348/559/215 | 358/562/169 | 1.17 (1.06–1.28) | 1.35 × 10−3 | 1.08 (0.79–1.49) | 5.94 × 10−1 | 1.39 (1.04–1.85) | 2.30 × 10−2 | 1.14 (0.95–1.35) | 1.58 × 10−1 | 1.15 (1.01–1.33) | 4.06 × 10−2 | |||
| rs7579899 | 349/559/214 | 355/565/169 | 1.16 (1.05–1.28) | 2.13 × 10−3 | 1.06 (0.78–1.47) | 6.79 × 10−1 | 1.37 (1.03–1.81) | 2.98 × 10−2 | 1.12 (0.94–1.33) | 2.04 × 10−1 | 1.15 (1.01–1.33) | 3.97 × 10−2 | |||
| Imputation | rs4953348a | 1.37 (1.27–1.48) | 2.77 × 10−14 | 1.78 (1.48–2.14) | 3.15 × 10−8 | 1.47 (1.15–1.88) | 2.41 × 10−3 | 1.25 (1.07–1.47) | 4.69 × 10−3 | 1.18 (1.04–1.35) | 8.66 × 10−3 | ||||
| rs4953346a | 1.37 (1.26–1.48) | 4.09 × 10−14 | 1.77 (1.47–2.13) | 4.29 × 10−8 | 1.47 (1.15–1.88) | 2.39 × 10−3 | 1.25 (1.07–1.46) | 5.18 × 10−3 | 1.18 (1.04–1.35) | 8.70 × 10−3 | |||||
| rs12620992a | 1.34 (1.24–1.45) | 4.94 × 10−13 | 1.69 (1.41–2.03) | 5.20 × 10−7 | 1.45 (1.13–1.85) | 3.56 × 10−3 | 1.21 (1.04–1.41) | 1.76 × 10−2 | 1.13 (0.99–1.29) | 3.14 × 10−2 | |||||
| rs12617313a | 1.32 (1.22–1.43) | 7.48 × 10−12 | 1.66 (1.39–2.00) | 5.30 × 10−7 | 1.51 (1.18–1.94) | 9.88 × 10−4 | 1.22 (1.04–1.42) | 1.31 × 10−2 | 1.16 (1.02–1.32) | 1.49 × 10−2 | |||||
| rs11125071a | 1.35 (1.24–1.46) | 3.58 × 10−13 | 1.66 (1.38–1.99) | 8.72 × 10−7 | 1.45 (1.13–1.85) | 2.79 × 10−3 | 1.23 (1.05–1.44) | 8.61 × 10−3 | 1.18 (1.04–1.34) | 7.44 × 10−3 | |||||
| rs10208823a | 1.34 (1.24–1.45) | 3.09 × 10−13 | 1.70 (1.42–2.04) | 4.04 × 10−7 | 1.44 (1.13–1.85) | 3.68 × 10−3 | 1.22 (1.04–1.43) | 1.31 × 10−2 | 1.13 (0.99–1.29) | 3.08 × 10−2 | |||||
| Validation | rs12617313b | 486/961/534 | 919/1529/635 | 1.28 (1.18–1.39) | 1.72 × 10−9 | 1.38 (1.17–1.63) | 1.63 × 10−4 | 1.47 (1.14–1.91) | 2.92 × 10−3 | 1.23 (1.04–1.44) | 1.26 × 10−2 | 1.15 (0.99–1.35) | 7.57 × 10−2 | 1.36 (1.08–1.72) | 8.07 × 10−3 |
| rs4953346b | 513/962/531 | 939/1546/650 | 1.24 (1.15–1.35) | 9.24 × 10−8 | 1.37 (1.16–1.62) | 1.68 × 10−4 | 1.38 (1.07–1.78) | 1.28 × 10−2 | 1.24 (1.06–1.46) | 6.89 × 10−3 | 1.09 (0.94–1.27) | 2.57 × 10−1 | 1.23 (0.98–1.54) | 7.59 × 10−2 | |
| rs4953348b | 519/968/528 | 932/1553/648 | 1.23 (1.13–1.33) | 5.02 × 10−7 | 1.35 (1.14–1.59) | 5.09 × 10−4 | 1.39 (1.07–1.79) | 1.20 × 10−2 | 1.24 (1.05–1.45) | 8.62 × 10−3 | 1.05 (0.90–1.22) | 5.10 × 10−1 | 1.27 (1.01–1.60) | 3.75 × 10−2 | |
| rs10208823b | 675/919/401 | 855/1524/724 | 1.22 (1.12–1.33) | 7.99 × 10−7 | 1.23 (1.05–1.45) | 1.13 × 10−2 | 1.34 (1.03–1.72) | 2.78 × 10−2 | 1.20 (1.03–1.41) | 2.13 × 10−2 | 1.13 (0.96–1.32) | 1.34 × 10−1 | 1.33 (1.05–1.67) | 1.48 × 10−2 | |
PLCO: Prostate, Lung, Colorectal and Ovarian cancer screening trial; ATBC: Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study; USKC: United States Kidney Cancer Case–Control Study; CEERCC: Central and Eastern European Renal Cell Cancer Case–Control study; CPS-II, Cancer Prevention Study II; All ORs were calculated adjusting for age, sex, smoking status, BMI, study center, family history of cancer (any type of cancer) and hypertension except for ORs in validation stage, where ORs were adjusted for sex and study. The numbers next to each study name correspond to the number of cases and number of controls, respectively.
aImputed SNPs.
bValidated SNPs by TaqMan.
