Abstract
Importance of the field
Fluorescence polarization (FP) is a homogeneous method that allows rapid and quantitative analysis of diverse molecular interactions and enzyme activities. This technique has been widely utilized in clinical and biomedical settings, including the diagnosis of certain diseases and monitoring therapeutic drug levels in body fluids. Recent developments in the field has been symbolized by the facile adoption of FP in high-throughput screening (HTS) and small molecule drug discovery of an increasing range of target classes.
Areas covered in this review
The article provides a brief overview on the theoretical foundation of FP, followed by updates on recent advancements in its application for various drug target classes, including G-protein coupled receptors (GPCRs), enzymes and protein-protein interactions (PPIs). The strengths and weaknesses of this method, practical considerations in assay design, novel applications, and future directions are also discussed.
What the reader will gain
The reader will be informed of the most recent advancements and future directions of FP application to small molecule screening.
Take home message
In addition to its continued utilization in high-throughput screening, FP has expanded into new disease and target areas and has been marked by increased use of labeled small molecule ligands for receptor binding studies.
Keywords: Competitive binding assay, drug discovery, fluorescence polarization, fluorescence anisotropy, high throughput screening, ligand displacement, enzyme assay
1. Introduction
Fluorescence polarization/fluorescence anisotropy (FP/FA) is a versatile solution-based technique that has been widely used to study molecular interactions, enzymatic activity, and nucleic acid hybridization. After its first theoretical description in 1926 by Perrin [1], the application has evolved from obtaining binding isotherms under carefully controlled settings to the study of small molecule-protein, antigen-antibody, and hormone-receptor binding in miniaturized automated settings. It was not until the mid 1990s that FP was adopted in high throughput screening to facilitate the drug discovery process, with its use being extended from direct interaction studies to complex enzymatic assays. Multiple reviews on the FP method have been published both around the turn of the century [2–5] and more recently [6, 7], covering its theoretical background, applications in basic research and drug discovery, and issues related to its use. This review aims to complement those articles and to highlight advancements of FP in several important drug target areas in the past 2 years (Table 1), and to extract general trends.
Table 1.
Articles from the past 2 years on FP application highlighted in this review
| Target/target type | Ligand/ligand type | Label | Reference |
|---|---|---|---|
| A2AAR/receptor | SCH442416/antagonist | Alexa Fluor 488 | [21] |
| CXCR4/receptor | T-22/peptide | Oregon Green 488 | [25] |
| Chymotrypsin and others/enzyme | BSA, others/protein | Epicocconone | [42] |
| Sfp/protein | CoA/co-substrate | BODIPY-TMR | [44] |
| AMP- and GMP-producing enzymes/enzyme | AMP, GMP/nucleotide | Alexa Fluor 633 | [55] |
| Kinase/enzyme | ADP/nucleotide | Alexa Fluor 633 | [57] |
| PI3 kinase/enzyme | ADP/nucleotide | Alexa Fluor 633 | [59] |
| TbCet1/enzyme | ADP/nucleotide | Alexa Fluor 633 | [60] |
| Hsp72 and Hsp90/protein | ADP/nucleotide | Alexa Fluor 633 | [61] |
| Gα mutants/protein | ADP/nucleotide | Alexa Fluor 633 | [62] |
| BRCT/protein | SRSTpSPTFNK/peptide | FITC and TAMRA | [69] |
| Plk1 polo-box domain/protein | GPMQSpTPLNG/peptide | FAM | [71, 72] |
| Thrombin/protein | T-15Ap, T-27Ap/aptamer | TAMRA | [73, 74] |
| Calmodulin/protein | W-7/antagonist | Cy5 | [85] |
| Gα/protein | RGS12 GoLoco motif/peptide | FITC and TAMRA | [86] |
| MIA/protein | AR54/peptide | Ru(bpy)3 | [92] |
| 5-HT2C/receptor | Serotonin analogs/compound | Cy3B | [99] |
| PKA and ROCK/kinase | Adc-Ahx-(D-Arg)6-D-Lys-NH2/peptide | TAMRA | [101] |
| FimH/protein | Mannoside/compound | FAM | [105] |
| Hsp90/protein | GA/compound | Cy3B | [109] |
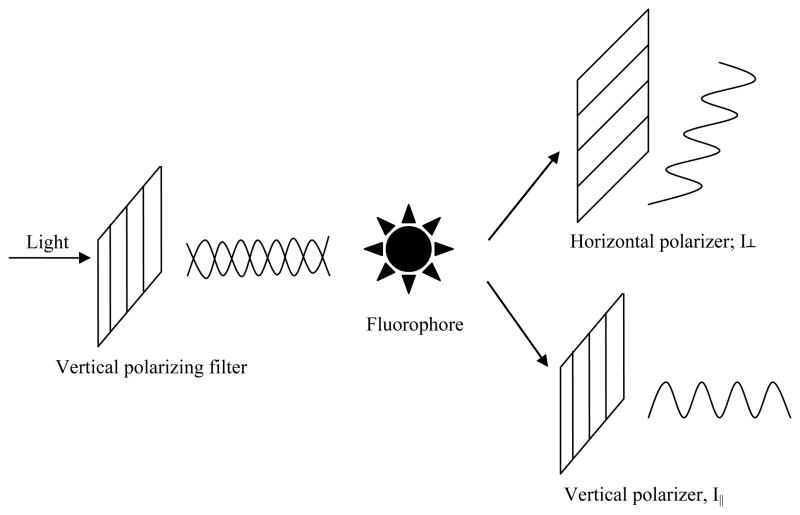
The principle of FP derives from the fact that the degree of polarization of a fluorophore is inversely related to its molecular rotation (Figure 1), itself being largely driven by Brownian motion [4]. Quantitatively, FP/FA is defined as the difference of the emission light intensity parallel (I||) and perpendicular (I⊥) to the excitation light plane normalized by the total fluorescence emission intensity (Equation 1 and 2, FP and FA are used interchangeably; see [2, 8] for discussion on the differences between the two terms)
Figure 1.
Basic principle of fluorescence polarization. A fluorophore is excited with light that is linearly polarized by passing through an excitation polarizing filter; the polarized fluorescence is measured through an emission polarizer either parallel or perpendicular to the exciting light’s plane of polarization. Two intensity measurements are obtained (I⊥ and I||) and used for the calculation of FA or FP.
| (Equation 1) |
and
| (Equation 2) |
It can be seen from Equations 1 and 2 that the FP value is independent of fluorophore concentration as it is not dependent on the absolute intensities of the emission light collected at either orientation. Such an independence of FP on the concentration of the fluorophore reagent (within the limits of instrument linearity and sensitivity) has largely been observed in a very broad spectrum of experimental settings and FP assay formats, and deviation from this relationship can serve as an indicator of fluorescence probe aggregation (anomalous FP increase and premature fluorescence intensity saturation upon increase of fluorophore concentration). On the other hand, the intrinsic fluorescence intensity (i.e., quantum yield) of a fluorophore may change upon binding to its cognate partner, thus resulting in significantly different contributions of the bound versus free forms of the fluorophore to the total fluorescence intensity of the sample, which in turn can complicate the interpretation of FP measurements [9, 10]. As instruments may have unequal sensitivity in detecting light in the perpendicular and the parallel orientations, a grating factor (commonly referred to as G factor) [6] has been introduced to correct for that bias in order to calculate absolute polarization values and for cases where data obtained from different instruments are to be compared [11].
Polarization relates a fluorophore’s lifetime (τ) (defined as the average time lapse between excitation and emission of the fluorophore) with its rotational relaxation time (μ), the latter being defined as the time it takes for a molecule to rotate through an approximate 68.5° angle after excitation [1] (Equation 3):
| (Equation 3) |
where R is the universal gas constant, V is the molar volume of the rotating molecule, T is the absolute temperature, and η is the solvent viscosity. It can be seen from Equation 3 that rotational relaxation time is dependent on molar volume provided that temperature and viscosity remain constant. Therefore, as molecular size of the fluorescent species is being altered through dissociation/breakdown or association/binding events, the degree of depolarization of plane polarized light changes accordingly, thereby directly affecting the FP value. While the FP technique thus explores the change in polarization of emitted light by a fluorescent reporter molecule as a function of the reporter’s size, approaches based on fluorescence lifetime measurements, not subject of the present review, are also gaining traction.
In the context of FP’s application in small molecule screening and drug development, the inhibition constant Ki is often desired, especially when data obtained under different experimental conditions are to be compared. As FP is often linearly proportional to the percent bound/free species and therefore can quantitatively determine IC50 (concentration at 50% inhibition), the corresponding Ki/Kd can be calculated using the appropriate versions of the Cheng-Prusoff equation [12–15]. Recently, two groups have incorporated derivations of Ki/Kd from IC50 data, including FP results, into web-based algorithm tools [16–19].
2. FP applications
The FP method has been applied to almost every protein class, including G-protein-coupled receptors (GPCRs), nuclear receptors, and enzymes. It has also been applied to the analysis of molecular interactions including protein-protein, protein-DNA, and protein-ligand binding events and, separately, has been used in multiple ways to provide readout for monitoring enzymatic reaction progress. FP applications can be classified based on target class or molecular interaction. These two classifications often cross each other as, for example, a nuclear receptor can be targeted for its interaction with ligand, co-regulator, or DNA [3].
