Abstract
Mechanical strength of nanofiber scaffolds formed by the self-assembling peptide RADA16-I or its derivatives is not very good and limits their application. To address this problem, we inserted spidroin uncrystalline motifs, which confer incomparable elasticity and hydrophobicity to spider silk GGAGGS or GPGGY, into the C-terminus of RADA16-I to newly design two peptides: R3 (n-RADARADARADARADA-GGAGGS-c) and R4 (n-RADARADARADARADA-GPGGY-c), and then observed the effect of these motifs on biophysical properties of the peptide. Atomic force microscopy, transmitting electron microscopy, and circular dichroism spectroscopy confirm that R3 and R4 display β-sheet structure and self-assemble into long nanofibers. Compared with R3, the β-sheet structure and nanofibers formed by R4 are more stable; they change to random coil and unordered aggregation at higher temperature. Rheology measurements indicate that novel peptides form hydrogel when induced by DMEM, and the storage modulus of R3 and R4 hydrogel is 0.5 times and 3 times higher than that of RADA16-I, respectively. Furthermore, R4 hydrogel remarkably promotes growth of liver cell L02 and liver cancer cell SMCC7721 compared with 2D culture, determined by MTT assay. Novel peptides still have potential as hydrophobic drug carriers; they can stabilize pyrene microcrystals in aqueous solution and deliver this into a lipophilic environment, identified by fluorescence emission spectra. Altogether, the spider fibroin motif GPGGY most effectively enhances mechanical strength and hydrophobicity of the peptide. This study provides a new method in the design of nanobiomaterials and helps us to understand the role of the amino acid sequence in nanofiber formation.
Keywords: uncrystalline motif, self-assembling peptide, β-sheet, nanofiber, mechanical strength, hydrophobic compound carrier
Introduction
Recently, the self-assembling peptide has attracted much attention, not only because of its scientific importance,1–3 but also because of its potential applications in tissue engineering, regenerative medicine,4–6 drug delivery,7,8 and nanotechnology.9–11 For example, the 16-residue peptide, RADARADARADARADA (RADA16-I) is able to self-assemble into well-ordered nanofibers and further form scaffolds, composed of interwoven nanofibers (~10 nm in diameter) with ~100 nm mesh pores and over 99% water content. The scaffold has a three-dimensional structure similar to naturally occurring extracellular matrix (ECM), and has been reported to support cell attachment, proliferation, and differentiation,12–15 and act as an effective hemostatic medicine.16
Furthermore, some self-assembling peptides, directly incorporating various functional motifs into RADA16-I, are designed to mimic particular ECM. These motifs include collagen (GGDGEA),17 laminin (GGPDSGR),18 fibronectin,19 osteopontin and osteogenic peptides,20 and myelo-regulatory peptides.21 These functionalized peptides also self-assemble into three-dimensional nanofiber scaffolds. However, the mechanical strength of these scaffolds is not very good and limits their application in tissue regeneration. There is urgent need to improve the physical performance of these nanomaterials.
As we know, the self-assembly process is ubiquitous in nature. One well-known example is silk assembly. The monomeric fibroin is only about one micrometer in length, but silk is over two kilometers in length. Spiders can produce silks with tremendous strength and flexibility through assembling spidroin building blocks.22 Spidroin contains highly repetitive amino acid motifs such as poly A, poly GA, GPGGX, and GGX (X: Y, A, S etc).23–25 Poly A and poly GA segments generally exhibit β-sheet structure and constitute the crystalline fraction of the protein. They are believed to be mainly responsible for the high tensile strength of spider silk. However, GPGGX and GGX motifs exist in the uncrystalline region, which can be elongated and recover. This region contributes to the incomparable elasticity, adhesion, and hydrophobicity of the fibers.26–28
In the present study, we integrated RADA16-I with spider fibroin uncrystalline motifs GPGGY and GGAGGS, respectively, and studied the mechanical strength of the new peptides, and their potential as hydrophobic drug carriers. The spider fibroin motif GPGGY enhances the storage modulus and hydrophobicity of the scaffolds effectively, and promotes cell growth notably.
Materials and methods
Materials
The peptides RADA16-I, R3, and R4 (theoretical mass = 1713, 2099 and 2144, respectively) were purchased from Shanghai Biotech Bioscience and Technology Co, Ltd, Shanghai, China. The N-terminus and C-terminus of peptides were protected by acetyl and amino groups, respectively. Peptide solutions at 6 mM or 1% (w/v) were prepared with ddH2O and diluted to different concentrations for further characterization before use. The human liver cell line L02 and liver cancer cell line SMCC7721 were purchased from the American Type Culture Collection ( Manassas, VA) and cultivated in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco®, Rockville, MD) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Pyrene (99%) (Sigma, St, Louis, MO) was recrystallized twice from ethanol before experimentation.
Projection formulas of peptides
To obtain projection formulas of the peptides, the software ICM-Pro (v3.4; Molsoft LLC, San Diego, CA) was used. N-Terminus and C-Terminus of peptides were set to “nter” and “cooh”, respectively.
Atomic force microscopy (AFM)
An aliquot of 1 μL peptide solution (100 μM) was placed on a freshly cleaved mica sheet and observed with AFM (SPI4000 Probe Station and SPA-400 SPM Unit, Seiko Instruments Inc, Chiba, Japan). Silicon tips (Si-DF20, Olympus Corp, Tokyo, Japan) with a cantilever length of 200 μm and a spring constant of 12 N/m were used. Typical scan parameters were as follows: Amplitude: ~1 V, integral gain: ~0.25 V, proportional gain: ~0.03 V, and scan speed: 0.83–1 Hz. The topographic images were recorded with 512 × 512 pixel resolution, in which the brightness of morphology increased with height. The real widths of the fibers were obtained by the following equation: W = Wobs − 2(2RtH–H2)1/2, where Rt is the radius of the AFM tip and H is the observed height.29
Transmission electron microscopy (TEM)
An aliquot of about 6 μL peptide solution (100 μM) was placed on the surface of a glass plate. A polyvinyl formal coated copper grid was dipped in the solution for about 5 seconds and then absorbed with filter paper to remove superfluous liquid. Then, a peptide-treated copper grid was tainted with 1% (w/v, pH = 6.5) phosphotungstic acid for negative staining. After air-drying, the bright-field images were taken with TEM (H-600, Hitachi Ltd, Tokyo, Japan) operating at 100 kV.
