Abstract
The global prevalence of severe Clostridium difficile infection highlights the profound clinical significance of clostridial glucosylating toxins1–4. Virulence is dependent on the autoactivation of a toxin cysteine protease5–9, which is promoted by the allosteric cofactor inositol hexakisphosphate (InsP6)10–17. Host mechanisms that protect against such exotoxins are poorly understood. It is increasingly appreciated that the pleiotropic functions attributed to nitric oxide (NO), including host immunity, are in large part mediated by S-nitrosylation of proteins18,19. Here we show that C. difficile toxins are S-nitrosylated by the infected host and that S-nitrosylation attenuates virulence by inhibiting toxin self-cleavage and cell entry. Notably, InsP6- and inositol pyrophosphate (InsP7)-induced conformational changes in the toxin enabled host S-nitrosothiols to transnitrosylate the toxin catalytic cysteine, which forms part of a structurally conserved nitrosylation motif. Moreover, treatment with exogenous InsP6 enhanced the therapeutic actions of oral S-nitrosothiols in mouse models of C. difficile infection. Allostery in bacterial proteins has thus been successfully exploited in the evolutionary development of nitrosothiol-based innate immunity and may provide an avenue to new therapeutic approaches.
C. difficile infection is the most prevalent cause of hospital-acquired infectious diarrhea and life-threatening colitis worldwide1–4. Two large exotoxins, TcdA (308 kDa) and TcdB (270 kDa), are secreted from the majority of C. difficile strains that cause intestinal disease in humans2–4. Cellular internalization of these exotoxins is dependent on the cytosolic cofactor InsP6, which activates an autocatalytic cysteine protease domain (CPD) to facilitate toxin self-cleavage9–17. InsP6 binds an allosteric site within the toxin CPD after its insertion into the plasma membrane. In a toxigenic disease model20,21, we found considerable cytopathology when we injected purified TcdA into ileal loops. TcdA induced marked intestinal mucosal damage, fluid secretion, neutrophil infiltration and transcription of proinflammatory genes, including Nos2 (encoding inducible nitric oxide synthase), Tnf (encoding tumor necrosis factor-α) and Il1b (encoding interleukin-1β) (Fig. 1a,b and Supplementary Fig. 1).
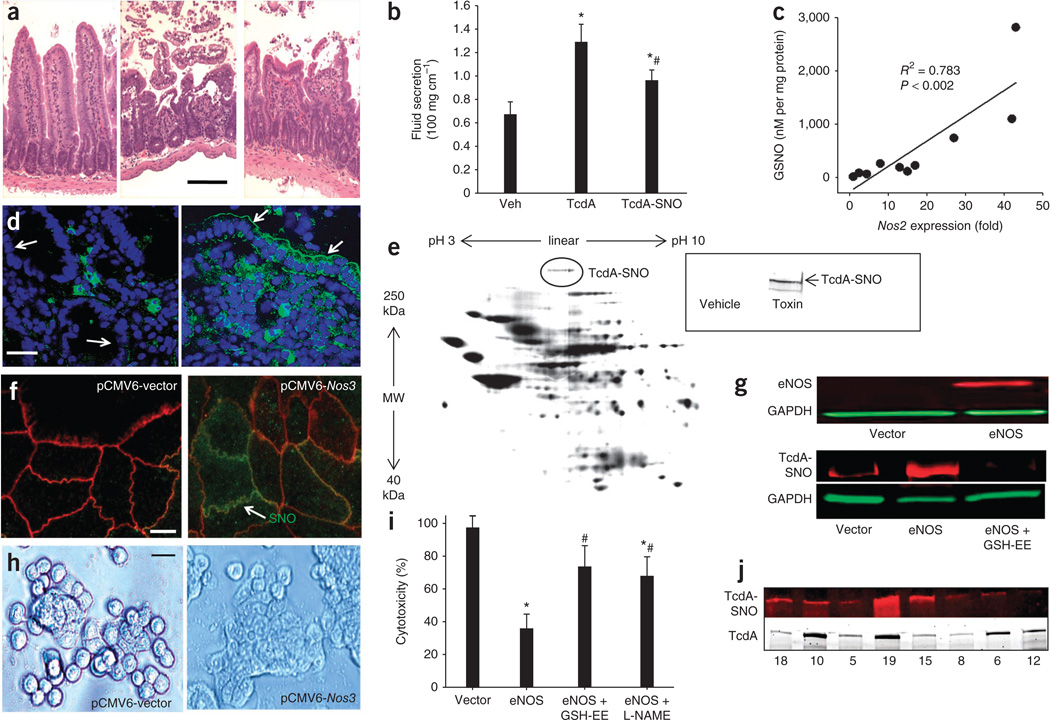
Figure 1.