2.1. G-protein coupled receptors (GPCR)
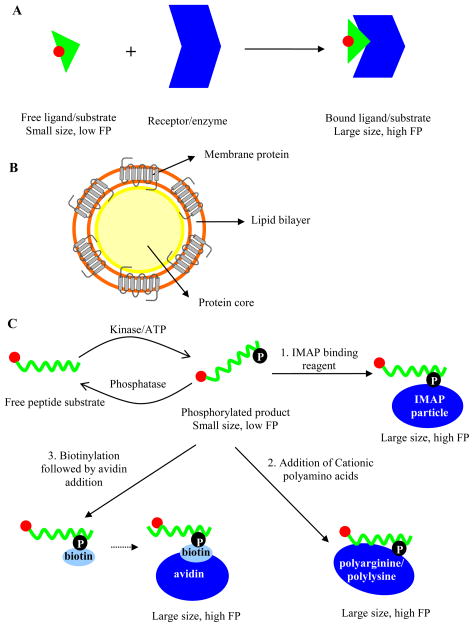
The GPCR super family comprises druggable transmembrane receptors that mediate signal transduction pathways [20]. Despite the availability of a number of known GPCRs (~800) and the prominent space they have taken among therapeutic targets, the application of FP has not been fully extended to these proteins. Isolation of sufficient GPCR with intact conformation and stability in soluble format and the design of a suitable fluorescent ligand are two major challenges that hamper the wide spread application of FP for GPCRs [21, 22]. As a result, a large portion of the GPCR assays for HTS and drug discovery still use radioligand binding assays due to the sensitivity that is afforded by the high radioligand specific activity, allowing the in vitro characterization of GPCRs expressed at low densities [23]. However, over the last decade, radioligand binding assays have been gradually replaced by FP for discovery of novel antagonist and agonists of GPCRs and determination of their binding affinities, with the added benefits of reduction in assay cost and health hazards. FP assay setup for GPCRs usually follows an increase in FP value upon binding of a fluorescently labeled ligand to its receptor (Figure 2A). In competition binding FP assays, the presence of unlabeled ligands or small molecule inhibitors of the interaction results in the displacement of the labeled ligand molecules, thereby increasing their tumbling motion which in turn can be detected as a decrease in FP value.
Figure 2.
A) Schematic illustration of FP principle in relation to receptor-ligand interaction; B) Illustration of the lipoparticle nanotechnology as in Jones et al. [25] (membrane proteins are captured on the surface of Gag core protein to produce a nanoparticle that serves as a GPCR depot, fluorescent ligand not shown); Schematic illustration of FP principle in relation to C1) the IMAP technology, C2) use of cationic polyamino acids, C3) biotinylation and addition of streptavidin. In the examples A and C1-C3, the rotation of a fluorophore-labeled ligand, substrate, or product is rapid (fluorophore denoted by a red circle throughout) whereas its motion is slowed down when it is bound to a larger entity (dark blue) and its FP value is correspondingly increased.
In an early report, Banks et al. suggested guidelines for successful FP assay design for GPCRs, including requirements for GPCR expression level, ligand binding affinity and fluorophore photophysical properties [24]. However, producing ample amounts of high-quality GPCR has remained challenging. In order to circumvent the difficulties associated with obtaining concentrated and stabilized receptors, Jones et al. utilized an lipoparticle nanotechnology (developed by Integral Molecular, Inc., Philadelphia, PA) to provide a cell-free system for the GPCRs [25]. The lipoparticle nanotechnology is built upon membrane-enveloped retroviruses whose core protein buds out of cell membrane and becomes enwrapped with the membrane protein of interest (Figure 2B) [26]. The large size of the resulting lipoparticles (150–200 nm) makes them an attractive choice for a binding partner of a fluorescently labeled ligand in an FP assay. The lipoparticles are generated at peak expression levels of the target protein, enabling capture of concentrated membrane protein, usually 10–100 fold enriched relative to cell or membrane preparations. In a direct FP binding assay using CXCR4 as a model receptor (Figure 2A), Oregon Green 488 labeled peptide ligand (T-22) was used as the cognate partner to bind CXCR4-coated lipoparticles, and a large assay window of 307 mP was obtained. The binding specificity of labeled T-22 to CXCR4 was confirmed through titration of an unlabeled version of T-22 and a control peptide, and it was demonstrated that ligand-receptor binding properties were not compromised. These results suggest that lipoparticle-based FP assays can be developed for other GPCRs incorporated into lipoparticles. An important caveat of this approach is worth noting: while the target protein may be expressed and isolated, key accessory proteins, such as binding partners that modulate the GPCR’s affinity towards its ligands, may be absent from the preparation, which in turn can lead to skewed assay results [27, 28].
Another recent advancement in this area is the successful design and utilization of a novel fluorescent tracer for the development of an FP assay to identify agonists and antagonists for A2A adenosine receptor (A2AAR) [21]. A2AAR is one of the four known subtypes of adenosine receptors (ARs), all of which are members of the GPCR superfamily [29]. FP or similar-type assays for this subset of GPCRs have been lacking due to the absence of appropriate ligand. The novel fluorescent tracer MRS5346 was based on the high-affinity A2AAR antagonist, SCH442416, and incorporated an Alexa Fluor 488 (AF488) label. The attachment point for the AF488 dye and the type and length of linker between it and SCH442416 were guided by ligand docking analysis utilizing the recently published X-ray structure of A2AAR. The MRS5346 displayed submicromolar affinity in radioligand membrane binding assays and was validated by testing several known A2AAR agonists and antagonists, with Ki values obtained by the FP assay being comparable to those obtained from the classical radioligand binding method. This report is the first example of a successfully configured FP assay for A2AAR.
2.2 Enzymes
The FP principle works well with popular enzymatic targets, such as kinase, protease, phosphatase, DNase and RNase. FP-based methods for the measurement of enzyme activity can be arbitrarily divided into two major categories: antibody-based and antibody-independent methods. Methods that are antibody-independent can be further differentiated as affinity-based or non affinity-based.
2.2.1 Antibody-free affinity-based methods
In affinity-based methods, in addition to a fluorophore that is used to label the reaction substrate, a chemical entity, such as nanoparticle, polymer, or large protein, is introduced into the system to effect a change in the size of the fluorescently labeled species. The addition serves to induce a change in the size of the labeled probe as a function of enzymatic reaction progress. Most of these affinity-based methods were originally designed for use with kinase reactions, with their utility being subsequently expanded to include other enzymes, such as proteases, phosphatases, and phosphodiesterases.
The Immobilized Metal Ion Affinity Particle (IMAP) technology is based on the utilization of a specific trivalent metal (MIII, most likely iron, see reference [30]) coordination complex as the binding partner for phosphate groups (Figure 2C1). The kinase-catalyzed attachment of a phosphate group onto the fluorescent peptide leads to its binding by the trivalent metal ions immobilized on nanoparticles. By complexing with the IMAP particles, the size of the peptide is increased, restricting its molecular mobility and leading to a high polarization upon excitation with plane-polarized light. The specificity of the IMAP particles toward only the phosphorylated peptide allows measurement of the degree of phosphorylation, and provides a platform that is largely independent of the sequence flanking the phosphorylated residue [31] (though important exceptions exist, such as in situations encountered with kinases that require a “primed” (pre-phosphorylated) substrate, or highly negatively charged substrates). Thus, the technology can be adopted for various kinases, phosphatases, and phosphodiesterases [32]. Because IMAP is used to directly quantitate the amount of phosphorylation product, and the concentration of fluorescently labeled peptide utilized in kinase assays is relatively high in order to match the enzyme kinetic parameter (KM), the fluorescence signal is strong, conferring more resistance to interference from autofluorescent library compounds [33]. IMAP-based HTS protocols [34] and additional evaluations of the technology [31, 35], have been described in detail elsewhere.
Similarly, cationic polyamino acids also present an attractive choice as large-size binding partners for the phophorylation product [36, 37]. Peptide substrates become more negatively charged upon phosphorylation, and cationic polyamino acids (i.e. polyarginine), under optimized conditions of ionic strength, bind more tightly to the phosphorylated peptide than to their unphosphorylated counterpart (provided the net charge of the peptide product is negative). Thus, FP increases to a greater extent when polyarginine is added to phosphorylated peptide, providing an assay window to detect the extent of kinase reaction [36] (Figure 2C2), and the method has been applied to both peptide substrates with net charge of near zero as well as those with higher negative charges. Additionally, this method has been expanded to detect kinase activity in real time and was found to be suitable for monitoring phosphatase and protease reactions [37]. While being less-frequently exploited, this cationic polyamino acid-based approach represents an overall cost-effective strategy relative to the IMAP technology.
Another affinity-based FP method capitalizes on the strong binding interaction between biotin and avidin/streptavidin. Jeong et al. described an FP protocol where a fluorescently labeled peptide substrate was first thiophosphorylated using ATPγS in lieu of ATP. The reaction was followed by covalent biotinylation of the nucleophilic thiophosphate sulfur with an iodoacetyl derivative of biotin [38]. The biotinylated fluorescent peptide product generated a high FP upon addition of streptavidin (Figure 2C3). Both the cationic polyamino acid-based and biotin-(strept)avidin-based FP methods are innovative, but they have respective limitations in their applications. For instance, issues related to the application of polyarginine include adverse effects on enzyme activity and precipitation of the biopolymer at high concentrations of negatively charged molecules [37]; in turn, compounds that interfere with the interaction of biotin and avidin can confound the interpretation of enzyme activity [39]. Lastly, assays utilizing ATPγS are generally more difficult to set up and use in detailed mode-of-inhibition kinetic studies because of the poor acceptance of this unnatural substrate by most kinases (leading to low turnover relative to ATP and a compressed range of possible concentrations that can be used to perform enzymology).