Circular dichroism (CD) spectroscopy
CD spectroscopy was performed with an AVIV Model 400 spectrometer (Aviv Biomedical Inc, Lakewood, NJ). The concentration of the samples was 100 μM. Readings were taken at 1 nm interval wavelengths from 190–260 nm. CD data were corrected by conversion to mean residue ellipticity to account for molecular weight and concentration. In order to further investigate the effects of different uncrystalline motifs on the secondary structure of peptides, the software CDPro (Colorado State University, Fort Collins, CO) was applied to estimate secondary structure contents. The peptides’ secondary structure fractions were calculated using the CONTINLL algorithm with comparison to a set of selected reference proteins (IBasis7 [SDP48], λ = 240 –190 nm).
Rheology
An aliquot of 100 μL, 3 mM peptide solution was placed in a 24-well plate. 600 μL DMEM medium or phosphate buffered saline was added around the peptides. About 15 hours later, the mechanical properties of the hydrogel were identified with a rheometer (AR2000, TA Instruments Inc, New Castle, DE). A 20 mm diameter 1° stainless steel cone with a 25 μm truncation was used and covered with a split solvent trap cover to protect solvent from volatilization. The elastic modulus (G′) as a function of time was measured with a frequency of 1 rad/s and constant strain of 0.5% at 25°C.
MTT assay
To monitor the influence of the modified peptides on cell viability, we applied MTT assay. 1.5 mM peptide solution was mixed with an equal volume of L02 or SMCC7721 cell suspension (1 × 105/mL), and 100 μL of the mixture per well was quickly inoculated into a 96-well dish. After 1, 2, and 3 days, cell viability was assessed with an Easy Reader 340 AT (SLT Lab Instruments GmbH, Salzburg, Austria). Absorbance was recorded at 570 nm. The Student’s t-test was performed and a 2D culture was used as a control. P < 0.05 was considered statistically significant.
Preparation of peptide-pyrene colloidal suspensions
Pyrene (PY) (2.47 mM) and peptide R4 or R3 (100 μM) were added into a vial. The samples were kept on a magnetic stirrer until equilibrium was reached. The Peptide-PY solution was deemed at equilibrium when its fluorescence spectrum did not change in 24 hours.
Liposome preparation and concentration determination
Phosphatidylcholine (PC) (1.4 g) was dissolved in chloroform to produce a PC thin film,30 and the film was subsequently resuspended in buffer A (25 mM Tris-HCl, 0.2 mM EDTA, pH 7.4) solution. The supernatant was filtered by 0.44 μm and 0.22 μm membrane filters in turn. The mass of the liposome was the difference between the mass of vesicle solution and that of buffer A, and the average concentration of liposome was 5.185 mM. It was diluted to 1.037 mM with buffer A before use.
Steady-state fluorescence measurements
Fluorescence spectra were recorded on a Hitachi F-7000 spectrofluorometer with a stir accessory. Excitation and emission slits were set to 10 nm and 2.5 nm, respectively. The excitation wavelength was set to 336 nm, and the emission fluorescence spectra were scanned from 350 nm – 650 nm with scan speed of 1200 nm/min.
Results
CD measurement of peptides R3 and R4
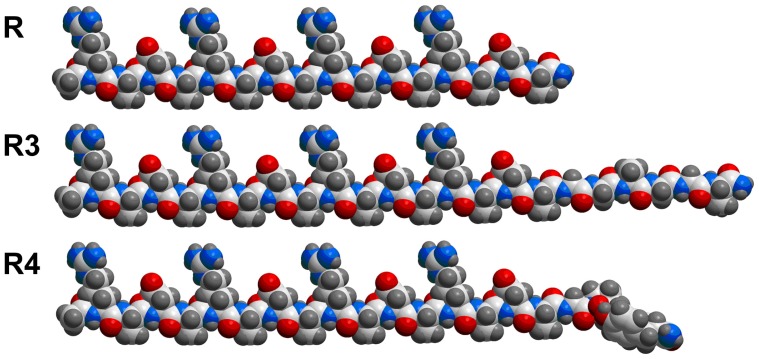
The projection formulas of peptides RADA16-I, R3, and R4 are shown in Figure 1. The 22-residue peptide R3 and the 21-residue peptide R4 are partially ionic-complementary, constructed by adding motifs GGAGGS and GPGGY to the C-terminus of the fully ionic-complementary peptide, RADA16-I. For peptide R4, there is a turn occurring at the proline site.
Figure 1.
Projection formula of peptides RADA16-I (R), R3, and R4.
Notes: Carbon atoms are white, oxygen atoms are red, nitrogen atoms are blue, and hydrogen atoms are gray. In this conformation, for RADA16-I, all the lysine and aspartic acid side chains face in one direction, and all alanine side chains face in the other direction to create two distinct faces: the hydrophobic and the hydrophilic. There is a turn at proline residue for R4.