To investigate the possibility that a more complex genetic architecture underlies the association with chromosome 2q21 (18), we imputed genotypes across the 120 kb surrounding rs9679290 (which included the two previously reported SNPs) using two publicly available reference data sets: 1000 Genomes Project March 2010 release (http://www.1000genomes.org/page.php) and Phase III HapMap (19,20). Of the imputed 304 SNPs tested by association analysis, we observed a promising new signal at rs4953348, which is highly correlated with rs9679290 (P = 2.77 × 10−14, per-allele OR = 1.37, 95% CI: 1.27–1.48) (Fig. 2A). We also note that we did not observe any new significant associations with rare variants with minor allele frequencies (MAF) less than 5% in our data set.
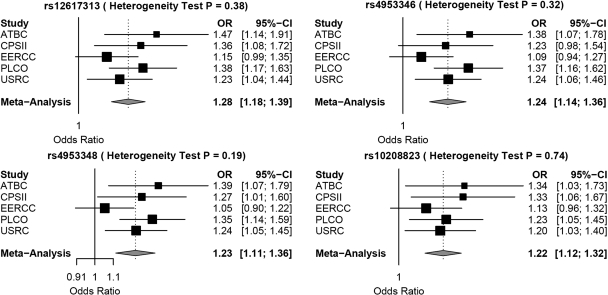
Four correlated SNPs (rs4953348, rs4953346, rs10208823 and rs12617313) among the top hits were genotyped in five studies [American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS-II) was added to PLCO, ATBC, CEERCC and USKC] (2481 cases and 4203 controls) to validate the association signals (Fig. 3). For the five studies combined, rs12617313 achieved genome-wide significance (P = 1.72 × 10−9) (Fig. 2B and Table 1).
Figure 3.
Forest plots and heterogeneity tests for the four validated EPAS1 SNPs.
A conditional analysis was performed to determine whether the effect observed for rs12617313 was independent of the previously reported markers in the GWAS, namely rs11894252 and rs7579899 (see Materials and Methods). When adjusted for rs12617313, the associations due to rs11894252 and rs7579899 were attenuated (from P = 1.35 × 10−3 and P = 2.12 × 10−3 to P = 4.09 × 10−1 and P = 4.69 × 10−1, respectively). When the two newly genotyped SNPs, rs12617313 and rs4953346, were evaluated in an analysis conditioned on rs11894252, the association signals remained notable and significant within the region (P = 3.08 × 10−4 and 2.70 × 10−4, respectively). A comparable finding was observed after conditioning for rs7579899 (data not shown).
To investigate whether the previously reported GWAS locus and our new markers were independent, we conducted an interaction test between rs11894252 and rs12617313, by fitting a logistic regression model that includes the main effects of both SNPs and their interaction term and covariates. The result showed that the interaction term was not significant (P = 0.45). Additionally, we performed an analysis of sets of SNPs to examine whether additional SNPs across this region might capture or explain the new signal we are reporting (rs4953346 and rs12617313) as well as the reported GWAS signal (marked by rs11894252). We fit a logistic regression model including a set of SNPs drawn from each of two regions of interest (rs12617313 and rs11894252) and six additional SNPs selected so that the pair-wise linkage disequilibrium (LD) (r2) among all the SNPs in the set was ≤0.2. None of the SNPs except rs12617313 (P = 0.007) showed a significant association (P < 0.1) with RCC risk. Similar analyses using r2 thresholds of 0.4 and 0.6 revealed similar results, suggesting two or more signals.
The two previously reported GWAS SNPs, rs11894252 and rs7579899, are strongly correlated (r2 = 1.0), but minimally correlated with the two SNPs that we identified by imputation, rs12617313 and rs4953346 (Fig. 2C; Supplementary Material, Table S1). In an analysis of 4203 controls, we used SequenceLDhot and identified strong evidence of a recombination hotspot separating the SNPs identified in the GWAS from the new SNPs reported here, rs12617313 and rs4953346 (Fig. 2C and Supplementary Material, Fig. S1).
To further investigate interactions between smoking status and genetic variants in EPAS1, which were reported by the previous GWAS (13), we conducted a series of pooled analyses stratified by smoking for the two sets of EPAS1 variants (see Materials and Methods). There was a notable interaction between the GWAS SNP (rs7579899) and smoking (P-interaction = 0.036) (Supplementary Material, Table S2). In contrast, no interaction was identified for the two new SNPs (rs12617313 and rs4953346) and smoking (P-interaction = 0.272 and 0.378, respectively).
Since 80% of clear cell RCCs are reported to have VHL somatic inactivation through either genetic or epigenetic mechanisms (21), the entire coding regions of VHL in 507 RCCs were sequenced to investigate whether common germline variants in EPAS1 were associated with VHL alterations in RCCs (see Materials and Methods). We observed that cases with germline EPAS1 variants in the new region were more likely to have tumor VHL alterations, with the strongest association observed for rs12617313 (P = 0.006, OR = 1.82, 95% CI, 1.19–2.80) (Supplementary Material, Table S3). Notably, the high-risk allele, A, was associated with VHL alterations. In contrast, germline EPAS1 variants identified by GWAS were not associated with VHL alterations in RCCs (P > 0.2). No change in results was observed after adjustment for stage or grade.