2.2.2 Antibody-free direct-label methods
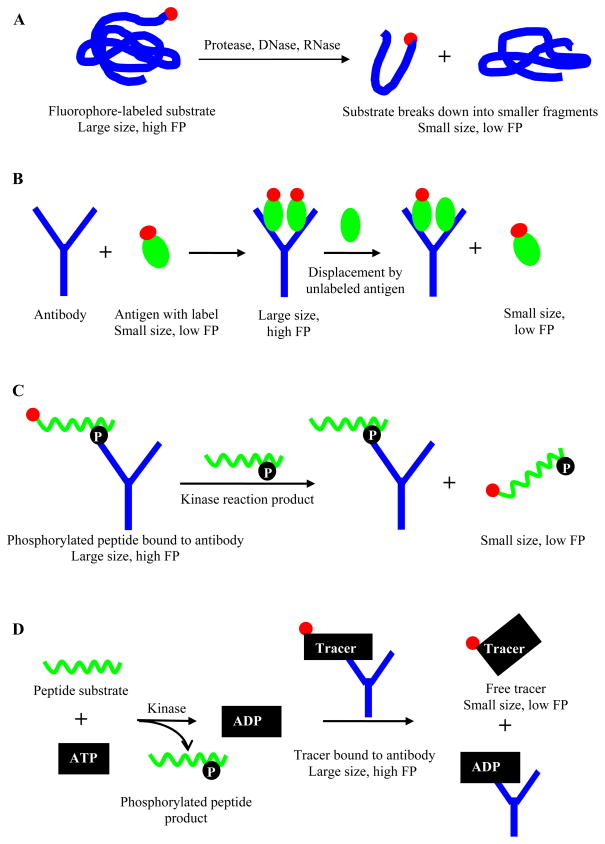
In non affinity-based methods, the substrate is usually labeled with a fluorophore and possesses a low rotational relaxation; thus the readout almost always follows a decrease in polarization value as the enzymatic reaction, such as a proteolytic or hydrolysis process, leads to the breakdown of the fluorescently labeled substrate and thus a decrease in its molecular size (Figure 3A). Early examples include the use of a fluorescein-labeled amylose for amylase activity in serum and urine [40], and a coumarin derivative labeled amide or peptide for testing the activities of trypsin, elastase, and chymotrypsin [41]. Recently, Cleemann et al. described a FP assay for protease activity using nonderivatized natural protein substrates that were labeled with a natural fluorophore, epicocconone [42], which binds proteins in a reversible manner, and the formation of an internal charge-transfer complex leads to high fluorescence in hydrophobic environment. The authors utilized the in situ labeling ability of epicocconone and found that the FP assay was capable of monitoring protein digestion using substrates of different molecular weights (3–77 kDa) and in a range of pH conditions. The epicocconone-based FP assay was also shown to allow measurements of enzyme kinetic parameters and inhibitor IC50s, and was amenable to HTS adoption.
Figure 3.
Schematic illustration of FP principle in relation to A) degradative enzymatic reactions (during hydrolysis, breakdown of fluorophore-labeled substrate into smaller molecules produces species with lower FP, which can be used to measure enzymatic activity ), B) FPIA (binding of a labeled antigen to its antibody leads to an increase in FP; displacement of labeled antigen by unlabeled antigen reduces the FP value, and the degree of FP reduction is correlated with the antigen concentration in unknown samples), C) competitive FPIA for kinase (displacement of a fluorescently labeled phosphopeptide tracer from phosphospecific antibodies by kinase reaction-generated phosphopeptide product (unlabeled) results in a decrease in FP, which can be used to measure kinase activity), and D) Transcreener™ assay (displacement of tracer bound to nucleotide-specific antibodies by kinase reaction-generated product leads to a lower FP value which correlates with the kinase activity). Red circle represents fluorophore.
2.2.3 Non-turnover Enzymatic Assays
In situations where enzymatic turnover assay cannot be realized, such as in cases of bimolecular reactions where one of the co-substrates is not readily accessible, an FP assay can be configured through direct binding of the protein with its substrate, in a design scheme similar to that applied in receptor-ligand binding (Figure 2A). In this case, the more readily available substrate is fluorescently labeled, and in the assay the FP value increases due to the formation of the larger enzyme-substrate complex. For instance, Sfp is a group II phosphopantetheinyl transferase (PPTase) from Bacillus subtilis, and is responsible for the transfer of the phosphopantetheinyl moiety from coenzyme A (CoA) to carrier proteins. Development of HTS assays to discover Sfp inhibitors as candidates for antibacterial drug development has been severely limited by the lack of knowledge regarding the preferred sequence of the phosphopantetheinyl acceptor region: to date, only one screening assay utilizing a specific consensus-sequence peptide has been described [43]. Capitalizing on the wide substrate tolerance of Sfp with respect to its CoA binding pocket, Duckworth et al. utilized a BODIPY-TMR labeled CoA derivative as the ligand for Sfp for the development of an FP HTS assay to identify PPTase inhibitors [44]. In the absence of inhibitors, the formation of the larger-size CoA-enzyme complex leads to a high FP value. In the presence of Sfp inhibitors, the labeled CoA is displaced into solution resulting in a low FP value. While this assay format allows for the facile set-up of HTS campaigns to address multiple phosphopantetheinyl transferases, the absence of carrier protein co-substrate in the present assay limits the utility of the method to the identification of only CoA-competitive inhibitors.
2.2.4 Fluorescence polarization immunoassay (FPIA)
First developed in the early 1960s [45], FPIA is an antibody-based technique that usually follows the competition between unlabeled antigen and labeled antigen for binding to an antibody. Upon the displacement of the labeled antigen by the unlabeled antigen, the rate of the tumbling motion is increased due to increased amount of free labeled antigen, resulting in a lower polarization (Figure 3B). Thus, FPIA method can be used to quantify antigen concentration in unknown samples as the analyte concentration is proportionally related to the degree of polarization change. Throughout the past decades, FPIA has been used to study antibody interactions with protein antigens [45], as well as for measurement of various small molecule analytes, such as hormones, toxins, antibiotics or drugs [46, 47]. FPIA is advantageous compared to other immunoassays, including radioimmunoassay or enzyme-linked immunosorbent assay (ELISA), as it does not require separation step to partition bound from unbound molecules [48].
In HTS, FPIA has been used to monitor reaction progress in multiple target settings, most prominent of which remain protein kinases. Kinases have remained highly popular in drug development due to their considerable diversity in humans (over 500 kinase genes have been identified) and their heavy involvement in a wide range of signaling pathways implicated in human diseases [33]. Early kinase assay methods have been universally based on the use of 32P-ATP [49] and as such present a good candidate for replacement with new-generation nonradioactive homogeneous platforms. FPIA represented an attractive alternative to replace radioactive biochemical assays in the early efforts for kinase assays [50–52]. Functionally, protein kinases transfer phosphate group from nucleoside triphosphate (primarily ATP) to their respective peptide substrates, generating phosphorylated peptides and nucleotide diphosphate. Early FPIA assays were designed to bind the phosphorylated peptide products using phosphospecific antibodies, making use of the corresponding reagents already developed for Western blot type studies. In direct FPIA, phosphorylation of a fluorescently labeled peptide substrate leads to its recognition by the phosphospecific antibody, and formation of the product-antibody complex results in a higher FP value compared to that associated with the free peptide substrate [53]. Direct FPIA kinase assay (Figure 3B) requires a substantial degree of substrate phosphorylation [33], and assay sensitivity can be improved by switching to a competitive FPIA format. In competitive FPIA, a fluorescently labeled phosphopeptide tracer representing the kinase reaction product is bound to an antibody, corresponding to a high FP value. Displacement of the tracer by increased amount of reaction-generated phosphopeptide product causes higher molecular mobility of the tracer and consequently, a decrease in FP value (Figure 3C).
Because the above competition assays require specific anti-phosphopeptide antibodies, and because the peptide substrates are variant molecules with each sequence only recognizable by one particular kinase, the development of a generic FPIA-based HTS assay to accommodate the diversity seen within the kinome has been hampered (although welcome exceptions exist where in recent years, PY20 or other generic pY antibodies have been described, which enable the testing of most tyrosine kinases; furthermore, small sets of antibodies that work with substrates which can be used by many serine/threonine kinases [54] have become commercially available). To circumvent the problems associated with peptide substrate-based kinase assays, the Transcreener™ FPIA platform (BellBrook Labs, Madison, WI) detects the invariant nucleotide product through high-affinity nucleotide-specific antibodies, obviating the need for kinase-specific antibody. The Transcreener™ assay is comprised of a red-shifted fluorophore labeled nucleotide, an antibody that binds to the nucleotide with high affinity and specificity [55, 56], and quench buffer used to stop the enzymatic reaction. Similar to competitive FPIA assays that utilize phosphospecific antibodies (Figure 3C), Transcreener™ FPIA assays follow a reduction in FP value as the nucleotides generated by reaction displace tracer bound by the antibodies (Figure 3D). The assay does not require high nucleotide consumption or a coupling enzyme reaction, facilitating the process of hit selection as a result of less compound interference and false positives [57]. Additionally, the design of the Transcreener™ assays allows the accommodation of kinase proteins whose native substrate is a protein acceptor instead of a nonphysiological peptide substrate, increasing the potential for identification of allosteric inhibitors [56]. The four different nucleotide (ADP, AMP/GMP, GDP and UDP)-based Transcreener™ assays allow a wide range of nucleotide-processing enzymes to be assayed in one universal format including kinases [58, 59], triphosphatase [60] and proteins possessing ATPase [61] or GTPase-accelerating protein (GAP) activity [62].