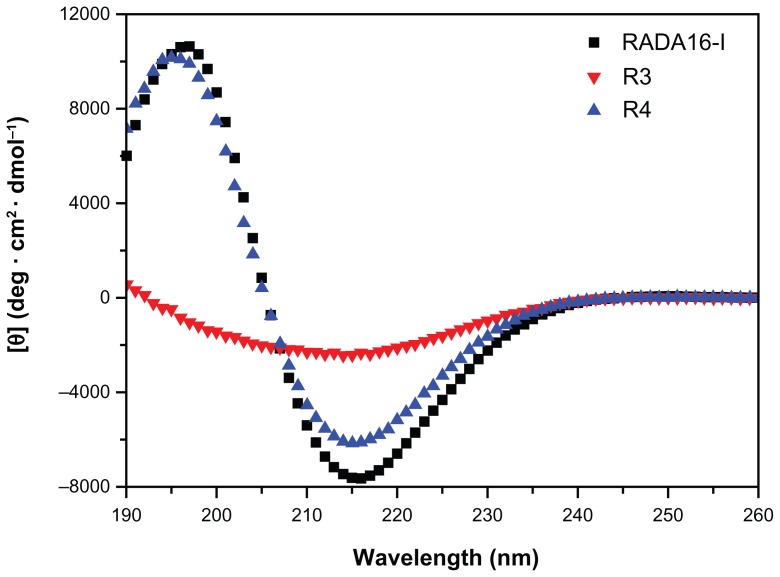
CD measurement of these new peptides demonstrates that R3 and R4 mainly form β-sheet structure at 25°C, and that the β-sheet content of R3 is lower than that of R4 and RADA16-I (Figure 2 and Table 1). Peptide R4 exhibits a typical β-sheet structure (minimum at 215 nm, maximum at 197 nm) (Figure 2), with 42.8% β-sheet content (Table 1; S (r) and S (d)) and 28.4% unordered structure (Table 1, Unrd). The CD spectra of R4 are similar to that of RADA16-I. However, the CD spectrum of peptide R3 becomes a wide valley at 190–240 nm, and the positive peak at about 197 nm disappears (Figure 2). The β-sheet content of R3 decreases to 37.8% (Table 1; S (r) + S (d)), and the unordered structure increases to about 40% (Table 1, Unrd). This suggests that the spidroin uncrystalline motif GPGGY is more helpful for forming β-sheet structure in peptides than the motif GGAGGS.
Figure 2.
CD spectra of peptides RADA16-I, R3 and R4 (100 μM) in Mill-Q water.
Notes: All the CD data were recorded at 25°C after incubation overnight at 4°C. Peptides RADA16-I (black) and R4 (blue) exhibit a typical β-sheet structure, while R3 (red) displays β-sheet structure that does not have positive peak.
Table 1.
Estimated structure fractions of different peptides
| Peptide (100 μM) | RMSD/NRMSD | Secondary structure fractions (%)* | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| H (r) | H (d) | S (r) | S (d) | Turn | Unrd | ||
| R3 | 0.029/0.021 | 0.5 | 3.1 | 25.0 | 12.7 | 18.7 | 40.0 |
| R4 | 0.070/0.043 | 4.0 | 3.5 | 28.9 | 13.9 | 21.3 | 28.4 |
| RADA16-I | 0.151/0.106 | 2.1 | 2.4 | 31.5 | 10.6 | 20.8 | 32.6 |
Note: Including 6 parts second structures.
Abbreviations: H (r), regular α-helix; H (d), distorted α-helix; S (r), regular β-strand; S (d), distorted β-strand (a partial but far-from-complete distortion of the regular β-strand, due to lack of some hydrogen bonds); Turn, β-turn structure; Unrd, unordered structure.
Self-assembling morphology of peptides R3 and R4
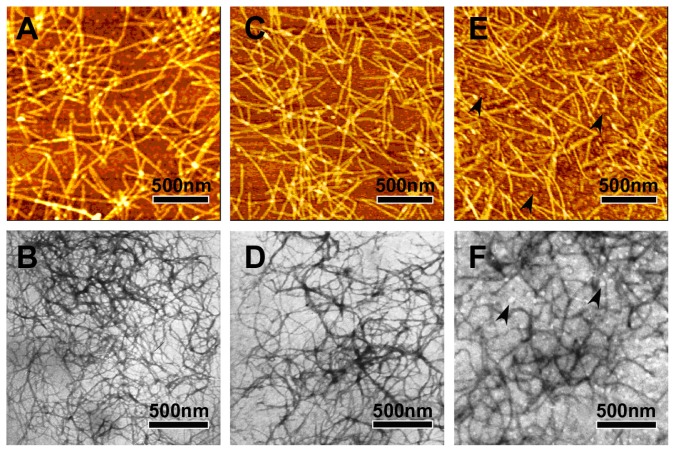
AFM and TEM measurements show that the peptides R3 and R4 are able to self-assemble into nanofibers at 25°C (Figure 3). The RADA16-I nanofibers (Figure 3A and B) are about 877.5 ± 172 nm in length, 19.8 ± 0.7 nm in width, and 1.1 ± 0.06 nm in height; those of R4 (Figure 3C and D) are about 884.4 ± 161 nm in length, 19.4 ± 0.68 nm in width, and 1.1 ± 0.06 nm in height; and those of R3 (Figure 3E and F) are about 871.2 ± 197.05 nm in length, 18.6 ± 0.86 nm in width, and 1.1 ± 0.07 nm in height. This suggests that nanofibers formed by peptides R3, R4, and RADA16-I are similar. However, peptide R3 also forms much smaller aggregates (Figure 3E and F, black arrows). This may be due to the higher unordered structure content of R3.
Figure 3.
The typical AFM (A, C and E) and TEM (B, D and F) images of 100 μM peptides RADA16-I (A and B), R4 (C and D) and R3 (E and F).
Notes: RADA16-I, R3, and R4 self-assemble into nanofibers that are about 1000 nm in length. Black arrows in E and F indicate aggregates.
Abbreviations: AFM, Atomic force microscopy; TEM, Transmission electron microscopy.