C. difficile toxins are S-nitrosylated in vivo. (a) H&E-stained sections of mouse ileum exposed to vehicle control (left), TcdA (middle) or TcdA-SNO (right) (10 µg toxin for 4 h; scale bar, 50 µm). (b) TcdA- or TcdA-SNO–induced fluid secretion in mouse ileal loops (10 µg for 4 h) (n = 4 per group; error bars show s.e.m.; P < 0.05 compared with vehicle (veh) control (*) and TcdA (#), respectively; analysis of variance (ANOVA) on ranks). (c) Tissue GSNO concentrations versus Nos2 mRNA expression levels in mouse ileal loops exposed to TcdA (10 µg) or vehicle for 4 h. (d) Anti-nitrosocysteine (SNO) immunofluorescence showing epithelial S-nitrosylation in human colitis (right) but not in histologically normal colon (left), where SNOs are largely confined to lamina propria cells (arrows illustrate brush border membrane; SNOs (green); DAPI nuclear counterstain (blue); scale bar, 20 µm). (e) SNO proteomics showing in situ S-nitrosylated proteins from TcdA-exposed mouse ileum labeled with BODIPY FL maleimide25. MW, molecular weight. Inset, biotin-switch assay using a C-terminus specific antibody to TcdA. (f) SNO immunofluorescence in Nos3-transfected Caco-2 cells. High SNO-protein levels (green) co-localize with membrane ZO-1 (red). Scale bar, 10 µm. (g) Top, immunoblot showing eNOS and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression using IR-680 (red) and IR-800 (green) antibody labels, respectively, in vector- and Nos3-transfected Caco-2 cells. Bottom, immunoblots for TcdA-SNO and GAPDH (using IR-680 (red) and IR-800 (green) antibody labels, respectively), after immunoprecipitation with nitrosocysteine-specific antibodies in vector- and Nos3-transfected Caco-2 cells exposed to TcdA. TcdA-exposed cells were also treated with GSH-ethyl ester (GSH-EE) to remove NO groups from proteins (control). (h) TcdB-induced cell rounding in vector (left) versus Nos3 (right) transfected Caco-2 cells (71.1 ± 14.9% versus 26.5 ± 8.2%, respectively ± s.e.m., n = 3; P < 0.05, Mann-Whitney U test on ranks) (scale bar, 25 µm). (i) MTT assay for cytotoxicity in vector and Nos3-transfected Caco-2 cells exposed to 3.7 nM TcdB for 10 min. Controls included cells treated with GSH-EE and L-NAME (n = 3; error bars show s.e.m.; P < 0.05 compared to vector control– (*) and Nos3- (#) transfected cells, respectively; Mann-Whitney U test on ranks). (j) Immunoblots showing TcdA-SNO (top; labeled with IR-680) in human stool (n = 8) samples positive for TcdA (bottom; labeled with IR-800). TcdA-SNO was first immunoprecipitated from stool samples with a nitrosocysteine-specific antibody, and samples were then probed for TcdA. Presence of toxin in the stool was independently confirmed by ELISA and immunocytotoxicity assay30 (Supplementary Fig. 4).
It has been suggested that Nos2-derived NO may limit C. difficile intoxication20,21, but the molecular mechanism is not known. To explore a potential role for S-nitrosylation, we first assayed tissues for S-nitrosoglutathione (GSNO), a small endogenous S-nitrosothiol (SNO) that constitutes a major source of NO bioactivity in the respiratory22 and gastrointestinal23 tracts. Using HgCl2-coupled photolysis-chemiluminescence22,24, we found a 12.1-fold increase in tissue GSNO concentration in TcdA versus buffer-injected loops (84 ± 29.1 versus 1,020 ± 475 nM mg−1 protein, respectively; P < 0.05; ± s.d.), which correlated with elevated Nos2 expression (Fig. 1c and Supplementary Fig. 1). The effects of GSNO are mediated primarily by targeted S-nitrosylation of proteins18. Immunofluorescence labeling of S-nitrosylated proteins (SNO-proteins) in tissue sections revealed high amounts of SNO-proteins within TcdA-exposed intestinal mucosa, and a similar SNO-protein accumulation was evident in biopsies from humans with active inflammation, suggesting pathophysiological relevance in human colitis (Fig. 1d).
We characterized the toxin-induced S-nitrosoproteome using an unbiased cysteine saturation fluorescence proteomics assay25. Endogenous protein species identified here as showing higher levels of S-nitrosylation after toxin exposure included several species that are known targets of GSNO or other endogenous nitrosylases, including hemoglobin, heat shock, cytoskeletal and various cell signaling proteins. C. difficile infection is therefore associated with increases in tissue S-nitrosylation that probably reflect toxin-induced proinflammatory responses. Unexpectedly, TcdA itself was also consistently identified as a molecular target of S-nitrosylation in inflamed tissues (Fig. 1e and Supplementary Fig. 2), and we confirmed this by a biotin-switch assay26 that specifically labels Cys-NO adducts (Fig. 1e).
Because there is no precedent for in situ S-nitrosylation of foreign proteins in host tissues, we established an in vitro model to examine the potential role of toxin S-nitrosylation. To recapitulate the epithelial SNO-protein accumulation observed during C. difficile infection, we transfected human Caco-2 colonocytes with a calcium-inducible endothelial nitric oxide synthase (eNOS) construct (pCMV6-Nos3) to transiently raise cellular SNO expression27 (Fig. 1f,g and Supplementary Fig. 3a). Both a biotin-switch assay and immunoblotting with an antibody to nitrosocysteine (SNO) showed that toxin recovered from toxin-exposed Nos3-transfected cells was S-nitrosylated and that S-nitrosylation was abolished by the membrane-permeable denitrosylating agent glutathione (GSH)-ethyl ester27 (Fig. 1g). Toxin-induced cell rounding and viability assays further showed that Nos3 transfection conferred significant protection against intoxication (Fig. 1h,i). Moreover, toxin-induced UDP glucosylation of Rho GTPases28 was markedly reduced in Nos3-transfected cells (Supplementary Fig. 3b,c), consistent with a SNO-based mechanism of protection that inhibits the toxin’s effects. Notably, this protection was significantly attenuated by GSH-ethyl ester and by the eNOS inhibitor N-(G)-nitro-L-arginine-methyl ester (L-NAME) (Fig. 1i). Mouse macrophages deficient in Nos2 were also more susceptible to toxin-induced glucosylation of Rac1 than were wild-type macrophages, which express elevated SNO29 (Supplementary Fig. 3d). Collectively, these data strongly suggest that toxin S-nitrosylation is a physiological mechanism of toxin inhibition.