2.3 Protein-protein interactions (PPIs)
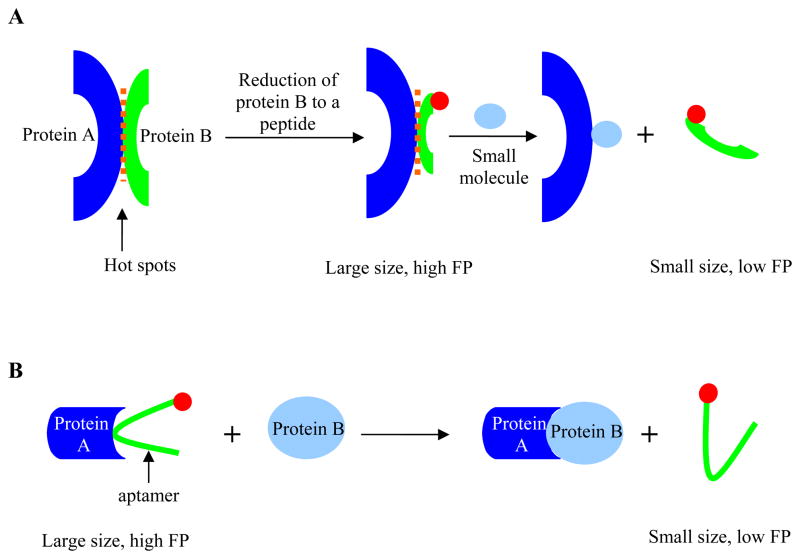
PPIs are emerging as novel therapeutic targets [63–65], and there are ample examples of PPI disrupters based on antibodies, proteins or peptides [66]. However, it is of interest to pursue “drug-like” small molecules as biochemical tools or potential compounds for therapeutic agents [64], to complement or replace the existing protein therapeutics. There are a number of methods to identify inhibitors of PPIs, such as virtual screening and structure based design, but HTS is the most common approach that can rapidly identify candidate small molecule inhibitors of PPIs [63]. The debate over whether PPIs are druggable or not remains, largely due to the fact that they are characterized by large and ill defined interfaces with variable contact points and lack of deep, well-defined binding cavities [67]. PPIs are complex and are governed by different physical features and forces [63], but occasionally, a small binding epitope, also referred to as a “hot spot”, can be identified for one of the binding partners. The concept of “hot spots” is based on the observation that the PPI free energy of binding is unevenly distributed across the protein interfaces, and the bulk of binding energy is contributed by a small subset of amino acid residues [68]. As a result, recent efforts have focused on utilization of truncated peptides containing such binding epitope and the search of “hot spots” PPI disrupters to increase the chance of inhibitor identification [69, 70]. The transfer of epitopes to smaller entities, such as peptides, affords both higher affinity binding and molecular size distinction between the smaller peptide entity and its binding protein partner, providing an opportunity for detection of PPIs by FP. In FP assays for PPIs that target the “hot spots”, a consensus peptide designed based on the “hot spots” interaction is labeled with a fluorophore, and in the absence of small molecule inhibitors, the formation of peptide-protein complex leads to a high FP value (Figure 4A), akin to the assay scheme utilized for GPCR-ligand interaction studies (Figure 2A). Small molecule inhibitors are detected as the peptide is displaced from its binding site, and the liberation of the fluorescently labeled peptide produces a low FP value due to its increased rotational mobility.
Figure 4.
Schematic illustration of FP principle in relation to A) “hot spots” PPIs (one of the protein binding partners can be truncated to a consensus peptide if a binding epitope is known/available; binding of the protein to the labeled peptide produces a high FP value, which decreases upon displacement of the labeled peptide by small molecule inhibitors), and B) fluorescently labeled aptamer for PPIs (displacement of fluorescently-labeled aptamer raised against one protein (denoted protein A) from its binding site by a cognate protein binding partner frees up the aptamer and produces a low FP value; the presence of allosteric inhibitors of protein A or inhibitors that specifically bind to protein B can be detected when the high FP value (aptamer remains bound to protein A) is retained). Red circle represents fluorophore.
Despite the increased use of FP on PPIs in high-throughput format, the challenge remains as initial hits are usually non-specific inhibitors or molecules that lack “drug-like” properties [63]. A welcome exception is the natural product thymoquinone and its synthetic derivative Poloxin, with the latter originally identified from a FP-based primary screen [71]. Polo-like kinase 1 (Plk1) has been suggested as a target for cancer therapy, with most available inhibitors acting as ATP competitors within the Plk1’s conserved ATP binding site [72]. In order to search for monospecific inhibitors, Reindl et al. conducted a screening campaign to target Plk1 polo-box domain (PBD) PPI which mediates intracellular localization of Plk1. The study utilized a FP assay for identification of small molecule inhibitors of protein-protein interactions, binding of the Plk1 PBD to a labeled phosphothreonine-containing peptide. In addition to the single-digit micromolar apparent IC50s obtained in the primary FP assay, both compounds were further shown to interfere with correct localization of Plk1, cause mitotic arrest, and induce apoptosis in cancer cells. Thus, thymoquinone and Poloxin, identified from a FP-based PPI screen, have subsequently been confirmed for their inhibitory activities and represent novel nonpeptidic inhibitors of Plk1 PBD.
In contrast to the “hot spots” approach and labeling a smaller protein binding partner, a different yet valuable approach is to tackle PPI assay development without any modification of the proteins. For example, in a recent study the interactions of α-thrombin with proteins were investigated by the use of TAMRA labeled aptamers [73, 74]. Aptamers are artificial nucleic acids designed to bind to their target with high specificity and affinity [75]. α-thrombin (protein A in Figure 4B) and a TAMRA labeled aptamer were mixed first and the formation of protein-aptamer complex led to a high FP value (Figure 4B). Upon the addition of HirF (protein B in Figure 4B), a sulfated C-terminal fragment of hirudin [76], the aptamer was displaced from its α-thrombin binding site, resulting in a decrease in FP value. Thus, both protein binding partners were utilized without modifications. Using this setup, small molecule inhibitors of PPIs can potentially be identified when high FP value is retained upon addition of protein B in the presence of inhibitor (i.e., the potential inhibitor prevents protein B from binding to protein A and from displacing the labeled aptamer). Such an approach may be limited to the identification of allosteric inhibitors of protein A or inhibitors that specifically bind to protein B as aptamer-competitive inhibitors would displace the aptamer from protein A’s active site and create low FP value. Despite its limited utility to small molecule HTS, this approach represents one of the few non-invasive methods to interrogate PPIs.
3. Strengths and drawbacks of FP
FP assays utilize single fluorescent label strategy and avoid the use of radioisotopes and filtration or separation steps. Thus, fewer reagents are generally needed, and the assay protocol is a simple mix-and-read (i.e. homogenous) with the assay reagents being overall inexpensive. Additionally, reagent equilibrium is not disturbed due to lack of separation step, and plates can often be repetitively measured as FP detection does not destroy samples. These features have led to the frequent use of FP assays in HTS.
As a ratiometric method, FP is relatively insensitive to absorptive interferences or inner filter effects, but can suffer from autofluorescence and light scattering [2, 48, 77] as these effects can confound sample FP calculation. They can be flagged as suspect by introducing kinetic read or a pre-read for background subtraction [48], or by fluorescence profiling of the libraries screened [78, 79]. Interference effects can also be reduced by using red-shifted probes (such as BODIPY® TMR [24] or Cy5 dyes [80]), and long lifetime fluorophores [79] (see Section 4 for examples).
In contrast to other binding methods whose design is based on differential saturation of protein binding sites, FP detects the fraction bound of the tracer. Thus, the requirement for a relative saturation of the tracer, as the signal window is directly proportional to the difference in fractions of tracer bound between the two control states, translates into the total amount of protein being required for an HTS to be strongly dependent on the Kd of the tracer-protein interaction and can, in certain cases, lead to protein production demand that cannot be readily met [50]. In addition, the ability of an FP assay to detect “weak” inhibitors is dependent on this tracer-receptor Kd, and it can be a problem for weak interactions [81], which in turn is typically overcome by configuring the assay in a TR-FRET (or radiometric) format [82]. Additionally, the necessary use of high concentrations of test compounds can lead to anomalous polarization through aggregation-based non-specific binding with the tracer molecule especially if the test compounds have hydrophobic moieties and tend to form micelle-like particles [83, 84]. A recent study highlights this issue: in a fluorescence probe competition experiment to evaluate calmodulin antagonists, anomalous probe polarization was observed at high compound concentrations (> 100 μM) [85]. The anomalous high FP value was consistent with nonspecific interactions between the fluorescent probe and compound aggregates. Thus, it was impossible to use high compound concentrations as they interfered with FP of the probe both in the presence and in the absence of the protein binder.
4. Practical consideration in FP assay design for HTS
Fluorophore nature can affect ligand binding affinity with target protein, protein function and, ultimately, FP assay success in ways that have been difficult to rationalize and predict. During the course of an FP assay development for inhibitors of the Gαi1 interaction with the GoLoco motif of the regulators of G-protein signaling (RGS), two red-shifted fluorophores were evaluated [86]. With an FP assay using a FITC labeled peptide of the same sequence already at hand, an attempt was made to reduce compound interference during HTS by pursuing a red-shifted fluorophore as the label. While the TAMRA-labeled peptide produced greater FP value compared to that from the FITC-peptide, the BODIPY Texas Red-labeled peptide failed to produce a titration curve with Gαi1. The lack of titration curve with BODIPY Texas Red labeled peptide was largely due to an adverse effect of the fluorophore on the peptide binding affinity and/or probe aggregation due to the hydrophobic nature of the fluorophore, and represents an example of the complexity associated with fluorophore selection for such assays.