In a re-sequence analysis of the 16 coding exons, the 5′ untranslated region (5′ UTR) and exon–intron junctions of EPAS1 (GenBank NM_001430) in 94 cases of RCCs (see Materials and Methods), we identified a common synonymous coding variant (c.1908T>C; N636N) in exon 12. This together with two novel 5′ UTR variants (c.-58insC and c.-140G>A) were confirmed in 100 CEPH (Centre d'Etudes du Polymorphisme Humain) controls. On the basis of the low MAF, they were not strongly correlated with the new SNPs described in our fine-mapping study (data not shown). The lack of observed coding variation is consistent with the high degree of coding sequence conservation of EPAS1 across species and with the paucity of EPAS1 common missense variants in the public SNP database (http://www.ncbi.nlm.gov/projects/SNP). Furthermore, nucleotide sequence alignment showed that the non-coding EPAS1 region containing the high-risk SNPs (rs12617313, rs4953346 and rs9679290) is evolutionarily conserved among species. As the strongest new signals were clustered in intron 1 of EPAS1, we searched for putative regulatory elements using ORegAnno and other public databases. We identified two transcription binding sites (p53 and CCCTC-binding factor) adjacent to the new high-risk variants and a distant iron-responsive element (IRE) in the 5′ UTR of the mRNA that regulates HIF2α (Fig. 2D). The new signals are not in LD with variants in the transcription-binding sites or IRE. In addition, microRNA public databases examined did not suggest that these are surrogate SNPs in strong LD with signals that map to microRNA coding sequences in chromosome 2p21.
DISCUSSION
Our fine-mapping study of the 2p21 locus, that harbors a plausible candidate gene for RCC risk, namely EPAS1, reveals a more complex genetic architecture underlying the association signal discovered in the initial GWAS (13). In this detailed analysis we investigated additional SNPs and integrated imputation data drawn from the 1000 Genome Project and the HapMap 3 data sets followed by testing of select variants. We observed a new set of SNPs (rs4953346, rs12617313 and rs9679290) that are not well correlated with the initial GWAS SNPs (rs11894252 and rs7579899). Interestingly, the signals decreased in the conditional analysis below the threshold for genome-wide significance, but still retained region-wide significance, namely after correction for the number SNPs examined in this region. As there is a recombination hotspot predicted to separate the two sets of SNPs, we suggest that there is evidence for a more complex haplotype. Further studies are needed to confirm the presence of a second independent locus or alternatively, a long-range haplotype with more than one functional element. We also recognize that synthetic association could underlie the signal and hence we investigated the possibility by testing associations using SNPs with MAF between 3 and 5% from our imputation and data, but no variants with MAF < 5% were identified among the top most significant association findings (18).
This region on 2p21 is particularly interesting because of the candidate gene that resides within the association interval identified in the initial GWAS. The role of EPAS1 variants in renal carcinogenesis is biologically possible because dysregulation of HIF2α has been implicated in RCC (14). Under normal oxygen conditions, HIFα is hydroxylated by a family of oxygen-dependent prolyl-hydroxylases (22) and the pVHL complex binds to the hydroxylated HIF1α or HIF2α subunits for ubiquitin-mediated degradation (23). Hypoxia or inactivation of pVHL causes HIFα subunits to accumulate and bind to HIF-response elements, increasing transcription of erythropoietin and anti-apoptotic and proliferative genes. It has been demonstrated that elimination of HIF2α is sufficient to suppress tumor growth in vivo and an apparent switch from HIF1α to HIF2α is associated with increased cellular atypia in pre-neoplastic kidney lesions (24).
To our knowledge, this is the first fine mapping, and the most comprehensive investigation of genetic variants associated with RCC risk in the EPAS1 region using genotyping, direct DNA sequencing and in silico studies. Our investigation took advantage of the opportunity to combine individual data from large, well-designed population-based, case–control and cohort studies. Although we did not identify significant statistical heterogeneity among the five data sets, there might be unobserved heterogeneity caused by the samples and the study design. Specifically, the controls in the CEERCC study were patients admitted to the same hospital during the same time period as the cases for a variety of diagnoses unrelated to RCC, while the controls in the four other studies were enrolled from the general population. However, it is important to note that despite the potential heterogeneity in the CEERCC study, the inclusion of this particular study did not affect our final results and conclusions. Moreover, examination of VHL gene alterations in tumor tissue and EPAS1 variants was conducted among only cases, which would eliminate the possibility of biases introduced by control selection.
Although the mechanism through which these variants modify RCC risk is unknown, our results suggest that the association is most likely mediated through regulation of EPAS1 or perhaps a distant target. Further sequence analysis of common and uncommon variants in this region is necessary to identify a comprehensive set of markers for follow-up functional analyses. Future investigations should include replications in other independent populations of both European and other continental backgrounds to dissect further the complex genetic signal observed in the chromosome 2p21 locus. Given the importance of HIF2α in carcinogenesis, common genetic variation in EPAS1 should be investigated in other cancers.
MATERIALS AND METHODS
Studies included in the analysis
PLCO Cancer Screening Trial
The PLCO Cancer Screening Trial is a large, randomized multicenter trial in the USA (Birmingham, AL; Denver, CO; Detroit, MI; Honolulu, HI; Marshfield, WI; Minneapolis, MN; Pittsburgh, PA; Salt Lake City, UT; St Louis, MO; and Washington, DC) of 155 000 men and women, designed to evaluate selected methods for early detection of these four cancers (25,26). PLCO enrollment began in 1993 and ended in 2001. Participants have been randomized to either a screening or control arm. Screening arm participants were asked to provide a blood sample at each screening visit and buccal samples were collected from the control arm participants. Incident cases of RCC (International Classification of Diseases ICDO, Ninth Revision, code 189.0) were ascertained from annual questionnaires. Medical and pathology records were obtained for all RCC cases identified from annual follow-up questionnaires, and trained medical record specialists abstracted the relevant clinical data. Study eligibility requirements consisted of informed consent, completion of the baseline questionnaire and no cancer prior to baseline. A total of 323 eligible cases were identified. Of the 118 975 eligible controls, 1918 had the necessary genotype data available. Cases exited at diagnosis date, whereas controls exited at the date of last follow-up. Eligible cases and 1598 controls were matched with incidence-density sampling at a case–control ratio of 1:5. Of the 1918 controls with the necessary genotype data, 1221 were selected as replacements for the 1598 matched controls. The matching factors included age at cohort entry (5-year intervals), year at cohort entry (1-year intervals), race, gender and time on study. The final population included a total of 323 cases and 1221 controls. The institutional review boards of the USA National Cancer Institute and the 10 study centers approved the trial. Participants provided written informed consent.