To illustrate more directly that S-nitrosylation of the exotoxin has cytoprotective effects, we assessed the activity of S-nitrosylated toxin (toxin-SNO) in pure form. Purified TcdA was S-nitrosylated by treatment with GSNO, and synthesis of toxin-SNO was confirmed by biotin-switch assay (Supplementary Fig. 4a). In vitro studies showed that toxin-SNO was significantly less cytotoxic compared with native toxin (Supplementary Fig. 4a), and the protective effects of S-nitrosylation were abolished by GSH or dithiothreitol, which denitrosylated the toxin. TcdA-SNO was also significantly less cytotoxic in vivo as assayed in the toxigenic-loop model (Fig. 1a,b and Supplementary Fig. 1). The pathophysiological relevance of these data was strengthened by finding S-nitrosylated toxin in stool samples from humans with C. difficile infection and by an inverse relationship between levels of toxin-SNO and stool cytotoxicity, as determined by an ultrasensitive immunocytotoxicity assay30 (Fig. 1j and Supplementary Fig. 4b,c). Thus, toxin S-nitrosylation seems to be both physiologically and functionally significant.
We sought to identify the S-nitrosylated residues responsible for inactivation of C. difficile toxins. Cysteine residues targeted by S-nitrosylation often conform to an acid-base consensus motif31. There are seven cysteine residues in TcdA and nine in TcdB, with four being conserved between the two toxins, including the catalytic cysteine7–9. Because the cysteine-histidine catalytic dyad conforms to a nitrosylation motif (see below)18,19, we explored whether the toxin CPD is a preferred target for post-translational modification by GSNO. Crystal CPD structures of TcdA15 and TcdB9 show a well-defined catalytic cleft that is separated from a positively charged InsP6 binding pocket abutting a β-hairpin fold (β-flap) (Fig. 2a and Supplementary Fig. 5). In silico docking of GSNO to the toxin CPD structures predicted excellent alignment of the S-NO bond with the active site cysteine, which forms part of an exposed acid-base S-nitrosylation consensus motif that is structurally conserved amongst diverse microbial cysteine proteases, with glutamic acid and histidine residues juxtaposing the catalytic Cys: Cys700 in TcdA, Cys698 in TcdB, Cys3568 in Vibrio cholerae RTX toxin (RTXVC)12,14 and Cys244 in gingipain R32 (Fig. 2b,c and Supplementary Figs. 6 and 7).
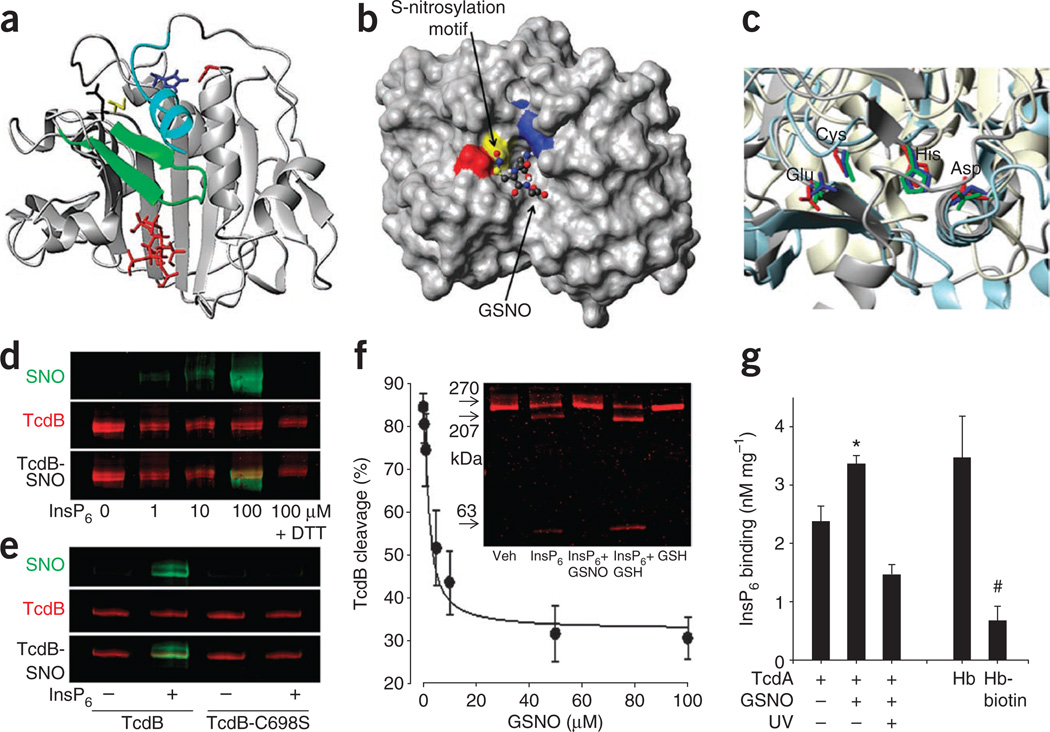
Figure 2.