Nevertheless, several important fluorophore properties need to be considered in FP assay design, such as lifetime, stability, quantum yield, and extinction coefficient (see [6, 22, 87, 88] for reviews on the subject). Fluorophore lifetime, along with ligand size and molecular weight change upon binding determine the dynamic range of FP assays. From simulation investigations [2] [89], it is evident that either coupling short-lived fluorophores with smaller ligands or attaching long-lived fluorophores to larger protein ligands can increase the chance of obtaining a useful FP assay window [90]. Long-lived fluorophores that have been applied in FP assays include those based on ruthenium (Ru) (τ ~ 500 ns) [91] and rhenium (Re (I)) (τ ~ 3 μs) [89] metal-ligand complexes. Their desirable features include inherent photostability, enhanced signal-to-noise ratio and higher resistance to autofluorescence [92]. Based on the aforementioned dependence of FP on fluorophore lifetime and ligand size, these metal-ligand complexes are particularly useful in FPIA where large antigens are present, such as human serum albumin (HSA) [89]. A recent report by Riechers et al. suggests that these long-lived fluorophores can be extended for binding studies of target protein with both high- and low-molecular weight ligands by immobilizing a known binding partner of the target protein [92]. In that study, a small peptide, AR54 (a known melanoma inhibitory activity (MIA) protein-binding peptide), was immobilized on the surface of a well plate while MIA was labeled with Ru(bpy)3-isothiocyanate. The mobility of MIA was severely restricted upon binding to the immobilized AR54 (high FP), and displacement of MIA from AR54 by an inhibitory compound led to a reduction in FP value. Drawbacks of using the long lifetime transition metal labels are that they limit the sensitivity of FP assays due to the complexes’ low quantum yields and low extinction coefficients [5, 89].
In addition to the fluorophore’s photophysical properties, conjugation chemistry (a term loosely combining properties such as linker length, linker rigidity, and precise attachment position of the fluorophore/linker to the rest of the probe) also influences the FP assay design success [93–98]. In the empirical process to obtain suitable probes using different linkers, including type, length, and rigidity, there are two main, and often conflicting, rules that need to be taken into consideration [11, 22]. Attachment points should be avoided where local rotational effect (“propeller effect”) of the ligand, even when bound to protein, is present; this requirement typically calls for the use of very short linker. On the other hand, synthetic probes whose binding affinities are interfered with upon the incorporation of the fluorophore should be re-designed, often by extending the linker portion. A recent study provides an example of the complexity associated with the development of a small molecule-based FP ligand. In it, fluorescent ligands were sought for the design of a direct FP binding assay (Figure 2A) to screen for agonists of the 5-HT2C receptor [99]. Molecular modeling of the serotonin-5-HT2C receptor interaction led to the identification of the aryl alcohol of serotonin as a convenient starting point for design of the fluorescent probe. A number of serotonin analogs were synthesized using several red-shifted dyes (Cy3B [100], TMR, EVOBlue™ 10) and different-length aminoalkyl linkers, and their binding affinities were established through radioligand binding and functional assays. It was found that, in addition to the type of dye, linker length also had a profound effect on binding affinity: the Cy3B-labeled serotonin analogue with the shortest linker length was found to be the best ligand, producing a robust increase in FP value and good assay window in receptor titration. In contrast, Cy3B-labeled ligands with increased length of aliphatic linker were associated with poor assay window and, somewhat surprisingly, with increased background FP value.
To manage the above-mentioned complexity, structural information in combination with molecular modeling is increasingly being used to guide the design of small molecule based tracers [21, 85, 99, 101]. Arai et al. utilized a small molecule tracer (Cy5-W-7), instead of a labeled peptide, to configure an FP-based binding assay (Figure 2A) for calmodulin (CaM) antagonists [85]. W-7 is a small molecule antagonist of CaM and has been shown to inhibit CaM-activated enzyme (such as calcineurin phosphatase) activity [85]. As outlined earlier, attaching fluorophores to small molecules frequently involves an extensive iterative process which includes testing a range of reaction schemes at multiple sites on the ligand molecule [50]. Steric hindrance may be introduced upon the addition of a fluorescent group to the ligand and this can lead to severe loss of affinity upon ligand binding to target protein [102]. With the aid of a previous structural study, Arial et al. selected the amine group of W-7 as the attachment point for fluorophore linkage. It was later determined that the binding affinity of W-7 to CaM was not only not disturbed upon incorporation of Cy5 but that improved binding affinity towards CaM was conferred relative to unlabeled W-7. This was attributed to the additional interaction between CaM and Cy5 or its linker region. Along with the Kecskés work on A2AAR ligands described in section 2.1 [21], this study is yet another example of the possibility to design small molecule-based tracers for FP assays.
5. Expert opinion
Properly configured FP assays provide quantitative measurement on the strength of molecular interactions and allow monitoring of protein enzymatic activity. High speed, simplicity, and relatively low cost combine to make FP an attractive assay technology. In addition to the developments of FP methods for testing of popular drug targets, there has been recent expansion of the application into new disease areas, new types of assay reactions, and new target types, as outlined below.
Carbohydrates play numerous roles in living organisms, serving as energy source, mediators of signaling, and structural building blocks. Limited understanding of carbohydrate-protein interactions and high polarity of carbohydrates may be responsible for the underrepresentation of this class of molecules as a source of therapeutics [103]. FP is applicable to the characterization of carbohydrate-protein interactions, but there have been limited reports of its use [104]. Recently, an FP competition binding assay was developed to determine affinities of a series of biarylmannosides to FimH [105], one of the subunits that constitute type 1 fimbriae, a bacterial surface lectin that mediates attachment to host tissues [106]. The carbohydrate binding pocket of type 1 fimbriae has been identified to locate only within the FimH subunit [107]. Because carbohydrates have been shown to block bacteria adhesion to animal cells in vitro [108], inhibition of FimH is considered a promising approach to prevent bacterial entry and infection. Carboxyfluorescein (FAM)-labeled mannoside was used to configure the FimH FP assay where displacement of the FAM-labeled mannoside by test compounds was expected to cause dose-dependent reduction in polarization (Figure 2A). The assay was used to support SAR during a structure-based drug design to yield biarylmannosides as the most potent antagonists of FimH reported to date.
A large category of diseases where there has been an overall paucity of FP assays is neglected tropical diseases, and a welcome change is a recent work to develop an FP assay targeting Hsp90 in the context of adult filarial worm lysates for development of therapeutics against lymphatic filariasis [109]. Hsp90 has been studied as a target in diseases such as cancer, and FP assays have been applied to evaluate purified Hsp90’s ATPase activity [61], as well as its interaction with other proteins [110]. As Hsp90 had been suggested as a possible target in lymphatic filariasis, the present work utilized a Cy3B labeled geldanamycin, a specific inhibitor of Hsp90, to configure a high-throughput FP assay which was further optimized for use with worm lysates and validated using unlabeled geldanamycin and other Hsp90 binders. The FP assay was shown to be sensitive to species-specific inhibitors and to distinguish binders of different terminal sites on Hsp90. This study illustrated that FP could be applied to probe protein-small molecule interactions in lysates in search of drug leads to treat a neglected tropical disease.
Looking forward, FP has the potential to be applied in novel ways to address the emerging field of epigenetics. To date, FP has been used to monitor the binding of a fluorescein-labeled histone peptide to the methyltransferase G9a in a simple non-turnover format [111]. However, the corresponding use of FP to configure assays for the typically weak (Kd > 5 μM) interactions involving the “readers” of the histone code, such as bromodomains and chromodomains, has not been demonstrated, highlighting the FP method’s limitations when weak interactions are considered.
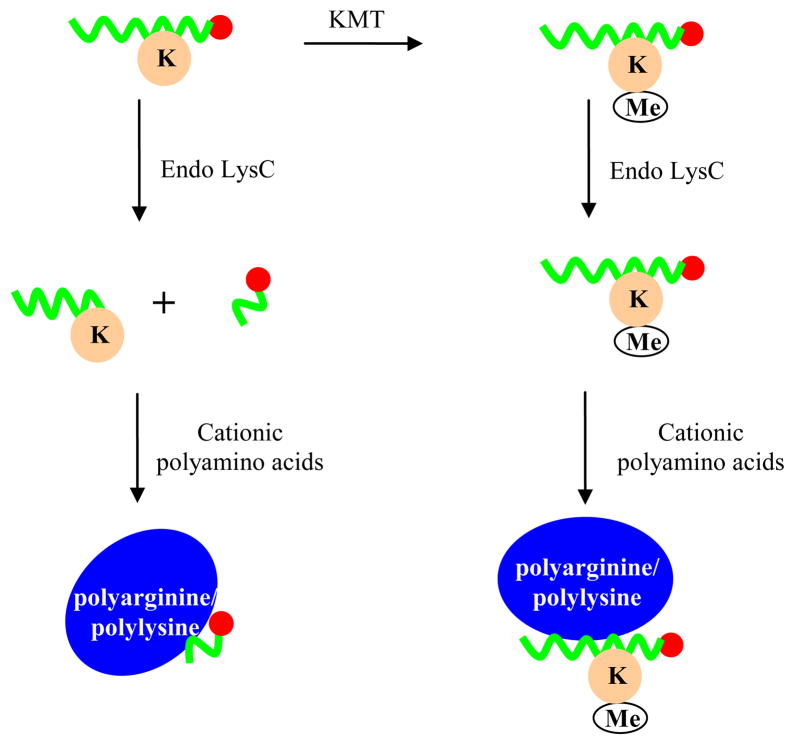
At the same time, it should be possible for FP to find multiple uses in the epigenetics field as a method to assay the enzymes involved in installation or removal of histone posttranslational modifications. Among different histone-modifying enzymes, protein methyltransferases are of interest due to their key role in transcriptional regulation and implication in diseases such as cancer [112]. Protein lysine methylation can be measured by assays that utilize radiolabeled S-adenosylmethionine [113], the ThioGlo coupled assays system [114], or the antibody-based AlphaScreen platform [115]. Despite this progress, superior assay methods are still being sought. Recently, a microfluidic capillary electrophoresis assay for enzymatic activity of methyltransferases was developed [116]. Because the methylation of lysine residues does not change the net charge of the histone peptide substrate, hence making the electrophoretic separation of product and substrate impossible, the methylation-sensitive endoproteinase Endo-LysC was employed to selectively cleave the unmethylated peptide into two fragments while leaving the methyllysine-containing product intact (Figure 5). The Endo-LysC-generated peptide fragments carried different net charges from the starting substrate, allowing their separation in the microfluidic electrophoretic instrument; placement of a fluorescent tag on one terminus of the peptide substrate enabled post-electrophoresis detection of methyltransferase reaction product (left intact after the methylation-specific protease treatment) and substrate (where the fluorophore became part of one of the small fragments generated by the protease).