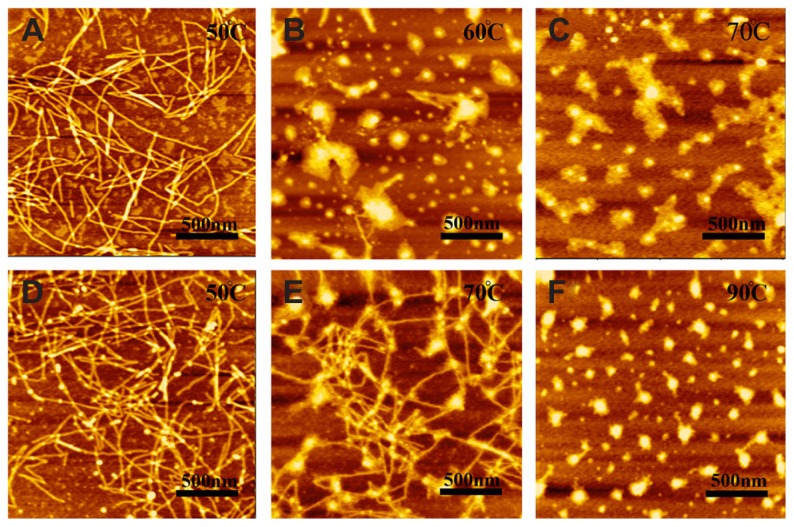
Stability of self-assembling peptides R3 and R4
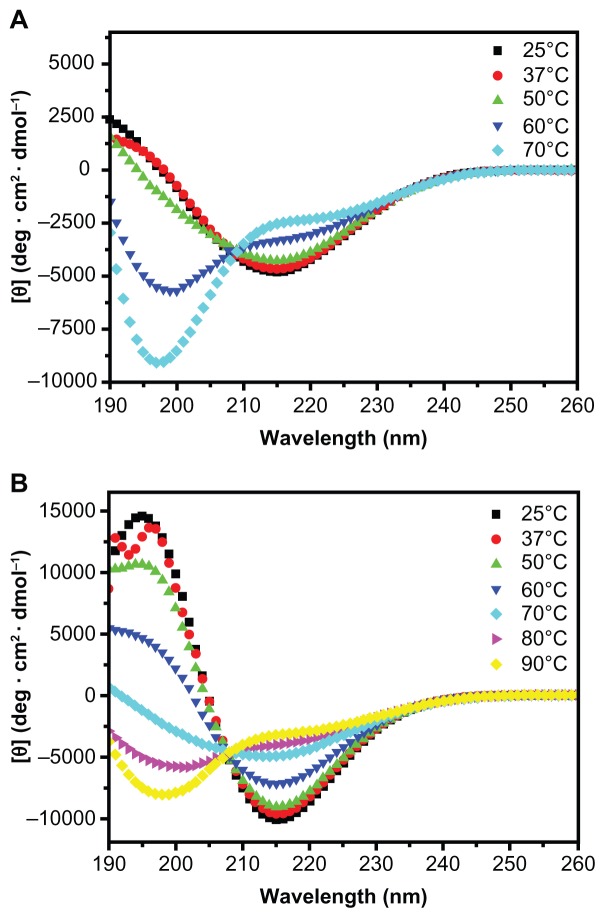
In order to further investigate the effect of motifs GPGGY and GGAGGS on β-sheet structure and self-assembly of peptides R3 and R4, we heated the peptide solutions up to 90°C and subsequently detected their secondary structure and self-assembling morphology using CD (Figure 4, Tables 2 and 3) and AFM (Figure 5).
Figure 4.
CD spectra of R3 and R4 (100 μM) after thermal denaturation.
Notes: A: CD spectra of R3 from 25°C to 70°C. B: CD spectra of R4 from 25°C to 90°C. Peptide R3 mainly forms unordered structure at 50°C, while peptide R4 forms unordered structure when heated up to 70°C.
Abbreviation: CD, Circular dichroism spectroscopy.
Table 2.
Estimated structure fractions of R3 after thermal denaturation
| Temperature (°C) | RMSD/NRMSD | Secondary structure fractions (%)* | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| H (r) | H (d) | S (r) | S (d) | Turn | Unrd | ||
| 25 | 0.028/0.031 | 2.2 | 4.4 | 23.3 | 12.2 | 19.9 | 38.0 |
| 37 | 0.018/0.021 | 2.0 | 4.3 | 22.9 | 12.1 | 20.0 | 38.8 |
| 50 | 0.022/0.027 | 1.7 | 4.1 | 22.4 | 11.6 | 18.6 | 41.6 |
| 60 | 0.026/0.025 | 0.8 | 2.9 | 17.7 | 9.6 | 14.9 | 54.1 |
| 70 | 0.045/0.032 | 0.4 | 2.4 | 14.5 | 7.8 | 11.6 | 63.4 |
Note: Including 6 parts second structures.
Abbreviations: H (r), regular α-helix; H (d), distorted α-helix; S (r), regular β-strand; S (d), distorted β-strand (a partial but far-from-complete distortion of the regular β-strand, due to lack of some hydrogen bonds); Turn, β-turn structure; Unrd, unordered structure.
Table 3.
Estimated structure fractions of R4 after thermal denaturation
| Temperature (°C) | RMSD/NRMSD | Secondary structure fractions (%)* | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| H (r) | H (d) | S (r) | S (d) | Turn | Unrd | ||
| 25 | 0.352/0.144 | 0.0 | 6.9 | 38.4 | 15.8 | 15.2 | 23.7 |
| 37 | 0.340/0.149 | 0.0 | 6.9 | 40.9 | 15.9 | 14.0 | 22.4 |
| 50 | 0.043/0.021 | 5.6 | 3.7 | 31.6 | 15.3 | 24.8 | 19.0 |
| 60 | 0.024/0.018 | 4.6 | 4.9 | 25.3 | 13.3 | 22.7 | 29.1 |
| 70 | 0.015/0.015 | 2.5 | 4.3 | 19.7 | 10.8 | 18.0 | 44.6 |
| 80 | 0.023/0.020 | 1.3 | 2.7 | 15.0 | 8.8 | 14.8 | 57.4 |
| 90 | 0.033/0.024 | 0.5 | 2.6 | 14.0 | 8.0 | 12.5 | 62.4 |
Note: Including 6 parts second structures.