ATBC study
The ATBC study is a randomized, double-blind, placebo-controlled, primary prevention trial that tested whether daily supplementation with α-tocopherol, β-carotene or both could reduce the incidence of lung or other cancers among male smokers when compared with those without intervention (27). The trial was registered as ClinicalTrials.gov number, NCT00342992 [ClinicalTrials.gov]. A total of 29 133 men between the ages of 50 and 69 years who smoked at least five cigarettes per day were recruited from southwestern Finland between 1 April 1985 and 30 June 1988, and randomly assigned to one of four intervention groups on the basis of a 2 × 2 factorial design. Men were excluded at the time of study entry if they had a history of cancer (other than non-melanoma skin cancer or carcinoma in situ), severe angina upon exertion, chronic renal insufficiency, cirrhosis of the liver, alcoholism, anticoagulant use, use of vitamin E (>20 mg/day), vitamin A (>20 000 IU/day) or β-carotene (>6 mg/day). Participants received α-tocopherol (50 mg/day) as d,l-α-tocopheryl acetate, β-carotene (20 mg/day) as all-trans-β-carotene, both supplements, or a placebo capsule daily for 5–8 years (median = 6.1 years). Follow-up of the ATBC study cohort after the intervention continued through the Finnish Cancer Registry. This study was approved by the IRBs of the US National Cancer Institute and the National Public Health Institute of Finland. Written informed consent was obtained from each participant. Cases were defined as subjects with incident RCC (International Classification of Diseases, Ninth Revision, code 189.0) diagnosed at least 5 years after their baseline blood collection (range = 5–12 years). Incident RCC cases, diagnosed by April 2006, were identified through the Finnish Cancer Registry that provides nearly 100% case ascertainment. Ninety percent of cases had histology confirmation. Control subjects were defined as subjects alive and cancer-free at the time of case diagnosis. Cases and controls were selected with a case–control ratio of 1:3 frequency matched by age at randomization (5-year intervals) and baseline blood draw date. The total of 191 cases and 460 controls were included in the final analyses.
CEERCC case–control study
The CEERCC study is a hospital-based case–control study conducted in: Russia (Moscow), Romania (Bucharest), Poland (Lodz) and Czech Republic (Prague, Olomouc, Ceske Budejovice and Brno) from August 1999 to January 2003 (28). A common protocol was used at each center to recruit consecutive newly diagnosed patients with RCC and a comparable group of hospital-based control subjects without RCC. Cases were newly diagnosed patients with histologically confirmed RCC between the ages of 20 and 79 years. The pathologic diagnosis of RCC was confirmed by an expert kidney pathologist at the National Cancer Institute. In order to minimize selection bias, we selected controls from patients admitted to the same hospital during the same time period as the cases for a variety of diagnoses unrelated to smoking or kidney cancer (except for benign prostatic hyperplasia) and matched to cases on age, sex and study center. Both cases and controls were residents of the study areas for at least 1 year at the time of recruitment. This study was approved by the institutional review boards and ethical committees of all participating study centers, the International Agency for Research on Cancer (Lyon, France) and the US National Cancer Institute. All study subjects provided written informed consent. A total of 1097 cases and 1555 controls were interviewed in person. The response rates ranged from 90 to 99% for cases and 90 to 96% for controls across the different study sites. Smoking status was defined as never, former or current smoker. Blood samples were collected in 99% of the study participants and stored at −80°C and shipped to the NCI on dry ice. DNA was extracted using standard procedures. Subjects without genotyping data were similar in age and known RCC risk factors to those genotyped (data not shown).
USKC case–control study
The US Kidney Cancer study is a population-based case–control study conducted in Detroit, MI and Chicago, IL from February 2002 to January 2007. Cases were residents of the study areas 20–79 years of age who were newly diagnosed with histologically confirmed RCC (ICD-O2 C64.9) (29). Controls were frequency-matched to cases by study center, race, age and sex. Controls aged 65 years and older were identified from Medicare files, and those under age 65 years were identified from Division of Motor Vehicle records. Written informed consent was obtained from all participants, and IRB approvals were obtained from all participating study centers and the US National Cancer Institute. Questionnaires were administered in person by trained interviewers (to elicit information on demographic background, consumption of tobacco, height and weight history, family history of cancer, medical and medication history and other exposures). Blood samples were also collected. A total of 1568 Caucasians (856 cases and 712 controls) were interviewed, yielding response rates of >64%. Of these subjects, blood samples were collected from 718 (83.9%) cases and 615 (86.4%) controls. DNA was extracted using standard procedures. Genotyping data were available for 99.9% of blood samples genotyped. Subjects without genotyping data were similar in age and known RCC risk factors to those genotyped (data not shown).