Figure 5.
Schematic illustration of proposed FP assay for lysine methyltransferase activity. Upon treatment of the methyltransferase reaction with endoproteinase-LysC (Endo LysC), any remaining unmethylated fluorescently labeled peptide substrate is cleaved into smaller fragments, and the fragment containing the fluorophore (red circle) carries a different net charge from the labeled methylated peptide product (Me = methyl group) which remains intact during Endo LysC treatment. Thus, a different degree of binding to oppositely charged polyamino acid molecules by these two labeled peptides is expected (example utilizing polylysine/polyarginine shown here assumes a negatively-charged peptide, although a similar effect can be obtained with a positively-charged peptide by using poly(glutamic acid)), which correlates with the extent of histone peptide methylation and methyltransferase activity, and in turn the changing ratio of these two peptide forms as a function of the methyltransferase reaction progress can be detected by the change in FP.
Despite the adequacy of this method, the separation was performed on a specialized instrument, which may limit the assay’s potential for drug screening and prevent its broader adoption [117]. To simplify this method, we envision that FP can be used as the detection technique instead of electrophoresis to reveal the different-charge peptides generated by the Endo-LysC treatment by following the principle described in section 2.2.1: under optimal ionic strength conditions, the labeled peptides carrying different net charges can be expected to bind to an opposite-charged poly-amino acid (polylysine/polyarginine or polyglutamic acid) to a different extent, thereby providing an FP window to quantitate the extent of histone peptide methylation (Figure 5). Thus, it could be possible for methyltransferase enzymatic activity to be monitored through FP, and subsequently, small molecule inhibitors can be detected by dose-dependent change in FP value.
Article highlights.
FP is a homogeneous method that affords quantitative measurement of binding interactions and enzymatic activities.
Since its adoption in HTS in the mid 1990s for a number of drug targets, FP has shown its value and place in small molecule-based drug discovery as a homogeneous, cheap, and simple assay used frequently in HTS campaigns.
FP technology has been implemented for a number of important drug targets and molecular interactions, and recent advancement of FP in those fields is highlighted in this article.
Advantages and limitations of FP, as well as factors to consider when designing FP assays, are also discussed.
Current trends and possible future directions for FP in novel target areas such as epigenetics are presented.
The box summarizes key points mentioned in the text
List of abbreviations
- A2AAR
A2A adenosine receptor
- ADP
adenosine diphosphate
- AMP
adenosine monophosphate
- ATP
adenosine triphosphate
- AR
adenosine receptor
- BODIPY
4,4-difluoro-4-bora-3a,4a-diaza-s-indacene or boron-dipyrromethene
- BSA
bovine serum albumin
- CoA
coenzyme A
- FP
fluorescence polarization
- FA
fluorescence anisotropy
- FAM
carboxyfluorescein
- FPIA
fluorescence polarization immunoassay
- GMP
guanosine monophosphate
- GDP
guanosine diphosphate
- GPCR
G-protein coupled receptor
- Hsp
heat shock protein
- HTS
high throughput screening
- IC50
concentration of inhibitor which causes 50% inhibition
- IMAP
immobilized metal ion affinity particle
- KMT
lysine methyltransferase
- PBD
polo-box domain
- Plk1
polo-like kinase 1
- PPI
protein-protein interaction
- SAR
structure-activity relationship
- TAMRA
carboxytetramethylrhodamine
References
- 1.Perrin F. Polarization of light of fluorescence, average life of molecules. J Phys Radium. 1926;7:390–401. [Google Scholar]
- 2.Owicki JC. Fluorescence polarization and anisotropy in high throughput screening: perspectives and primer. J Biomol Screen. 2000;5(5):297–306. doi: 10.1177/108705710000500501. [DOI] [PubMed] [Google Scholar]
- 3.Burke TJ, Loniello KR, Beebe JA, et al. Development and application of fluorescence polarization assays in drug discovery. Comb Chem High Throughput Screen. 2003;6(3):183–94. doi: 10.2174/138620703106298365. [DOI] [PubMed] [Google Scholar]
- 4.Jameson DM, Croney JC. Fluorescence polarization: past, present and future. Comb Chem High Throughput Screen. 2003;6(3):167–73. doi: 10.2174/138620703106298347. [DOI] [PubMed] [Google Scholar]
- 5.Nasir MS, Jolley ME. Fluorescence polarization: an analytical tool for immunoassay and drug discovery. Comb Chem High Throughput Screen. 1999;2(4):177–90. [PubMed] [Google Scholar]
- 6.Jameson DM, Ross JA. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem Rev. 2010;110(5):2685–708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gradinaru CC, Marushchak DO, Samim M, et al. Fluorescence anisotropy: from single molecules to live cells. Analyst. 2010;135(3):452–9. doi: 10.1039/b920242k. [DOI] [PubMed] [Google Scholar]
- 8.Lakowicz JR. Principles of fluorescence spectroscopy. Kluwer Academic/Plenum; New York: 1999. [Google Scholar]
- 9.Dandliker WB, Hsu ML, Levin J, et al. Equilibrium and kinetic inhibition assays based upon fluorescence polarization. Methods Enzymol. 1981;74(Pt C):3–28. doi: 10.1016/0076-6879(81)74003-5. [DOI] [PubMed] [Google Scholar]
- 10.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10(6):607–12. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 11.Moerke NJ. Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Current protocols in chemical biology. 2009;1:1–15. doi: 10.1002/9780470559277.ch090102. [DOI] [PubMed] [Google Scholar]
- 12.Cheng YC, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 percent inhibition (I50) of an enzymatic reaction. Biochemical Pharmacology. 1973;22:3099–108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 13.Molecular Probes The Handbook. Invitrogen. [Google Scholar]
- 14.Munson PJ, Rodbard D. An exact correction to the “Cheng-Prusoff” correction. J Recept Res. 1988;8(1–4):533–46. doi: 10.3109/10799898809049010. [DOI] [PubMed] [Google Scholar]
- 15.Swillens S. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol Pharmacol. 1995;47(6):1197–203. [PubMed] [Google Scholar]
- 16.Cer RZ, Mudunuri U, Stephens R, et al. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37(Web Server issue):W441–5. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolovska-Coleska Z, Wang R, Fang X, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332(2):261–73. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 18.http://botdb.abcc.nccifcrf.gov/toxin/kiConverter.jsp
- 19.http://sw16.im.med.umich.edu/software/calc_ki/
- 20.Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol Endocrinol. 24(1):261–74. doi: 10.1210/me.2009-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kecskes M, Kumar TS, Yoo L, et al. Novel Alexa Fluor-488 labeled antagonist of the A(2A) adenosine receptor: Application to a fluorescence polarization-based receptor binding assay. Biochem Pharmacol. 2010;80(4):506–11. doi: 10.1016/j.bcp.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leopoldo M, Lacivita E, Berardi F, et al. Developments in fluorescent probes for receptor research. Drug Discov Today. 2009;14(13–14):706–12. doi: 10.1016/j.drudis.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Davenport AP, Russell FD. Radioligand binding assays: theory and practice. In: Mather SJ, editor. Current Directions in Radiopharmaceutical Research and Development. 1996. pp. 169–79. [Google Scholar]
- 24.Banks P, Gosselin M, Prystay L. Impact of a red-shifted dye label for high throughput fluorescence polarization assays of G protein-coupled receptors. J Biomol Screen. 2000;5(5):329–34. doi: 10.1177/108705710000500504. [DOI] [PubMed] [Google Scholar]
- 25.Jones JW, Greene TA, Grygon CA, et al. Cell-free assay of G-protein-coupled receptors using fluorescence polarization. J Biomol Screen. 2008;13(5):424–9. doi: 10.1177/1087057108318332. [DOI] [PubMed] [Google Scholar]
- 26.www.integralmolecular.com
- 27.Huwiler KG, De Rosier T, Hanson B, et al. A fluorescence anisotropy assay for the muscarinic M1 G-protein-coupled receptor. Assay Drug Dev Technol. 2010;8(3):356–66. doi: 10.1089/adt.2009.0257. [DOI] [PubMed] [Google Scholar]
- 28.Lee PH, Miller SC, van Staden C, et al. Development of a homogeneous high-throughput live-cell G-protein-coupled receptor binding assay. J Biomol Screen. 2008;13(8):748–54. doi: 10.1177/1087057108317835. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5(3):247–64. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Zhang Y, Sportsman JR. A fluorescence polarization assay for cyclic nucleotide phosphodiesterases. J Biomol Screen. 2002;7(3):215–22. doi: 10.1177/108705710200700305. [DOI] [PubMed] [Google Scholar]
- 31.Sportsman JR, Gaudet EA, Boge A. Immobilized metal ion affinity-based fluorescence polarization (IMAP): advances in kinase screening. Assay Drug Dev Technol. 2004;2(2):205–14. doi: 10.1089/154065804323056549. [DOI] [PubMed] [Google Scholar]
- 32.www.moleculardevices.com