Toxin S-nitrosylation is allosterically regulated by inositolphosphate. (a) N-terminus extended (cyan) CPD model for TcdB (based on 3PA8.pdb9 and 3FZY.pdb11 crystal structures), shows the β-flap (green) abutting the bound allosteric ligand InsP6 (red). (b) Alignment of GSNO at CPD active site. Surface rendering of the TcdB CPD (3PA8.pdb)9 showing the exposed S-nitrosylation consensus motif Glu743 (red)-Cys698 (yellow)-His653 (blue). GSNO docking shows the SNO group to be aligned with the catalytic Cys. (c) Crystal structures of TcdA (red)15, Vibrio cholerae RTX toxin (RTXVC; blue)11 and gingipain33 (green). Structurally conserved residues, constituting a nitrosylation motif, are shown in the respective crystal structures. (d) Assessment of TcdB S-nitrosylation (using SNO-specific (IR800) and TcdB-specific (IR680) antibodies) and effect of InsP6 (100 µM). DTT (1 mM) is used to remove NO from proteins (control). (e) Assessment of TcdB S-nitrosylation (as in d) for native and Cys698Ser mutant TcdB in the presence and absence of InsP6 cofactor. (f) Effect of GSNO on InsP6-induced TcdB autocleavage. Inset, GelCode Blue staining of unprocessed TcdB (270 kDa) and TcdB cleavage products (207 and 63 kDa) in the presence and absence of GSNO (100 µM) and GSH (100 µM). (g) Effect of S-nitrosylation on InsP6 binding. Tritiated InsP6 binding to TcdA (Bmax and Kd values are shown in Supplementary Fig. 8a). SNO photolysis is used to eliminate SNO. InsP6 binding to deoxygenated hemoglobin (Hb), which is inhibited by primary amide biotinylation, is provided as a positive control (n = 3; error bars show s.e.m.; *P < 0.05, #P < 0.05, compared to untreated TcdA and hemoglobin, respectively; Mann-Whitney U test on ranks).
S-nitrosylation is often subject to allosteric regulation, and the prototypic example of hemoglobin provides a precedent where S-nitrosylation is modulated allosterically by both organic and inorganic phosphates, including InsP6. As a corollary, we hypothesized that InsP6 binding to the CPD may regulate S-nitrosylation. Multiple lines of experimental evidence indeed confirmed this and suggest that the inositolphosphate-bound (that is, enzymatically active) form of the toxin is preferentially S-nitrosylated: (i) InsP6, and the more potent membrane-associated allosteric cofactor InsP7 (ref. 33), potentiated S-nitrosylation of TcdB by GSNO (Fig. 2d and Supplementary Fig. 8), and this effect was abolished by mutation of Cys698 to serine in TcdB (Fig. 2e); (ii) standard in vitro and real-time toxin cleavage assays showed that GSNO rapidly inhibited InsP6-induced toxin self-cleavage with a half-maximal inhibitory concentration (IC50) of 12.9 ± 4.2 µM for TcdB (Fig. 2f and Supplementary Fig. 9a); (iii) cysteine-specific cyanylation of C. difficile toxins using nitro-thiocyanobenzoic acid (NTCB)-based cleavage assays34 identified TcdA Cys700 and TcdB Cys698 as principal S-nitrosylation sites (Supplementary Fig. 9b); (iv) S-nitrosylated CPD peptide fragments were identified by mass spectrometry only after InsP6 and GSNO co-treatment (Supplementary Fig. 10); (v) S-nitrosylation prevents the intracellular release of the toxin N-terminus effector domain (as evidenced by SNO-mediated inhibition of TcdB autocleavage; Supplementary Fig. 11), which is dependent on inositolphosphate binding; and (vi) GSNO treatment of toxin markedly inhibited InsP6 binding to the CPD, and removal of SNO by photolysis restored InsP6 binding (Fig. 2g). This ‘linkage’, incurred without major conformational change in the toxin (Supplementary Fig. 12), suggests that S-nitrosylation of the active-site cysteine regulates communication with the InsP6 binding pocket, possibly by disordering the β-flap12,14,15. Taken together with earlier work35, these results raise the idea that crosstalk between inositolphosphate- and NO-based signaling may be a widespread phenomenon.
In control studies, we confirmed that cell membrane binding and in vitro UDP-glucosyltransferase activity of toxin-SNO was not diminished compared with native toxin (Supplementary Fig. 11c) and that InsP6 binding affinity for TcdA (Kd of 62.6 ± 7.0 nM) is in close agreement with the activation constant (AC50) for the CPD15 (Fig. 2g and Supplementary Fig. 8a). Thus, GSNO attenuates the toxin by a dual orthosteric and allosteric mechanism of action: InsP6 enables S-nitrosylation of the CPD, which, in turn, displaces the allosteric activator. Notably, the toxin cysteine protease structurally resembles the protease domain of caspases, which are subject to S-nitrosylation19. Inhibition of pro-caspase-3 by S-nitrosylation also entails both orthosteric (active site) and allosteric mechanisms36. Procaspase-3 is maintained in the S-nitrosylated and inactive state in the mitochondria but not in the cytosol37. Our new findings (Fig. 1d,f) suggest that membrane compartments may provide privileged access to allosteric cofactors33 that may act as determinants of subcellular S-nitrosylation.