American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS-II)
The American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS-II) was established in 1992; the cohort includes >86 000 men and 97 000 women from 21 US states who completed a mailed questionnaire in 1992 (30). At baseline, the cohort was 97% Caucasian and the median age of participants was 63 years (range: 40–92 years). After 1997, follow-up questionnaires were sent to surviving cohort members every other year to update exposure information and to ascertain occurrence of new cases of cancer; a >90% response rate has been achieved for each follow-up questionnaire. Incident cancers are verified through medical records, state cancer registries or death certificates. From 1998 to 2001, blood samples were collected from 39 376 cohort members, and from 2001 to 2002 an additional 70 004 cohort members provided buccal cell samples. The CPS-II RCC cases and controls included in the study consist of non-Hispanic Caucasian participants who were cancer-free at enrollment (except for non-melanoma skin cancer) with stored blood or buccal samples, and cases were diagnosed with RCC following enrollment. For some RCC cases, collection of biological samples occurred after cancer diagnosis or immediately preceding cancer diagnosis. Subjects with no history of RCC who were scanned as controls in previous GWAS projects investigating cancers of the bladder, lung and prostate were selected as controls for this study. A total of 203 RCC cases and 448 controls were included in the study.
SNP selection and genotyping
We surveyed selected SNPs with MAF > 3% from the HapMap Project using Haploview software to estimate LD across the region. We selected a total of 59 SNPs across the genomic region of EPAS1 (chromosome 2p21) to cover 120 kb of genomic sequence. These SNPs were also genotyped in the NCI Core Genotyping Facility (CGF) in 280 control samples from the Human Diversity Panel that includes 76 African/African Americans, 66 Caucasians, 49 Native American/Hispanics and 89 Pacific Rim Asians (31). All selected SNPs for actual genotyping had a minor allele frequency >5%.
Methods for the three genotype assays (TaqMan, Illumina OPA and Illumina Infinium) can be found at http://variantgps.nci.nih.gov/home.cfm. Genotyping of cases and controls was conducted at CGF. DNA from cases and controls were blinded and randomized on polymerase chain reaction (PCR) plates to avoid any potential bias, and duplicate genotyping was performed for a randomly selected 5% of the total series for quality control. All SNPs and assay information are reported in the NCI SNP500 Cancer database at http://snp500cancer.nci.nih.gov/home.cfm (32).
Quality control assessment
Systematic quality control was conducted separately for the CEERCC, USKC, PLCO and ATBC data sets before merging the four data sets, which included quality control steps specific for the performance of different arrays at distinct times. For SNP assays, exclusions included those with <90% completion rate and those with extreme deviation from fitness for Hardy–Weinberg equilibrium (P < 1 × 10−7). Monomorphic assays observed in either cases or controls only and SNPs with alleles ambiguously coded (AT- and CG-coding alleles) were excluded. Comparable quality control metrics were applied to the data sets, and following sample and SNP exclusions, genotype data for up to 59 SNPs were available for a total of 2278 cases and 3719 controls from the GWAS scans and iSELECT. The genotype frequencies among controls showed no deviation from the expected Hardy–Weinberg equilibrium proportions (P > 0.05). The genotyping completion rates were between 98 and 100% for all reported SNPs.
GWAS scan data
Three thousand three hundred and thirty-one previously scanned samples (317 K BeadChips) at the Centre Nationale Genotypage (CNG), Paris, France, from The NCI/IARC study in central Europe were included (13). The final participant count for the association analysis was 2573 samples. The total number of SNPs used was 15. One thousand two hundred and twenty-seven cases' and 2868 controls' previously scanned samples (on 550 or 610 BeadChips) at the Core Genotyping Facility from PLCO, USKC and ATBC were included (13). Participant exclusion criteria are as previously described (13). For the known 50 duplicate pairs, concordance was 99.95%. The final participant count for the association analysis was 2278 cases and 3719 controls. Fifteen SNPs were available for analysis in one or more studies.
iSELECT
Custom Infinium® (iSelect™) assay including 44 SNPs were used to genotyping at the Core Genotyping Facility: samples from 299 cases and 1193 controls from PLCO, and 199 cases and 466 controls from ATBC. Participants were excluded on the basis of unanticipated inter-study duplicates, incompletion rates <94% as per the quality control groups, abnormal heterozygosity values of <25% or >35%, expected duplicates, abnormal X-chromosome heterozygosity and phenotype exclusions (due to ineligibility or incomplete information) (N = 31). Four hundred and eighty-five cases and 1641 controls were available for analysis.
Merging data sets
The post-quality-control data sets were merged, normalizing strand differences when necessary. No incompatible encodings were detected, and the final data set contained a total of 59 SNPs (after excluding monomorphic and ambiguously coded AT and CG SNPs) for 2278 cases and 3719 controls.
TaqMan
TaqMan genotyping assays (ABI) for replication were optimized for four of six SNPs in the new notable region identified using imputation (Table 1). TaqMan assays for replication were genotyped in CGF. Concordance of known duplicates was greater than 99%.
Statistical methods
An association between each SNP and case–control status was tested using a logistic regression model adjusting for age, sex, smoking status, body mass index, study center, family history of cancer (any type of cancer) and hypertension for the initial genotyping analysis (PLCO, ATBC, CEERCC, USKC), and adjusting for sex and study for the validation analysis (CPS-II was added to PLCO, ATBC, CEERCC, USKC). We did not adjust for the same covariates in both stages of analyses since we have missing data for several covariates [hypertension, family history, smoking and body mass index (BMI)] in the CPS-II, which was used in the validation study. Assuming standard additive genetic model, per allele-copy OR was calculated using a likelihood ratio test with one degree of freedom.