- 33.Zaman GJ, Garritsen A, de Boer T, et al. Fluorescence assays for high-throughput screening of protein kinases. Comb Chem High Throughput Screen. 2003;6(4):313–20. doi: 10.2174/138620703106298563. [DOI] [PubMed] [Google Scholar]
- 34.Sharlow ER, Leimgruber S, Yellow-Duke A, et al. Development, validation and implementation of immobilized metal affinity for phosphochemicals (IMAP)-based high-throughput screening assays for low-molecular-weight compound libraries. Nat Protoc. 2008;3(8):1350–63. doi: 10.1038/nprot.2008.111. [DOI] [PubMed] [Google Scholar]
- 35.Beasley JR, Dunn DA, Walker TL, et al. Evaluation of compound interference in immobilized metal ion affinity-based fluorescence polarization detection with a four million member compound collection. Assay Drug Dev Technol. 2003;1(3):455–9. doi: 10.1089/154065803322163768. [DOI] [PubMed] [Google Scholar]
- 36.Coffin J, Latev M, Bi X, et al. Detection of phosphopeptides by fluorescence polarization in the presence of cationic polyamino acids: application to kinase assays. Anal Biochem. 2000;278(2):206–12. doi: 10.1006/abio.1999.4438. [DOI] [PubMed] [Google Scholar]
- 37.Simeonov A, Bi X, Nikiforov TT. Enzyme assays by fluorescence polarization in the presence of polyarginine: study of kinase, phosphatase, and protease reactions. Anal Biochem. 2002;304(2):193–9. doi: 10.1006/abio.2002.5599. [DOI] [PubMed] [Google Scholar]
- 38.Jeong S, Nikiforov TT. Kinase assay based on thiophosphorylation and biotinylation. Biotechniques. 1999;27(6):1232–8. doi: 10.2144/99276rr01. [DOI] [PubMed] [Google Scholar]
- 39.Levine LM, Michener ML, Toth MV, et al. Measurement of specific protease activity utilizing fluorescence polarization. Anal Biochem. 1997;247(1):83–8. doi: 10.1006/abio.1997.2047. [DOI] [PubMed] [Google Scholar]
- 40.Hofman M, Shaffar M. Fluorescence depolarization assay for quantifying alpha-amylase in serum and urine. Clin Chem. 1985;31(9):1478–80. [PubMed] [Google Scholar]
- 41.Zimmerman M, Ashe B, Yurewicz EC, et al. Sensitive assays for trypsin, elastase, and chymotrypsin using new fluorogenic substrates. Anal Biochem. 1977;78(1):47–51. doi: 10.1016/0003-2697(77)90006-9. [DOI] [PubMed] [Google Scholar]
- 42.Cleemann F, Karuso P. Fluorescence anisotropy assay for the traceless kinetic analysis of protein digestion. Anal Chem. 2008;80(11):4170–4. doi: 10.1021/ac7025783. [DOI] [PubMed] [Google Scholar]
- 43.Yasgar A, Foley TL, Jadhav A, et al. A strategy to discover inhibitors of Bacillus subtilis surfactin-type phosphopantetheinyl transferase. Mol Biosyst. 2010;6(2):365–75. doi: 10.1039/b913291k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duckworth BP, Aldrich CC. Development of a high-throughput fluorescence polarization assay for the discovery of phosphopantetheinyl transferase inhibitors. Anal Biochem. 2010;403(1–2):13–9. doi: 10.1016/j.ab.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Dandliker WB, Feigen GA. Quantification of the antigen-antibody reaction by the polarization of fluorescence. Biochem Biophys Res Commun. 1961;5:299–304. doi: 10.1016/0006-291x(61)90167-x. [DOI] [PubMed] [Google Scholar]
- 46.Smith DS, Eremin SA. Fluorescence polarization immunoassays and related methods for simple, high-throughput screening of small molecules. Anal Bioanal Chem. 2008;391(5):1499–507. doi: 10.1007/s00216-008-1897-z. [DOI] [PubMed] [Google Scholar]
- 47.Westphal K, Weinbrenner A, Giessmann T, et al. Oral bioavailability of digoxin is enhanced by talinolol: evidence for involvement of intestinal P-glycoprotein. Clin Pharmacol Ther. 2000;68(1):6–12. doi: 10.1067/mcp.2000.107579. [DOI] [PubMed] [Google Scholar]
- 48.Invitrogen. Fluorescence Polarization. 4. 2006. Technical Resource Guide. [Google Scholar]
- 49.Jia Y, Quinn CM, Kwak S, et al. Current in vitro kinase assay technologies: the quest for a universal format. Curr Drug Discov Technol. 2008;5(1):59–69. doi: 10.2174/157016308783769414. [DOI] [PubMed] [Google Scholar]
- 50.Parker GJ, Law TL, Lenoch FJ, et al. Development of high throughput screening assays using fluorescence polarization: nuclear receptor-ligand-binding and kinase/phosphatase assays. J Biomol Screen. 2000;5(2):77–88. doi: 10.1177/108705710000500204. [DOI] [PubMed] [Google Scholar]
- 51.Turek-Etienne TC, Small EC, Soh SC, et al. Evaluation of fluorescent compound interference in 4 fluorescence polarization assays: 2 kinases, 1 protease, and 1 phosphatase. J Biomol Screen. 2003;8(2):176–84. doi: 10.1177/1087057103252304. [DOI] [PubMed] [Google Scholar]
- 52.Fowler A, Swift D, Longman E, et al. An evaluation of fluorescence polarization and lifetime discriminated polarization for high throughput screening of serine/threonine kinases. Anal Biochem. 2002;308(2):223–31. doi: 10.1016/s0003-2697(02)00245-2. [DOI] [PubMed] [Google Scholar]
- 53.Seethala R, Menzel R. A homogeneous, fluorescence polarization assay for src-family tyrosine kinases. Anal Biochem. 1997;253(2):210–8. doi: 10.1006/abio.1997.2365. [DOI] [PubMed] [Google Scholar]
- 54.Ross H, Armstrong CG, Cohen P. A non-radioactive method for the assay of many serine/threonine-specific protein kinases. Biochem J. 2002;366(Pt 3):977–81. doi: 10.1042/BJ20020786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staeben M, Kleman-Leyer KM, Kopp AL, et al. Development and validation of a transcreener assay for detection of AMP- and GMP-producing enzymes. Assay Drug Dev Technol. 2010;8(3):344–55. doi: 10.1089/adt.2009.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowery RG, Kleman-Leyer K. Transcreener: screening enzymes involved in covalent regulation. Expert Opin Ther Targets. 2006;10(1):179–90. doi: 10.1517/14728222.10.1.179. [DOI] [PubMed] [Google Scholar]
- 57.Kleman-Leyer KM, Klink TA, Kopp AL, et al. Characterization and optimization of a red-shifted fluorescence polarization ADP detection assay. Assay Drug Dev Technol. 2009;7(1):56–67. doi: 10.1089/adt.2008.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huss KL, Blonigen PE, Campbell RM. Development of a Transcreener kinase assay for protein kinase A and demonstration of concordance of data with a filter-binding assay format. J Biomol Screen. 2007;12(4):578–84. doi: 10.1177/1087057107300221. [DOI] [PubMed] [Google Scholar]
- 59.Klink TA, Kleman-Leyer KM, Kopp A, et al. Evaluating PI3 kinase isoforms using Transcreener ADP assays. J Biomol Screen. 2008;13(6):476–85. doi: 10.1177/1087057108319864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antczak C, Shum D, Radu C, et al. Development and validation of a high-density fluorescence polarization-based assay for the trypanosoma RNA triphosphatase TbCet1. Comb Chem High Throughput Screen. 2009;12(3):258–68. doi: 10.2174/138620709787581729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rowlands M, McAndrew C, Prodromou C, et al. Detection of the ATPase activity of the molecular chaperones Hsp90 and Hsp72 using the TranscreenerTM ADP assay kit. J Biomol Screen. 2010;15(3):279–86. doi: 10.1177/1087057109360253. [DOI] [PubMed] [Google Scholar]
- 62.Zielinski T, Kimple AJ, Hutsell SQ, et al. Two Galpha(i1) rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J Biomol Screen. 2009;14(10):1195–206. doi: 10.1177/1087057109347473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma SK, Ramsey TM, Bair KW. Protein-protein interactions: lessons learned. Curr Med Chem Anticancer Agents. 2002;2(2):311–30. doi: 10.2174/1568011023354191. [DOI] [PubMed] [Google Scholar]
- 64.Berg T. Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed Engl. 2003;42(22):2462–81. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 65.Arkin M. Protein-protein interactions and cancer: small molecules going in for the kill. Curr Opin Chem Biol. 2005;9(3):317–24. doi: 10.1016/j.cbpa.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 66.Cochran AG. Antagonists of protein-protein interactions. Chem Biol. 2000;7(4):R85–94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 67.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3(4):301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 68.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 69.Simeonov A, Yasgar A, Jadhav A, et al. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT-phosphoprotein interaction. Anal Biochem. 2008;375(1):60–70. doi: 10.1016/j.ab.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trosset JY, Dalvit C, Knapp S, et al. Inhibition of protein-protein interactions: the discovery of druglike beta-catenin inhibitors by combining virtual and biophysical screening. Proteins. 2006;64(1):60–7. doi: 10.1002/prot.20955. [DOI] [PubMed] [Google Scholar]
- 71.Reindl W, Strebhardt K, Berg T. A high-throughput assay based on fluorescence polarization for inhibitors of the polo-box domain of polo-like kinase 1. Anal Biochem. 2008;383(2):205–9. doi: 10.1016/j.ab.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 72.Reindl W, Yuan J, Kramer A, et al. Inhibition of polo-like kinase 1 by blocking polo-box domain-dependent protein-protein interactions. Chem Biol. 2008;15(5):459–66. doi: 10.1016/j.chembiol.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 73.Parekh P, Martin J, Chen Y, et al. Using aptamers to study protein-protein interactions. Adv Biochem Engin/Biotechnol. 2008;110:177–94. doi: 10.1007/10_2008_104. [DOI] [PubMed] [Google Scholar]