Abbreviations: H (r), regular α-helix; H (d), distorted α-helix; S (r), regular β-strand; S (d), distorted β-strand (a partial but far-from-complete distortion of the regular β-strand, due to lack of some hydrogen bonds); Turn, β-turn structure; Unrd, unordered structure.
Figure 5.
AFM images of 100 μM peptides R3 (A–C) and R4 (D–F) after thermal denaturation.
Note: R3 undergoes morphological changes from nanofibers (A) to aggregations (C) at 60°C, while R4 displays this change at 90°C.
Abbreviation: AFM, Atomic force microscopy.
CD measurement illustrates that an increase in temperature results in a structural transition from β-sheet to unordered structure, and that the β-sheet structure of R4 is more stable than that of R3. Though the β-sheet structure of R3 remains stable below 50°C (30.2%) (Table 2; S (r) and S (d)), when heated up to 70°C, the β-sheet content of R3 decreases to 22.2%, and the unordered structure rapidly increases to 63.4% (Figure 4A and Table 2). However, R4 still mainly forms β-sheet structure at 70°C (Figure 4B and Table 3).
The temperature-dependent self-assembling behavior of peptides R3 and R4 shows that the nanostructure of R4 is also more stable than that of R3 (Figure 5). At 50°C, both R3 and R4 form nanofibers that are about 1000 nm in length (Figure 5A and D). However, when heated to 60°C, peptide R3 almost completely self-assembles into unordered aggregation (Figure 5B), while peptide R4 forms nanofibers even at 70°C (Figure 5E). When heated to 90°C, the self-assembled structure of R4 changes into aggregates (Figure 5F). Previous study31 has shown that RADA16-I exhibits β-sheet structure and forms unordered aggregates at 70°C, indicating that the nanostructure of R4 is more stable than that of RADA16-I.
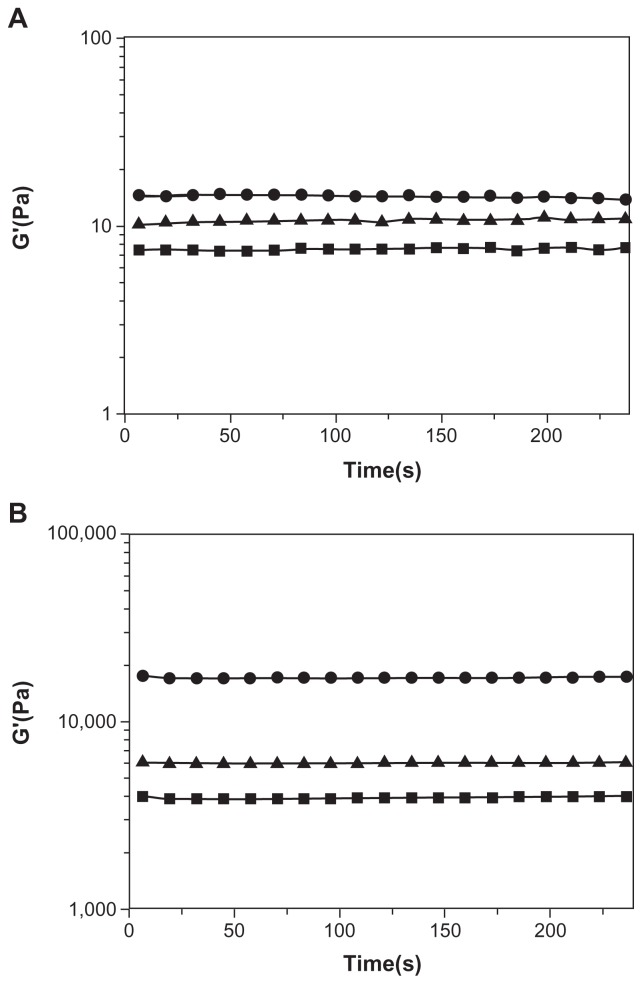
Mechanical strength of peptide hydrogel
Since spidroin uncrystalline motifs are critical to the notable elasticity of spider fibers, we examined the effect of the motifs GPGGY and GGAGGS on mechanical strength of the peptides. Rheology measurement shows that, though they all have low mechanical strength in water (Millipore, Billerica, MA), the storage modulus of peptide R4 (Figure 6B) is enhanced more than that of peptides R3 and RADA16-I when exposed to DMEM medium (Figure 6A). The storage modulus of RADA16-I, R3, and R4 peptide solution is about 7 Pa, 10 Pa, and 14 Pa, respectively (Figure 6A). However, when exposed to DMEM medium, all three peptides form hydrogel, and their storage modulus increases to 4000 Pa, 6000 Pa, and 17000 Pa, respectively (Figure 6B). R4 hydrogel is 2 times stronger than R3 hydrogel, and 3 times stronger than RADA16-I hydrogel. The uncrystalline motif GPGGY remarkably improves the mechanical strength of the peptide.
Figure 6.
Rheological results of RADA16-I (■), R3 (▴) and R4 (●) at 3 Mm.
Notes: A: The storage modulus of RADA16-I, R3, and R4 in aqueous solutions. B: The storage modulus of RADA16-I, R3, and R4 after they have formed hydrogels in DMEM medium. The storage modulus, G′, is plotted against oscillatory frequency on log-log scales. The mechanical strength of R4 hydrogel is 3 times greater than that of RADA16-I and R3 hydrogels.