To find associations on a finer scale, we imputed genotypes of all untyped SNPs (with MAF > 3%) in the region spanning from 46 358 466 to 46 479 285 bp on chromosome 2, using two publicly available reference data sets from 1000 Genomes Project (March 2010 release; Build 36) and HapMap Phase 3. Both the tagSNPs and GWAS SNPs with MAF > 3% are used for imputation. We used the software Impute v2 (33,34). This method uses a hidden Markov model for the joint distributions of each individual's missing and observed genotype data. The distributions are obtained by conditioning on phased haplotypes observed from reference data. In applying the imputation method specifically to our data, we used CEU panels (European Ancestry) both from 1000 Genomes and HapMap Phase 3, which yielded 304 new SNPs in addition to our 59 SNPs in the region. We performed imputation analyses separately on each of four studies. For conducting association tests on imputed data, we used SNPTEST software (33) with the option ‘Missing data likelihood score method’, which takes into account genotype uncertainty.
Given multiple centers and countries were used in our study, the potential for population stratification exists. Before conducting combined analysis, genetic heterogeneity of the effect size was evaluated using the meta-analysis Q-statistics and was not significant (35). Heterogeneity of genotype frequencies between centers was evaluated by using the likelihood ratio test. We found no evidence of heterogeneity across study centers. Moreover, no evidence of population stratification was apparent from a principal component analysis of genome-wide association study conducted in these populations (8), and the likelihood of this is small among European populations (36).
Conditional analysis was performed to assess the independence of association for two sets of SNPs not highly correlated. We performed an association test for one locus after adjusting for the other locus (and vice versa) in addition to the above set of covariates and concluded that the signals were dependent (or independent) if the significances were decreased (or unchanged) after adjusting for each other. For testing the interaction between smoking status and SNP, we used a likelihood ratio test with two degrees of freedom, treating SNP as a trend and a smoking status as a categorical variable with three levels.
Sequence analysis of EPAS1
We randomly selected the 94 RCC cases from the USA with >2 μg of genomic DNA. The genomic sequence analysis of EPAS1 (NM_001430) was conducted in 94 DNA samples by direct PCR sequencing. Primers used to sequence the 16 coding exons, 5′ UTR and intron–exon junctions of HIF2A were previously described (37). We sequenced 100 CEPH DNAs to determine whether a variant identified was a polymorphism. To confirm that the observed alterations did not arise as artifacts during the PCR or sequencing steps, we independently re-amplified and re-sequenced the corresponding regions in all 94 RCC DNA samples.
Transcription binding sites and microRNA sequence analyses
Analysis of transcription-factor binding site in the genomic regions with high-risk SNPs was conducted using the Cister program (38). In addition, using the UCSC genome browser (http://genome.ucsc.edu/), we examined the genomic sequence of EPAS1 to identify regulatory elements (39). A p53 binding site (161123-13) 5 kb upstream to rs9679290 was located in the GIS CHIP-PET track (40) and a CTCF binding site (OREG0014595) 3 kb downstream from rs9679290 was displayed in the ORegAnno track under the ‘Expression and Regulation’ tab (41). The search for predicted miRNA sequences in the new genomic EPAS1 region was conducted using miRBase database (42).
Comparative genomics
The genomic conservation of the high-risk SNPs and its adjacent genomic region was examined in the conservation track under the ‘Comparative Genomics’ tab of the UCSC genome browser (http://genome.ucsc.edu). The EPAS1 intronic sequences in human, rhesus monkey, horse, armadillo, dog and mouse were extracted from UCSC genome browser and multiple nucleotide sequence alignments were re-generated using ClustalW2.
EPAS1 SNPs and VHL alteration status association study
Cases were selected from the CEERC case–control study from whom we collected frozen RCC tumor biopsies (n = 507) (29). Tumor DNA extraction and PCR of VHL coding sequences, endonuclease scanning and sequencing were performed as previously described (21). Tumor and patient characteristics such as clinical stage, Fuhrman nuclear grade (I–IV), node stage (N0, N1, N2–3), BMI (<25, 25–35, >35 kg/m2) and smoking status were considered as categorical variables. Smoking status was defined as status 2 years before the interview. Participants who were smoking in the 24 months prior to the interview were classified as current smokers. Metastasis (M0, M1) self-reported hypertension (no/yes), family history of cancer or kidney cancer (no/yes), sex and age at diagnosis (<50 and ≥50 years) were analyzed as dichotomous variables. VHL somatic inactivation was considered as a dichotomous variable per case (no/yes). Genetic alterations were sequence changes that occurred within exons 1–3 leading to an altered amino acid sequence or truncated VHL protein including deletions, insertions, missense, nonsense and putative splice site mutations. The prevalence of cases with a VHL alteration was calculated by dividing the number of cases with that type of alteration by the total number of cases analyzed in the group.
For univariate analyses, χ2-tests were applied to contingency table (2 × 2) analysis to test for differences between the proportion of cases with or without a particular alteration subtype within each group. Trend tests were used to analyze associations between categorical variables and cases with and without alterations. Ordered logistic regression was used for multivariate analyses to evaluate associations between categorical variables and case subgroups, initially adjusting for all variables that were associated with VHL inactivation in univariate analyses (P < 0.20). With the exception of sex, age and country, only tobacco smoking and fruit intake remained in multivariate models because their inclusion changed risk estimates by at least 10%.
Associations between SNPs and genetic alteration category were estimated using both additive and dominant models. Risk per allele and trends were calculated using logistic regression. Analyses were conducted using STATA 10.0 (Stata Corporation, TX, USA) and statistical tests were two-sided.
Detection of recombination hotspot
To identify recombination hotspots in the region, we used PHASE v2.1 to estimate haplotypes from 1000 Genomes and HapMap CEU data. Resultant haplotypes and background recombination rates were used as direct input for SequenceLDhot, a program that uses the approximate marginal likelihood method and calculates likelihood ratio statistics at a set of possible hotspots (43,44).