- 74.Cao Z, Tan W. Molecular aptamers for real-time protein-protein interaction study. Chemistry. 2005;11(15):4502–8. doi: 10.1002/chem.200400983. [DOI] [PubMed] [Google Scholar]
- 75.Blank M, Blind M. Aptamers as tools for target validation. Curr Opin Chem Biol. 2005;9(4):336–42. doi: 10.1016/j.cbpa.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 76.Rydel TJ, Tulinsky A, Bode W, et al. Refined structure of the hirudin-thrombin complex. J Mol Biol. 1991;221(2):583–601. doi: 10.1016/0022-2836(91)80074-5. [DOI] [PubMed] [Google Scholar]
- 77.Pope AJ, Haupts UM, Moore KJ. Homogeneous fluorescence readouts for miniaturized high-throughput screening: theory and practice. Drug Discov Today. 1999;4(8):350–62. doi: 10.1016/s1359-6446(99)01340-9. [DOI] [PubMed] [Google Scholar]
- 78.Simeonov A, Jadhav A, Thomas CJ, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51(8):2363–71. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 79.Gribbon P, Sewing A. Fluorescence readouts in HTS: no gain without pain? Drug Discov Today. 2003;8(22):1035–43. doi: 10.1016/s1359-6446(03)02895-2. [DOI] [PubMed] [Google Scholar]
- 80.Turek-Etienne TC, Lei M, Terracciano JS, et al. Use of red-shifted dyes in a fluorescence polarization AKT kinase assay for detection of biological activity in natural product extracts. J Biomol Screen. 2004;9(1):52–61. doi: 10.1177/1087057103259346. [DOI] [PubMed] [Google Scholar]
- 81.Huang X. Fluorescence polarization competition assay: the range of resolvable inhibitor potency is limited by the affinity of the fluorescent ligand. J Biomol Screen. 2003;8(1):34–8. doi: 10.1177/1087057102239666. [DOI] [PubMed] [Google Scholar]
- 82.Vogel KW, Marks BD, Kupcho KR, et al. Facile conversion of FP to TR-FRET assays using terbium chelates: nuclear receptor competitive binding assays as examples. Letters in Drug Design & Discovery. 2008;5:416–22. [Google Scholar]
- 83.Shoichet BK. Screening in a spirit haunted world. Drug Discov Today. 2006;11(13–14):607–15. doi: 10.1016/j.drudis.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seidler J, McGovern SL, Doman TN, et al. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J Med Chem. 2003;46(21):4477–86. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 85.Arai T, Yatabe M, Furui M, et al. A fluorescence polarization-based assay for the identification and evaluation of calmodulin antagonists. Anal Biochem. 2010;405(2):147–52. doi: 10.1016/j.ab.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 86.Kimple AJ, Yasgar A, Hughes M, et al. A high throughput fluorescence polarization assay for inhibitors of the GoLoco motif/G-alpha interaction. Comb Chem High Throughput Screen. 2008;11(5):396–409. doi: 10.2174/138620708784534770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS Chem Biol. 2008;3(3):142–55. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kricka LJ. Stains, labels and detection strategies for nucleic acids assays. Ann Clin Biochem. 2002;39(Pt 2):114–29. doi: 10.1258/0004563021901865. [DOI] [PubMed] [Google Scholar]
- 89.Guo XQ, Castellano FN, Li L, et al. Use of a long-lifetime Re(I) complex in fluorescence polarization immunoassays of high-molecular-weight analytes. Anal Chem. 1998;70(3):632–7. doi: 10.1021/ac970827k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banks P, Gosselin M, Prystay L. Fluorescence polarization assays for high throughput screening of G protein-coupled receptors. J Biomol Screen. 2000;5(3):159–68. doi: 10.1177/108705710000500308. [DOI] [PubMed] [Google Scholar]
- 91.Terpetschnig E, Szmacinski H, Lakowicz JR. Fluorescence polarization immunoassay of a high-molecular-weight antigen based on a long-lifetime Ru-ligand complex. Anal Biochem. 1995;227(1):140–7. doi: 10.1006/abio.1995.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Riechers A, Schmidt J, Konig B, et al. Heterogeneous transition metal-based fluorescence polarization (HTFP) assay for probing protein interactions. Biotechniques. 2009;47(4):837–44. doi: 10.2144/000113223. [DOI] [PubMed] [Google Scholar]
- 93.Brinkley M. A brief survey of methods for preparing protein conjugates with dyes, haptens, and cross-linking reagents. Bioconjug Chem. 1992;3(1):2–13. doi: 10.1021/bc00013a001. [DOI] [PubMed] [Google Scholar]
- 94.Fischer R, Mader O, Jung G, et al. Extending the applicability of carboxyfluorescein in solid-phase synthesis. Bioconjug Chem. 2003;14(3):653–60. doi: 10.1021/bc025658b. [DOI] [PubMed] [Google Scholar]
- 95.Weber PJ, Bader JE, Folkers G, et al. A fast and inexpensive method for N-terminal fluorescein-labeling of peptides. Bioorg Med Chem Lett. 1998;8(6):597–600. doi: 10.1016/s0960-894x(98)00084-5. [DOI] [PubMed] [Google Scholar]
- 96.Jameson DM, Sawyer WH. Fluorescence anisotropy applied to biomolecular interactions. Methods Enzymol. 1995;246:283–300. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- 97.Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24(22):4535–42. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chadwick PM, Durrant I. Labeling of oligonucleotides with fluorescein. Mol Biotechnol. 1997;7(3):299–301. doi: 10.1007/BF02740820. [DOI] [PubMed] [Google Scholar]
- 99.Cornelius P, Lee E, Lin W, et al. Design, synthesis, and pharmacology of fluorescently labeled analogs of serotonin: application to screening of the 5-HT2C receptor. J Biomol Screen. 2009;14(4):360–70. doi: 10.1177/1087057109331804. [DOI] [PubMed] [Google Scholar]
- 100.Cooper M, Ebner A, Briggs M, et al. Cy3B: improving the performance of cyanine dyes. J Fluoresc. 2004;14(2):145–50. doi: 10.1023/b:jofl.0000016286.62641.59. [DOI] [PubMed] [Google Scholar]
- 101.Vaasa A, Viil I, Enkvist E, et al. High-affinity bisubstrate probe for fluorescence anisotropy binding/displacement assays with protein kinases PKA and ROCK. Anal Biochem. 2009;385(1):85–93. doi: 10.1016/j.ab.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 102.Allen M, Reeves J, Mellor G. High throughput fluorescence polarization: a homogeneous alternative to radioligand binding for cell surface receptors. J Biomol Screen. 2000;5(2):63–9. doi: 10.1177/108705710000500202. [DOI] [PubMed] [Google Scholar]
- 103.Ernst B, Magnani JL. From carbohydrate leads to glycomimetic drugs. Nat Rev Drug Discov. 2009;8(8):661–77. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kakehi K, Oda Y, Kinoshita M. Fluorescence polarization: analysis of carbohydrate-protein interaction. Anal Biochem. 2001;297(2):111–6. doi: 10.1006/abio.2001.5309. [DOI] [PubMed] [Google Scholar]
- 105.Han Z, Pinkner JS, Ford B, et al. Structure-based drug design and optimization of mannoside bacterial FimH antagonists. J Med Chem. 2010;53(12):4779–92. doi: 10.1021/jm100438s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharon N. Carbohydrates as future anti-adhesion drugs for infectious diseases. Biochim Biophys Acta. 2006;1760(4):527–37. doi: 10.1016/j.bbagen.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 107.Schembri MA, Kjaergaard K, Sokurenko EV, et al. Molecular characterization of the Escherichia coli FimH adhesin. J Infect Dis. 2001;183 (Suppl 1):S28–31. doi: 10.1086/318847. [DOI] [PubMed] [Google Scholar]
- 108.Sharon N, Ofek I. Safe as mother’s milk: carbohydrates as future anti-adhesion drugs for bacterial diseases. Glycoconj J. 2000;17(7–9):659–64. doi: 10.1023/a:1011091029973. [DOI] [PubMed] [Google Scholar]
- 109.Taldone T, Gillan V, Sun W, et al. Assay strategies for the discovery and validation of therapeutics targeting Brugia pahangi Hsp90. PLoS Negl Trop Dis. 2010;4(6):e714. doi: 10.1371/journal.pntd.0000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yi F, Regan L. A novel class of small molecule inhibitors of Hsp90. ACS Chem Biol. 2008;3(10):645–54. doi: 10.1021/cb800162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu F, Chen X, Allali-Hassani A, et al. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J Med Chem. 2009;52(24):7950–3. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8(9):724–32. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 113.Gowher H, Zhang X, Cheng X, et al. Avidin plate assay system for enzymatic characterization of a histone lysine methyltransferase. Anal Biochem. 2005;342(2):287–91. doi: 10.1016/j.ab.2005.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Collazo E, Couture JF, Bulfer S, et al. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342(1):86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 115.Quinn AM, Allali-Hassani A, Vedadi M, et al. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors. Mol Biosyst. 2010;6(5):782–8. doi: 10.1039/b921912a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wigle TJ, Provencher LM, Norris JL, et al. Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis. Chem Biol. 2010;17(7):695–704. doi: 10.1016/j.chembiol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bissinger EM, Jung M. No taste for methyl: methylation sensitive proteolysis in drug screening. Chem Biol. 2010;17(7):677–8. doi: 10.1016/j.chembiol.2010.07.001. [DOI] [PubMed] [Google Scholar]