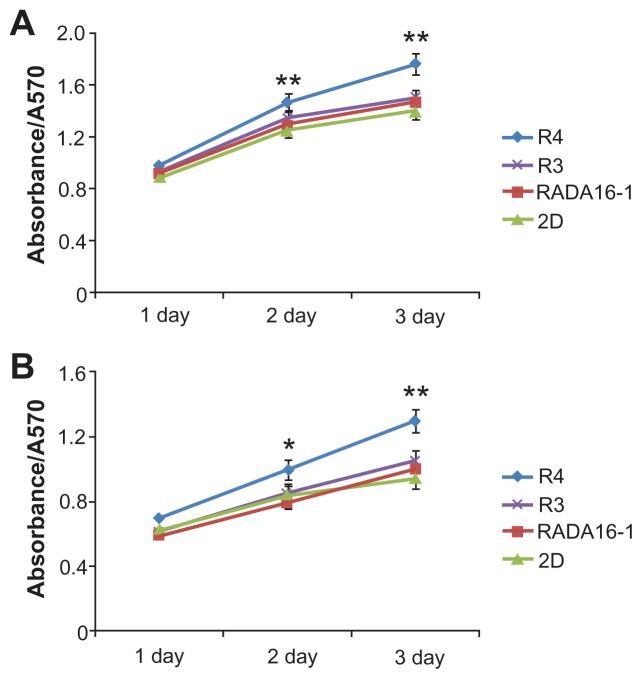
Furthermore, MTT assay (Figure 7) demonstrates that the modified peptides R4 and R3 are not only noncytotoxic but also promote cell growth. Compared with 2D culture, R4 hydrogel significantly enhances proliferation of liver cells L02 (Figure 7A) and liver cancer cells SMCC7721 (Figure 7B). The promoting role of peptide R3 is not as remarkable as that of R4. Maybe this is because the mechanical strength of R4 is much greater.
Figure 7.
Cell viabilty in 3D culture, determined by MTT assay.
Notes: (A) L02 cells. (B) SMCC7721 cells. *P < 0.05; **P < 0.01 vs 2D culture. R4 hydrogel remarkably promotes cell growth.
Peptides R3 and R4 as potential carriers of hydrophobic compounds
Spidroin uncrystalline motifs GGAGGS and GPGGY are hydrophobic. Therefore, the self-assembling peptides R3 and R4 are more hydrophobic and may be able to pack hydrophobic compounds and transfer them into liposome vesicles. We employed PY as a hydrophobic compound model, and PC vesicles as cell mimics. In the presence of peptides R3 or R4, R3-PY (Figure 8-2) and R4-PY ( Figure 8-3) solutions form stable colloidal suspensions after being stirred for 24 hours, while the control sample without peptide remains transparent, with PY crystals floating on the top or precipitating at the bottom (Figure 8-1). The formation of colloidal suspension suggests that PY is stabilized in water via the peptides.
Figure 8.

Pyrene (2.47 mM) in water (1) and with R3 (2, 100 μM), or R4 (3, 100 μM) in aqueous solution after stirring for 24 hours.
Abbreviations: PY, (pyrene); PC, phosphatidylcholine vesicles.
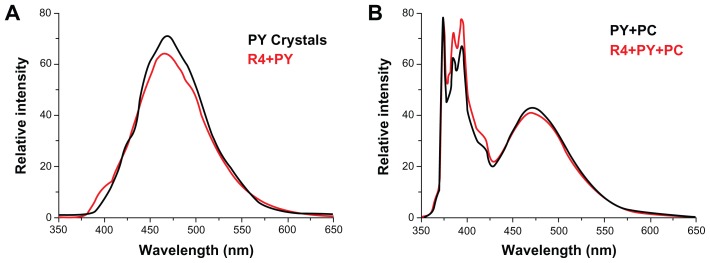
The emission fluorescence spectra of PY (Figure 9) demonstrate that PY encapsulated with peptide R4 can be transferred into PC vesicles. PY crystals and the R4-PY solution (Figure 9A) only exhibit pyrene excimer emission (emission band at 470 nm) without visible monomer emission (five emission peaks between 370 nm and 400 nm), while PY in liposome and the R4-PY solution mixed with the liposome (Figure 9B) display both pyrene monomer and excimer emission. The ratio of intensities of the first peak (I1, 373 nm) and the third peak (I3, 385 nm) of pyrene monomer (I1/I3) for PY in R4-PY solution mixed with the liposome is 1.16,32–34 indicating that PY was released from the peptide encapsulation and transferred into the hydrophobic lipid bilayer. Similar data were acquired with R3-PY solution (data not shown).
Figure 9.
Steady-state fluorescence emission spectra of pyrene at equilibrium.
Notes: (A) solid pyrene crystals (black) and R4-PY solution (red, PY = 2.47 mM, R4 = 100 μM). (B) Pyrene in PC vesicles (black, PY = 65.3 μM, PC = 1.037 mM), and R4-PY solution mixed with PC vesicles (red, PY = 42.5 μM, R4 = 100 μM, PC = 1.037 mM). λex = 336 nm. Self-assembling peptide R4 transfers hydrophobic compound pyrene into liposome vesicle.
Abbreviations: PY, pyrene; PC, phosphatidylcholine vesicles.
Discussion
In this study, we found that the new peptide R4 integrated with spider fibroin uncrystalline motif GPGGY tends to form more stable β-sheet structure and nanofibers compared with the peptide R3 integrated with GGAGGS. Mechanical strength of R4 hydrogel is much higher than that of RADA16-I and R3 hydrogels, and promotes cell growth remarkably. Furthermore, these peptides can stabilize PY in aqueous solutions and deliver it to liposome vesicles. Altogether, spider fibroin motif GPGGY effectively enhances mechanical strength and hydrophobicity of the peptide.