SUPPLEMENTARY MATERIAL
FUNDING
This research was supported in part by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, DHHS, Bethesda, MD, USA, and by European Commission INCOCOPERNICUS grant IC15-CT96-0313.
CONTRIBUTORS
J.R.T., S.S.H., M.Y., M.T. and S.J.C. contributed to the design and execution of the study. J.R.T., S.S.H. and S.J.C. wrote the first draft of the manuscript.
S.S.H., N.C., M.Y., R.P., L.E.M., P.R., M.P.P., M.J., G.S., V.G., J.D.M., P.B. and C.C. participated in the statistical analysis. J.R.T., M.H.W. and O.T. were involved in SNP selection, sequencing of EPAS1 and in silico studies. L.E.M., N.R., W.H.H. and J.R.T. were involved in the sequencing of the VHL gene. L.E.M., P.B., P.B., D.Z., V.B., V.J., V.M., L.F., M.N., N.S.D., D.M., J.S.C., N.R., A.B. and W.H.C. were responsible for data and sample collection in the Central and Eastern European case–control study. J.R.T., R.P., M.P.P., N.R., D.M., S.W. and J.V. were responsible for the data and sample selection in the ATBC study. J.R.T., R.P., M.P.P., N.R., J.M., R.B. and C.B. were responsible for the data and sample collection in the PLCO study. L.E.M., K.S., F.D., N.R., J.S.C. and W.H.C. were responsible for the data and sample collection of the kidney cancer case–control study in the US. V.S., M.J.T., W.R.D. and S.G. were responsible for the data and sample collection in the CPS-II study. L.B., M.Y. and S.J.C. were responsible for the genotyping. All authors participated in the writing and revision of the manuscript and approved the final version.
WEB RESOURCES
The URLs for data presented herein are as follows: 1000 Genomes Project March 2010 release (http://www.1000genomes.org/page.php), SNP database (http://www.ncbi.nlm.gov/projects/SNP/), NCI SNP500 Cancer database (http://snp500cancer.nci.nih.gov/home.cfm), UCSC genome browser (http://genome.ucsc.edu) and Genotype assays (TaqMan, Illumina OPA and Illumina Infinium) (http://variantgps.nci.nih.gov/home.cfm).
Supplementary Material
ACKNOWLEDGMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Conflict of Interest statement. The authors do not have any conflicts of interest.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M.J. Cancer statistics, 2009. CA Cancer J. Clin. 2009;59:225–249. doi: 10.3322/caac.20006. doi:10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Chow W.H., Dong L.M., Devesa S.S. Epidemiology and risk factors for kidney cancer. Nat. Rev. Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. doi:10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ljungberg B., Cowan N.C., Hanbury D.C., Hora M., Kuczyk M.A., Merseburger A.S., Patard J.J., Mulders P.F.A., Sinescu I.C. EAU Guidelines on Renal Cell Carcinoma: the 2010 update. Eur. Urol. 2010;58:398–406. doi: 10.1016/j.eururo.2010.06.032. doi:10.1016/j.eururo.2010.06.032. [DOI] [PubMed] [Google Scholar]
- 4.Yu M.C., Mack T.M., Hanisch R., Cicioni C., Henderson B.E. Cigarette smoking, obesity, diuretic use, and coffee consumption as risk factors for renal cell carcinoma. J. Natl Cancer Inst. 1986;77:351–356. [PubMed] [Google Scholar]
- 5.Chow W.H., Gridley G., Fraumeni J.F., Järvholm B. Obesity, hypertension, and the risk of kidney cancer in men. N. Engl. J. Med. 2000;343:1305–1311. doi: 10.1056/NEJM200011023431804. doi:10.1056/NEJM200011023431804. [DOI] [PubMed] [Google Scholar]
- 6.Linehan W.M., Pinto P.A., Bratslavsky G., Pfaffenroth E., Merino M., Vocke C.D., Toro J.R., Bottaro D., Neckers L., Schmidt L.S., et al. Hereditary kidney cancer: unique opportunity for disease-based therapy. Cancer. 2009;115:2252–2261. doi: 10.1002/cncr.24230. doi:10.1002/cncr.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomlinson I.P., Alam N.A., Rowan A.J., Barclay E., Jaeger E.E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. doi:10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 8.Wei M.H., Toure O., Glenn G.M., Pithukpakorn M., Neckers L., Stolle C., Choyke P., Grubb R., Middelton L., Turner M.L., et al. Novel mutations in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer. J. Med. Genet. 2006;43:18–27. doi: 10.1136/jmg.2005.033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlehofer B., Pommer W., Mellemgaard A., Stewart J.H., McCredie M., Niwa S., Lindblad P., Mandel J.S., McLaughlin J.K., Wahrendorf J. International renal cell cancer study. VI. The role of medical and family history. Int. J. Cancer. 1996;66:723–726. doi: 10.1002/(SICI)1097-0215(19960611)66:6<723::AID-IJC2>3.0.CO;2-1. doi:10.1002/(SICI)1097-0215(19960611)66:6<723::AID-IJC2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Hung R.J., Moore L., Boffetta P., Feng B.J., Toro J.R., Rothman N., Zaridze D., Navratilova M., Bencko V., Janout V., et al. Family history and the risk of kidney cancer: a multicenter case control study in Central Europe. Cancer Epidemiol. Biomarkers Prev. 2007;16:1287. doi: 10.1158/1055-9965.EPI-06-0963. doi:10.1158/1055-9965.EPI-06-0963. [DOI] [PubMed] [Google Scholar]
- 11.McLaughlin J.K., Mandel J.S., Blot W.J., Schuman L.M., Mehl E.S., Fraumeni J.F., Jr A population-based case–control study of renal cell carcinoma. J. Natl Cancer Inst. 1984;72:275–284. [PubMed] [Google Scholar]
- 12.Chung C.C., Chanock S.J. Current status of genome-wide association studies in cancer. Hum. Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. doi:10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- 13.Purdue M.P., Johansson M., Zelenika D., Toro J.R., Scelo G., Moore L.E., Prokhortchouk E., Wu X., Kiemeney L.A., Gaborieau V. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13. 3. Nat. Genet. 2011;43:60–65. doi: 10.1038/ng.723. doi:10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaelin W.G., Jr, Ratcliffe P.J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. doi:10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Eltzschig H.K., El Kasmi K.C., Eckle T. The HIF2A gene in familial erythrocytosis. N. Engl. J. Med. 2008;358:1962–1968. doi: 10.1056/NEJMc080274. [DOI] [PubMed] [Google Scholar]