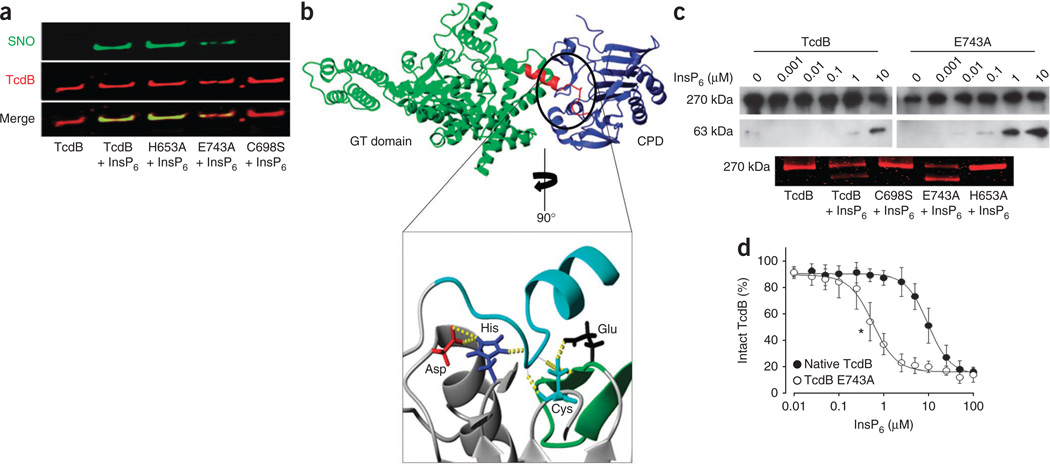
Consensus motifs for S-nitrosylation may influence the nucleophilicity of thiol or the binding of nitrosylating agents18,31,37. Disruption of the CPD-containing motif for S-nitrosylation (Fig. 2b,c and Supplementary Figs. 6 and 7) by mutagenesis of Glu743 to alanine diminished the ability of GSNO to transnitrosylate TcdB (Fig. 3a). By contrast, mutation of the catalytic His653 to alanine in TcdB had little effect on S-nitrosylation (Fig. 3a). This observation is consistent with the unusually large distances (>6Å) between the catalytic cysteines and histidines in toxin CPDs15. Rather than increasing nucleophilicity of the catalytic cysteine, His653 is likely to play a part in substrate orientation, assisted by a catalytic aspartic acid that may stabilize the histidine imidazolium ring (Fig. 3b and Supplementary Figs. 13 and 14). Mutagenesis of His653 to alanine in TcdB confirmed its role in catalytic activity (Fig. 3c). Cysteine and histidine CPD mutants were also significantly less cytotoxic than wild-type protein, confirming previous reports7,13,16. Glu743 in the cysteine protease active site is predicted to modulate cysteine thiolate reactivity (in addition to docking of GSNO) via hydrogen-bonding interactions (Fig. 3b and Supplementary Fig. 14). Mutagenesis of Glu743 in TcdB greatly enhanced toxin self-cleavage in the presence of inositolphosphate cofactor (Fig. 3c,d). Thus, Glu743 regulates the allosteric switch38 induced by InsP6 on the one hand (which may restrict toxin self-cleavage in the gut lumen where InsP6 is derived from dietary sources), and facilitates efficient S-nitrosylation on the other hand.
Figure 3.
A catalytic-site motif for S-nitrosylation. (a) Effect of InsP6 on S-nitrosylation of the TcdB catalytic site and TcdB catalytic-site mutants (His653Ala, Glu743Ala and Cys698Ser). GSNO (100 µM) and InsP6 (100 µM) were added for 10 min at 37 °C. (b) Extensive hydrogen bonding within the active site of the N-terminus RTXVC CPD11 (inset, yellow dotted lines) interconnects a catalytic-site S-nitrosylation motif that is conserved in TcdA and TcdB (Supplementary Figs. 7 and 14). The substrate domain (autocleavage site) is shown in turquoise (inset) and in red (top), where it bridges TcdB glucosyltransferase (GT) and cysteine protease (blue) domains, as shown in the combined crystal structure model, described further in Supplementary Figure 13. (c) Allosteric effect of InsP6 on catalytic activity of native TcdB versus catalytic-site mutants. Top, immunoblot showing autocleavage of native TcdB versus Glu743Ala mutant in the presence of InsP6. Bottom, GelCode Blue staining showing allosteric influence of InsP6 on catalytic activity of native TcdB versus catalytic-site mutants (C698S, E743A and H653A). (d) Autocleavage of native TcdB versus E743A mutant with increasing InsP6 concentrations (n = 3; error bars show s.e.m. *P < 001 for the lowest significant InsP6 concentration, Mann-Whitney U test for ranks).