Fourteen years ago, peptide RADA16-I was first designed and synthesized by mimicking the amphiphilic segment EAEAKAKAEAEAKAKA (EAKA16-II) of the naturally occurring yeast protein, Zuotin,35 and its nanofiber scaffold was then applied to 3D cell culture. Consequently, several functionalized self-assembling peptides were designed for particular cell growth, such as (RADA16-I)- SKPPTSS (PFSSTKT) (bone marrow homing peptide motif), which were used for neural stem cell proliferation and differentiation. 15 However, the normal microenvironment of cells is complex, and composition and strength of the extracellular matrix is different for different cell types. Previous work has shown that chondrocytes can grow in KLD12 hydrogel but not in RADA16-I scaffolds, which have lower strength.36 With addition of the spider fibroin uncrystalline motif GPGGY, the mechanical strength of the hydrogel is enhanced significantly, indicating that R4 hydrogel may also foster chondrocyte division – except for liver cells – and can be used to promote cartilage tissue repair.
Affinitive aggregation among ionic bridge bonds that form between salt ions in the DMEM medium and charged side chains of peptides, and affinity among uncrystalline hydrophobic motifs may be the main contributors to the mechanical strength of peptides. Due to the hydrophobic motifs, the mechanical strength of R3 and R4 hydrogels is greater than that of RADA16-I hydrogel. Moreover, the rigid turn of proline in motif GPGGY may prevent peptide R4 from forming unordered structure, and the hydrophobic phenyl ring of tyrosine may promote affinitive aggregation of R4. Therefore, the self-assembling peptide R4 forms more stable nanofibers and displays greater mechanical strength than the peptides R3 and RADA16-I.
Previous study has reported that the amphiphilic self-assembling peptide EAEAKAKAEAEAKAKA (EAK16-II) can encapsulate and stabilize hydrophobic compounds in aqueous solution and deliver them into liposome vesicles.7 In this process, the hydrophobic interaction between peptide and compound is critical. Therefore, we should improve the hydrophobicity of peptides, keeping their self-assembly properties at the same time. Our work demonstrates that peptide R4, integrating uncrystalline motif GPGGY with RADA16-I, self-assembles into long nanofibers and has higher affinity to hydrophobic compounds. R4 is able to deliver PY into PC vesicles, suggesting that it is a potential carrier of hydrophobic compounds.
This study provides a new method for designing nanobiomaterials, and illustrates that the nanostructures and macroscopic properties (mechanical strength, etcetera) of peptides can be controlled by adjustment of amino acid sequence. We will further investigate the application of new peptides in tissue engineering and regenerative medicine. Moreover, it will be interesting to modify peptide sequences in order to optimize the subtle balance between hydrophobic and hydrophilic residues, and to enhance the loading and release capacities of the peptide-based delivery system. With this knowledge, more biomaterials with desirable properties can be designed from these self-assembling peptides.
Acknowledgments
We acknowledge Zhaoyang Ye, Fushan Tang, and Jing Wang for their helpful suggestions, and also Zhihua Xing for her help in AFM and CD operations. This work was supported by Chinese National 985 Project of the Education Ministry and by National Natural Science Foundation of China (Grant No. 30770232).
Footnotes
Disclaimer
The authors declare that they have no conflict of interest in this work.
References
- 1.Zhao X, Zhang S. Self-assembling nanopeptides become a new type of biomaterial. Adv Polym Sci. 2006;203:145–170. [Google Scholar]
- 2.Zhang S. Fabrication of novel biomaterials through molecular self-assembly. Nat Biotechnol. 2003;21(10):1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 3.Mira H, Vilar M, Esteve V, et al. Ionic self-complementarity induces amyloid-like fibril formation in an isolated domain of a plant copper metallochaperone protein. BMC Struct Biol. 2004;4:7. doi: 10.1186/1472-6807-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelain F, Bottai D, Vescovi A, Zhang S. Designer self-assembling Peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS One. 2006;1:e119. doi: 10.1371/journal.pone.0000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Gelain F, Zhao X. Designer self-assembling peptide nanofiber scaffolds for 3D tissue cell cultures. Semin Cancer Biol. 2005;15(5):413–420. doi: 10.1016/j.semcancer.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Schachner M. Neurobiology. Nervous engineering. Nature. 2000;405(6788):747–748. doi: 10.1038/35015648. [DOI] [PubMed] [Google Scholar]
- 7.Keyes-Baig C, Duhamel J, Fung SY, Bezaire J, Chen P. Self-assembling peptide as a potential carrier of hydrophobic compounds. J Am Chem Soc. 2004;126(24):7522–7532. doi: 10.1021/ja0381297. [DOI] [PubMed] [Google Scholar]
- 8.Wang M, Law M, Duhamel J, Chen P. Interaction of a self-assembling peptide with oligonucleotides: complexation and aggregation. Biophys J. 2007;93(7):2477–2490. doi: 10.1529/biophysj.106.102624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Designer short peptide surfactants stabilize G protein-coupled receptor bovine rhodopsin. Proc Natl Acad Sci U S A. 2006;103(47):17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiley P, Zhao X, Vaughn M, Baldo MA, Bruce BD, Zhang S. Self-assembling peptide detergents stabilize isolated photosystem I on a dry surface for an extended time. PLoS Biol. 2005;3(7):e230. doi: 10.1371/journal.pbio.0030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu F, Chen Y, Tang C, et al. De novo design of a bolaamphiphilic peptide with only natural amino acids. Macromol Biosci. 2008;8(11):1053–1059. doi: 10.1002/mabi.200800180. [DOI] [PubMed] [Google Scholar]