- 16.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X.P., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. doi:10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beall C.M., Cavalleri G.L., Deng L., Elston R.C., Gao Y., Knight J., Li C., Li J.C., Liang Y., McCormack M. Natural selection on EPAS1 (HIF2 α) associated with low hemoglobin concentration inTibetan highlanders. Proc. Natl Acad. Sci. USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. doi:10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickson S.P., Wang K., Krantz I., Hakonarson H., Goldstein D.B. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. doi:10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. doi:10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nickerson M.L., Jaeger E., Shi Y., Durocher J.A., Mahurkar S., Zaridze D., Matveev V., Janout V., Kollarova H., Bencko V., et al. Improved identification of von Hippel–Lindau gene alterations in clear cell renal tumors. Clin. Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. doi:10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamura T., Koepp D.M., Conrad M.N., Skowyra D., Moreland R.J., Iliopoulos O., Lane W.S., Kaelin W.G., Elledge S.J., Conaway R.C. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. doi:10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 23.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J.M., Lane W.S., Kaelin W.G., Jr HIF targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. doi:10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 24.Kondo K., Kim W.Y., Lechpammer M., Kaelin W.G., Jr Inhibition of HIF2 is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:e83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prorok P.C., Andriole G.L., Bresalier R.S., Buys S.S., Chia D., David Crawford E., Fogel R., Gelmann E.P., Gilbert F., Hasson M.A. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial. Controlled Clin. Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. doi:10.1016/S0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 26.Hayes R.B., Sigurdson A., Moore L., Peters U., Huang W.Y., Pinsky P., Reding D., Gelmann E.P., Rothman N., Pfeiffer R.M. Methods for etiologic and early marker investigations in the PLCO trial. Mutat. Res. 2005;592:147–154. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Heinonen O.P., Albanes D. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N. Engl. J. Med. 1994;330:1029–1035. doi: 10.1056/NEJM199404143301501. doi:10.1056/NEJM199404143301501. [DOI] [PubMed] [Google Scholar]
- 28.Moore L.E., Brennan P., Karami S., Menashe I., Berndt S.I., Dong L.M., Meisner A., Yeager M., Chanock S., Colt J., et al. Apolipoprotein E/C1 locus variants modify renal cell carcinoma risk. Cancer Res. 2009;69:8001–8008. doi: 10.1158/0008-5472.CAN-09-1734. doi:10.1158/0008-5472.CAN-09-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karami S., Schwartz K., Purdue M.P., Davis F.G., Ruterbusch J.J., Munuo S.S., Wacholder S., Graubard B.I., Colt J.S., Chow W.H. Family history of cancer and renal cell cancer risk in Caucasians and African Americans. Br. J. Cancer. 2010;102:1676–1680. doi: 10.1038/sj.bjc.6605680. doi:10.1038/sj.bjc.6605680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calle E.E., Rodriguez C., Jacobs E.J., Almon M.L., Chao A., McCullough M.L., Feigelson H.S., Thun M.J. The American Cancer Society Cancer Prevention Study II Nutrition Cohort. Cancer. 2002;94:2490–2501. doi: 10.1002/cncr.101970. doi:10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 31.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. doi:10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 32.Packer B.R., Yeager M., Burdett L., Welch R., Beerman M., Qi L., Sicotte H., Staats B., Acharya M., Crenshaw A. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. doi:10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. doi:10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 34.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;535:e1000529. doi: 10.1371/journal.pgen.1000529. doi:10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DerSimonian R., Laird N. Meta-analysis in clinical trials* 1. Control Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. doi:10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 36.Wacholder S., Rothman N., Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol. Biomarkers Prev. 2002;11:513–520. [PubMed] [Google Scholar]
- 37.Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. doi:10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 38.Frith M.C., Hansen U., Weng Z. Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics. 2001;17:878–889. doi: 10.1093/bioinformatics/17.10.878. doi:10.1093/bioinformatics/17.10.878. [DOI] [PubMed] [Google Scholar]
- 39.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y.T., Weng Z. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. doi:10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 41.Griffith O.L., Montgomery S.B., Bernier B., Chu B., Kasaian K., Aerts S., Mahony S., Sleumer M.C., Bilenky M., Haeussler M. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–D113. doi: 10.1093/nar/gkm967. doi:10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S., Grocock R.J., Van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. doi:10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics. 2006;22:3061–3066. doi: 10.1093/bioinformatics/btl540. doi:10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 44.Crawford D.C., Bhangale T., Li N., Hellenthal G., Rieder M.J., Nickerson D.A., Stephens M. Evidence for substantial finescale variation in recombination rates across the human genome. Nat. Genet. 2004;36:700–706. doi: 10.1038/ng1376. doi:10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.