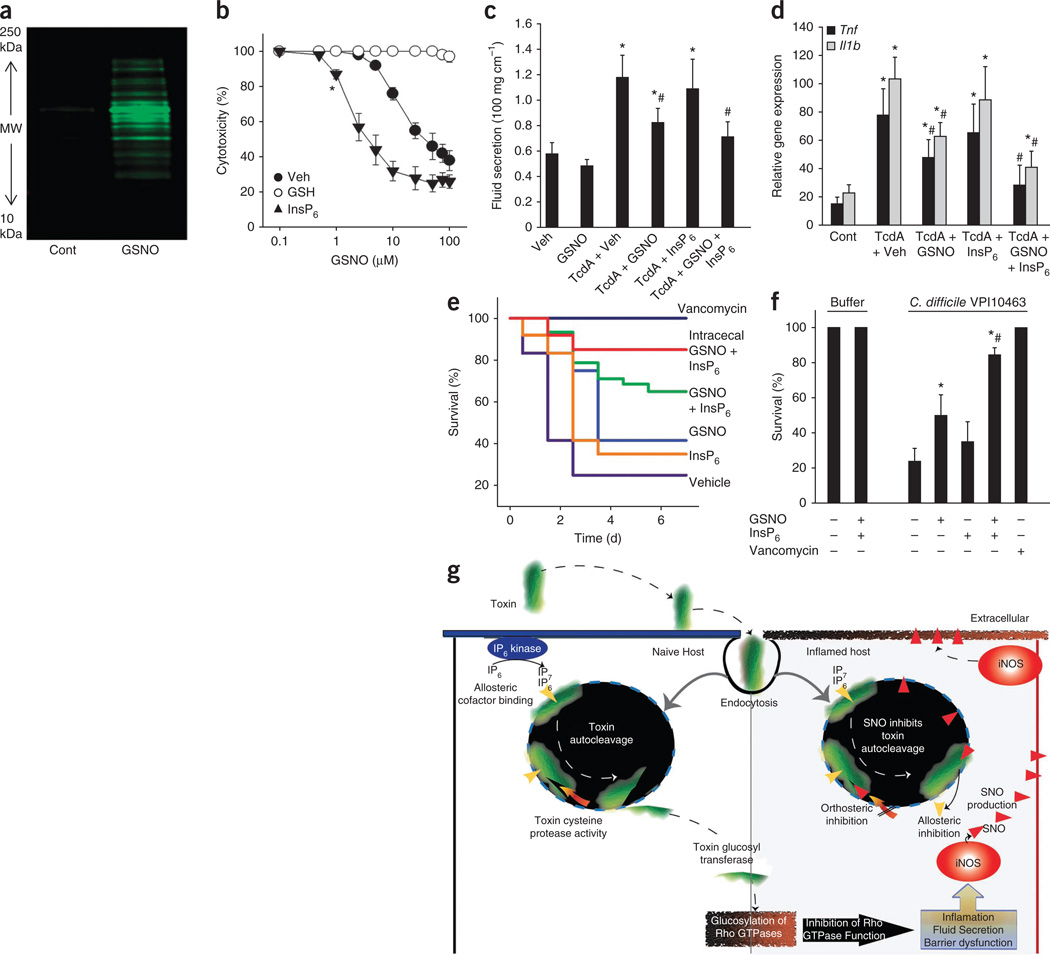
Metronidazole and vancomycin can effectively treat C. difficile infection, but the association of these drugs with high relapse rates represents a major health problem1–4. These considerations necessitate the development of alternative therapeutic strategies. The observation that S-nitrosylation inhibits toxin self-cleavage and that GSNO probably serves as the endogenous S-nitrosylating agent prompted us to test whether GSNO can be exploited therapeutically to protect against C. difficile infection. Exogenous GSNO efficiently S-nitrosylated epithelial cell proteins in culture (Fig. 4a) and protected against Tcd cytotoxicity (IC50 of 57.9 ± 13.7 and 46.3 ± 7.1 µM for TcdA and TcdB, respectively) (Fig. 4b). Inhibition of toxin activity by GSNO was reversed by denitrosylating agents (GSH or dithiothreitol), and cytoprotection by GSNO was greatly enhanced by inositolphosphate cofactor, which lowered the IC50 into the low micromolar range (Fig. 4b and Supplementary Fig. 15a). Using the toxigenic-loop model, we confirmed that exogenous GSNO injected intralumenally reduced TcdA-induced inflammation, fluid secretion and histopathology (Fig. 4c,d and Supplementary Fig. 15b). In addition, intraluminal InsP6 potentiated the therapeutic actions of exogenous S-nitrosothiol in vivo (Fig. 4c,d). We also tested the therapeutic efficacy of GSNO in a mouse infectious disease model that closely mimics the human disease1. Kaplan-Meier survival plots of infected mice showed a survival benefit of oral GSNO that was markedly enhanced by oral InsP6 (Fig. 4e). Direct delivery of GSNO and InsP6 into the cecum produced an even greater therapeutic effect (84.4 ± 4.1% survival with GSNO and InsP6 versus 50.0 ± 11.7% survival with GSNO, respectively; P < 0.05; ± s.e.m.) (Fig. 4e,f). These combined benefits of GSNO and InsP6 identify the toxin as a primary locus of GSNO action and may thus form the basis of a new treatment for unchecked C. difficile infection.
Figure 4.