- 12.Ellis-Behnke RG, Liang YX, You SW, et al. Nano neuro knitting: peptide nanofiber scaffold for brain repair and axon regeneration with functional return of vision. Proc Natl Acad Sci U S A. 2006;103(13):5054–5059. doi: 10.1073/pnas.0600559103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi K, Wang G, Liu Z, Feng Z, Huang B, Zhao X. Influence of a self-assembling peptide, RADA16, compared with collagen I and Matrigel on the malignant phenotype of human breast-cancer cells in 3D cultures and in vivo. Macromol Biosci. 2009;9(5):437–443. doi: 10.1002/mabi.200800262. [DOI] [PubMed] [Google Scholar]
- 14.Bokhari MA, Akay G, Zhang S, Birch MA. The enhancement of osteoblast growth and differentiation in vitro on a peptide hydrogel-polyHIPE polymer hybrid material. Biomaterials. 2005;26(25):5198–5208. doi: 10.1016/j.biomaterials.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 15.Nowakowski GS, Dooner MS, Valinski HM, Mihaliak AM, Quesenberry PJ, Becker PS. A specific heptapeptide from a phage display peptide library homes to bone marrow and binds to primitive hematopoietic stem cells. Stem Cells. 2004;22(6):1030–1038. doi: 10.1634/stemcells.22-6-1030. [DOI] [PubMed] [Google Scholar]
- 16.Ellis-Behnke RG, Liang YX, Tay DK, et al. Nano hemostat solution: immediate hemostasis at the nanoscale. Nanomedicine. 2006;2(4):207–215. doi: 10.1016/j.nano.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Knight CG, Morton LF, Peachey AR, Tuckwell DS, Farndale RW, Barnes MJ. The collagen-binding A-domains of integrins alpha(1) beta(1) and alpha(2)beta(1) recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J Biol Chem. 2000;275(1):35–40. doi: 10.1074/jbc.275.1.35. [DOI] [PubMed] [Google Scholar]
- 18.Ranieri JP, Bellamkonda R, Bekos EJ, Vargo TG, Gardella JA, Aebischer P. Neuronal cell attachment to fluorinated ethylene propylene films with covalently immobilized laminin oligopeptides YIGSR and IKVAVII. J Biomed Mater Res. 1995;29(6):779–785. doi: 10.1002/jbm.820290614. [DOI] [PubMed] [Google Scholar]
- 19.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238(4826):491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 20.Greenberg Z, Gavish H, Muhlrad A, et al. Isolation of osteogenic growth peptide from osteoblastic MC3T3 E1 cell cultures and demonstration of osteogenic growth peptide binding proteins. J Cell Biochem. 1997;65(3):359–367. [PubMed] [Google Scholar]
- 21.Mikhailova A, Fonina L, Kirilina E, Gur’yanov S, Efremov M, Petrov R. Peculiarities of immunocorrective effects of the bone marrow regulatory peptides (myelopeptides) Regul Pept. 2003;114(2–3):183–187. doi: 10.1016/s0167-0115(03)00123-x. [DOI] [PubMed] [Google Scholar]
- 22.Dicko C, Knight D, Kenney JM, Vollrath F. Structural conformation of spidroin in solution: a synchrotron radiation circular dichroism study. Biomacromolecules. 2004;5(3):758–767. doi: 10.1021/bm034373e. [DOI] [PubMed] [Google Scholar]
- 23.Colgin MA, Lewis RV. Spider minor ampullate silk proteins contain new repetitive sequences and highly conserved non-silk-like “spacer regions”. Protein Sci. 1998;7(3):667–672. doi: 10.1002/pro.5560070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24(2–3):271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 25.Parkhe AD, Seeley SK, Gardner K, Thompson L, Lewis RV. Structural studies of spider silk proteins in the fiber. J Mol Recognit. 1997;10(1):1–6. doi: 10.1002/(SICI)1099-1352(199701/02)10:1<1::AID-JMR338>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Simmons A, Ray E, Jelinski LW. Solid-state 13C NMR of nephila clavipes dragline silk establishes structure and identity of crystalline regions. Macromolecules. 1994;27(18):5235–5237. [Google Scholar]
- 27.Lawrence BA, Vierra CA, Moore AM. Molecular and mechanical properties of major ampullate silk of the black widow spider, Latrodectus hesperus. Biomacromolecules. 2004;5(3):689–695. doi: 10.1021/bm0342640. [DOI] [PubMed] [Google Scholar]
- 28.Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science. 2001;291(5513):2603–2605. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- 29.Hong Y, Legge RL, Zhang S, Chen P. Effect of amino acid sequence and pH on nanofiber formation of self-assembling peptides EAK16-II and EAK16-IV. Biomacromolecules. 2003;4(5):1433–1442. doi: 10.1021/bm0341374. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Szoka FC. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35(18):5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 31.Ye Z, Zhang H, Luo H, et al. Temperature and pH effects on biophysical and morphological properties of self-assembling peptide RADA16-I. J Pept Sci. 2008;14(2):152–162. doi: 10.1002/psc.988. [DOI] [PubMed] [Google Scholar]
- 32.L’Heureux GP, Fragata M. Localization of pyrene in small unilamellar phosphatidylcholine vesicles. Biophys Chem. 1988;30(3):293–301. doi: 10.1016/0301-4622(88)85024-5. [DOI] [PubMed] [Google Scholar]
- 33.Kalyanasundaram K, Thomas JK. Environmental effects on vibronic band intensities in pyrene monomer fluorescence and their application in studies of micellar systems. J Am Chem Soc. 1977;99(7):2039–2044. [Google Scholar]
- 34.Li M, Jiang M, Wu C. Fluorescence and light-scattering studies on the formation of stable colloidal nanoparticles made of sodium sulfonated polystyrene ionomers. J Polym Sci [B] 1997;35(10):1593–1599. [Google Scholar]
- 35.Zhang S, Lockshin C, Herbert A, Winter E, Rich A. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J. 1992;11(10):3787–3796. doi: 10.1002/j.1460-2075.1992.tb05464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kisiday J, Jin M, Kurz B, et al. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc Natl Acad Sci U S A. 2002;99(15):9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]