GSNO-based therapy for C. difficile infection. (a) Biotin-switch assay showing increased protein S-nitrosylation in Caco-2 cells treated with GSNO (100 µM for 30 min). (b) Dose-response curves for GSNO-mediated inhibition of TcdB (3.7 nM; 10 min) in the absence (closed circles) or presence of GSH (1 mM; open circles) and InsP6 (100 µM; filled triangles) (n = 3; error bars show s.e.m.; *P < 0.05 for the lowest significant GSNO concentration, Mann-Whitney U test for ranks). (c) TcdA induced fluid secretion in mouse ileal loops (10 µg for 4 h) in the presence (and absence) of luminal GSNO (10 mg per kg in 0.1 ml) and/or InsP6 (1 mM) (n > 6 group; error bars show s.e.m.; P < 0.05 compared with vehicle control (*) and TcdA-vehicle (#), respectively; ANOVA on ranks). (d) TcdA induced Tnf and Il1b mRNA expression in mouse ileal loops in the presence (and absence) of GSNO and/or InsP6 (as in c) (n > 6 group; error bars show s.e.m.; P < 0.05 compared with vehicle control (*) and TcdA-vehicle (#), respectively; ANOVA on ranks). (e) Kaplan-Meier survival plots of mice orally gavaged with 1 × 106 C. difficile VPI10463 and GSNO (10 mg per kg per day); GSNO-InsP6 (10 and 0.25 mg per kg per day), InsP6 (0.25 mg per kg per day) or vancomycin (50 mg per kg per day). GSNO-InsP6 (10 and 0.25 mg per kg per day) was also delivered continuously by intracecal catheter (n = 12 per group). (f) Mice were orally gavaged with C. difficile VPI10463, and GSNO, GSNO-InsP6, InsP6 or vancomycin was delivered continuously by intracecal catheter (as in e). Survival is shown for day 4 after infection (n = 12; error bars show s.e.m.; P < 0.05 compared with vehicle control (*) and GSNO (#) respectively; ANOVA on ranks). (g) Schematic diagram of cellular intoxication by C. difficile exotoxins and mechanism for inhibition by host-generated GSNO. Intoxication results from autocleavage and cytosolic entry of the glucosyltransferase domain, whereas inhibition (orthosteric and allosteric) in the host is mediated by S-nitrosylation (red triangles) of the membrane-associated toxin CPD.
S-nitrosylation is already known to suppress parasitic and viral pathogenesis by inhibiting cysteine proteases39. However, host S-nitrosylation has not previously been shown to directly inhibit microbial exotoxin activity, and the dual protective mechanism identified here (catalytic-site inhibition and displacement of allosteric activator) is novel (Fig. 4g). Given the recent descriptions of a large number of microbial proteases (multifunctional autoprocessing RTX toxins (CPDMARTX) and CPDadh, a cysteine peptidase family homologous to MARTX)5,8 that show close structural similarly to the C. difficile toxins and probably require an active-site cysteine for host cell adhesion and entry, S-nitrosylation may represent a common host defense mechanism. This notion is supported by similar GSNO docking results to the CPD active site in C. difficile toxins and RTXVC, a member of the MARTX family.
Our results may be viewed in a broader context in which hyper- and hyponitrosylation of specific proteins represent disease-modifying events18,37. A major challenge in NO therapeutics is to selectively and specifically control the nitrosylation state of proteins that are identified with pathophysiology. Previous studies have indicated that allosteric mechanisms may be crucial in governing NO specificity18,40, and we now provide a potential therapeutic context for this principle by showing that allosteric modulation of S-nitrosylation can be used to treat C. difficile infection in mice. More generally, improved understanding of allosteric mechanisms may suggest new therapeutic approaches to regulating molecular targets of nitrosylation.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Eli & Edith Broad Foundation, the John S. Dunn Gulf Coast Consortium for Chemical Genomics Robert A. Welch Collaborative Grant Program, the Howard Hughes Medical Institute and grants from the US National Institutes of Health National Institute of Allergy and Infectious Diseases (R01AI088748, N01AI30050), National Institute of Diabetes and Digestive and Kidney Diseases (R01DK084509, K01DK076549; R21-DK078032-01), National Heart, Lung, and Blood Institute (R01-HL059130, R01-HL091876, R01-HL095463, P01-HL075443-06A, NO1-HV-00245) and 1UL1RR029876-01. We thank D. Powell, S. Weinman, C.S. Schein and G. Prestwich for their critiques.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS
T.C.S. designed the study, performed InsP6, GSNO and cytotoxicity assays, BIACORE analysis, and wrote the paper; P.U. performed the toxin S-nitrosylation and InsP6 binding studies; N.O. and W.B. performed the toxin structural modeling and molecular docking simulations; I.P. performed the SNO immunofluorescence; K.A., A.C. and V.A. performed toxin autocleavage, InsP7 phosphorylation and UDP-glucosylation assays; A.G.T. performed animal toxin studies; R.D.E. performed the mass spectrometry; J.E.W. performed the cysteine saturation labeling studies; M.L. provided clinical specimens; R.K. performed the CD spectral analysis; L.S., W.N. and H.F. developed the toxin mutants, performed InsP6 cleavage and stool cytotoxicity assays and animals studies; B.H., A.H. and J.S.S. performed or oversaw the measurements of GSNO and SNO proteins; J.S.S. assisted with the study design and writing of the paper; C.P. prepared holotoxins, performed animal toxin studies and assisted with study design and manuscript editing.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemedicine/.
References
- 1.Chen X, et al. A mouse model of Clostridium difficile–associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Savidge TC, et al. Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology. 2003;125:413–420. doi: 10.1016/s0016-5085(03)00902-8. [DOI] [PubMed] [Google Scholar]
- 3.Lyras D, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuehne SA, et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 5.Satchell KJ. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect. Immun. 2007;75:5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheahan KL, Cordero CL, Fullner Satchell KJ. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J. 2007;26:2552–2561. doi: 10.1038/sj.emboj.7601700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egerer M, et al. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 2007;282:25314–25321. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- 8.Pei J, Lupardus PJ, Garcia KC, Grishin NV. CPDadh: a new peptidase family homologous to the cysteine protease domain in bacterial MARTX toxins. Protein Sci. 2009;18:856–862. doi: 10.1002/pro.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puri AW, et al. Rational design of inhibitors and activity-based probes targeting Clostridium difficile virulence factor TcdB. Chem. Biol. 2010;17:1201–1211. doi: 10.1016/j.chembiol.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reineke J, et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- 11.Prochazkova K, Fullner Satchell KJ. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin. J. Biol. Chem. 2008;283:23656–23664. doi: 10.1074/jbc.M803334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule–induced allosteric activation of the Vibrio cholera RTX cysteine protease domain. Science. 2008;322:265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egerer M, Giesemann T, Herrmann C, Aktories K. Auto-catalytic processing of Clostridium difficile toxin B-binding of inositol hexakisphosphate. J. Biol. Chem. 2009;284:3389–3395. doi: 10.1074/jbc.M806002200. [DOI] [PubMed] [Google Scholar]
- 14.Prochazkova K, et al. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholera at multiple sites. J. Biol. Chem. 2009;284:26557–26568. doi: 10.1074/jbc.M109.025510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruitt RN, Chagot B, Cover M, Chazin WJ, Lacy DB. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing in Clostridium difficile toxin A. J. Biol. Chem. 2009;284:21934–21940. doi: 10.1074/jbc.M109.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreimeyer I, et al. Autoproteolytic cleavage mediates cytotoxicity of Clostridium difficile toxin A. Naunyn Schmiedebergs Arch. Pharmacol. 2011;383:253–262. doi: 10.1007/s00210-010-0574-x. [DOI] [PubMed] [Google Scholar]
- 17.Guttenberg G, et al. Clostridal glucosylating toxins: inositol hexakisphosphate-dependent processing of Closterium sordellii lethal toxin and Clostridium novyi α-toxin. J. Biol. Chem. 2011;286:14779–14786. doi: 10.1074/jbc.M110.200691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 19.Foster MW, Hess DT, Stamler JS. S-nitrosylation in health and disease-a current perspective. Trends Mol. Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu B, Pothoulakis C, Castagliuolo I, Nikulasson Z, LaMont JT. Nitric oxide inhibits rat intestinal secretion by Clostridium difficile toxin A but not Vibrio cholerae enterotoxin. Gastroenterology. 1996;111:409–418. doi: 10.1053/gast.1996.v111.pm8690206. [DOI] [PubMed] [Google Scholar]
- 21.Ng J, et al. Clostridium difficile toxin–induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology. 2010;139:542–552. doi: 10.1053/j.gastro.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Que LG, et al. Protection from experimental asthma by an endogenous bronchodilator. Science. 2005;308:1618–1621. doi: 10.1126/science.1108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savidge TC, et al. Enteric glia regulate intestinal barrier function and inflammation via release of S-nitrosoglutathione. Gastroenterology. 2007;132:1344–1358. doi: 10.1053/j.gastro.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 24.Hausladen A, et al. Assessment of nitric oxide signals by triodide chemiluminescence. Proc. Natl. Acad. Sci. USA. 2007;104:2157–2162. doi: 10.1073/pnas.0611191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiktorowicz J, et al. Quantification of cysteinyl S-nitrosylation by fluorescence in unbiased proteomic studies. Biochemistry. 2011;50:5601–5614. doi: 10.1021/bi200008b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffrey SR, Snyder SH. The biotin-switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 27.Gow AJ, et al. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J. Biol. Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- 28.Popoff MR, Geny B. Multifaceted role of Rho, Rac, Cdc42 and Ras in intercellular junctions, lessons from toxins. Biochim. Biophys. Acta. 2009;88:797–812. doi: 10.1016/j.bbamem.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 30.He X, et al. An ultrasensitive rapid immunocytotoxicity assay for detecting Clostridium difficile toxins. J. Microbiol. Methods. 2009;78:97–100. doi: 10.1016/j.mimet.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J. Mol. Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eichinger A, et al. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty A, et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang HY, Speicher DW. Identification of alternative products and optimization of 2-nitro-5-thiocyanatobenzoic acid cyanylation and cleavage at cysteine residues. Anal. Biochem. 2004;334:48–61. doi: 10.1016/j.ab.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 35.McMahon TJ, et al. Functional coupling of oxygen binding and vasoactivity in S-nitrosohemoglobin. J. Biol. Chem. 2000;275:16738–16745. doi: 10.1074/jbc.M000532200. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto A, Comatas KE, Liu L, Stamler JS. Screening for nitric oxide–dependent protein-protein interactions. Science. 2003;301:657–661. doi: 10.1126/science.1079319. [DOI] [PubMed] [Google Scholar]
- 37.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saura M, et al. An antiviral mechanism of nitric oxide: Inhibition of a viral protease. Immunity. 1999;10:21–28. doi: 10.1016/S1074-7613(00)80003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.