Abstract
The formation of mRNPs controls the interaction of the translation and degradation machinery with individual mRNAs. The yeast Scd6 protein and its orthologs regulate translation and mRNA degradation in yeast, C. elegans, D. melanogaster, and humans by an unknown mechanism. We demonstrate that Scd6 represses translation by binding the eIF4G subunit of eIF4F in a manner dependent on its RGG domain, thereby forming an mRNP repressed for translation initiation. Strikingly, several other RGG domain-containing proteins in yeast co-purify with eIF4E/G and we demonstrate that two such proteins, Npl3 and Sbp1, also directly bind eIF4G and repress translation in a manner dependent on their RGG-motifs. These observations identify the mechanism of Scd6 function through its RGG-motif and indicate that eIF4G plays an important role as a scaffolding protein for the recruitment of translation repressors.
INTRODUCTION
The control of gene expression requires the proper regulation of translation and mRNA degradation. Translation and mRNA degradation are inversely related due to the dual nature of the cap and poly(A) tail structures in promoting translation initiation, and as targets of the mRNA degradation machinery. Translation initiation is promoted by the binding of the eIF4F complex to the cap structure thereby allowing the eIF4G subunit to serve as a scaffold to recruit the 43S pre-initiation multi-factor complex (MFC), which includes the small ribosomal subunit, the initiator tRNA, and additional initiation factors (reviewed in Sonenberg and Hinnebusch, 2009). Translation initiation is also enhanced by the poly(A) tail, in part through interactions of the poly(A) binding protein (Pab1) with eIF4G (Tarun and Sachs, 1996). Conversely, mRNA degradation is generally initiated by deadenylation followed by decapping and 5′ to 3′ degradation (Parker and Song, 2004; Garneau et al., 2007). Decapping is enhanced by inhibition of translation initiation and loss of the eIF4F cap-binding complex, which presumably increases the availability of the cap to the decapping enzyme (Schwartz and Parker, 1999; 2000).
The transition of mRNAs between translation and decapping by the Dcp1/Dcp2 decapping enzyme is promoted by a conserved set of decapping activator proteins including Dhh1/Rck, Pat1, Scd6/RAP55, the Lsm1-7 complex, and in metazoans Ge-1 (reviewed in Franks and Lykke-Andersen, 2008). These proteins physically interact with each other, and can both directly inhibit translation, and/or stimulate the decapping enzyme (Coller and Parker, 2005; Nissan et al., 2010; Tritschler et al., 2009). In addition, in metazoans, the NudT16 protein functions as a second decapping enzyme but how its activity is regulated remains to be determined (Song et al., 2010). Some of these translation repressor/decapping activator proteins also function in the storage and/or transport of mRNAs and can accumulate with translationally repressed mRNAs in dynamic cytoplasmic RNP granules referred to as P-bodies (reviewed in Eulalio et al., 2007; Anderson and Kedersha, 2009; Parker and Sheth, 2007). An unresolved issue is how translation repressor/decapping activator proteins repress translation and can both target mRNAs for decapping or mRNA storage.
A highly conserved component of the translation repression/mRNA decay machinery is the Scd6 protein family, whose orthologs in Drosophila (TraI), nematodes (CAR-1), humans (RAP55), and plants (DCP5) are involved in translation repression and mRNA storage, and accumulate in cytoplasmic mRNP granules related to P-bodies (Xu and Chua, 2009; Boag et al., 2005; Wilhelm et al., 2005; Barbee et al., 2006; Tanaka et al., 2006; Tritschler et al., 2007). Moreover, the planaria ortholog of Scd6 is required for maintenance of stem cell potential, possibly through the storage of mRNAs (Wang et al., 2010). In yeast and Arabidopsis, Scd6 and its plant ortholog (DCP5) promote mRNA decapping (Decourty et al., 2008; Xu and Chua, 2009). In contrast, in plasmodium the Scd6 ortholog is required for the stable storage of maternal mRNAs (Mair et al., 2010).
The Scd6 family members have a conserved domain organization with an N terminal Lsm domain, a central FDF motif, which interacts with a conserved RNA helicase referred to as Dhh1 in yeast (Tritschler et al., 2009), and a C terminal RGG domain. Both yeast and plant Scd6 family members can bind the Dcp2 decapping enzyme but are not sufficient to directly activate decapping activity (Nissan et al., 2010; Xu and Chua, 2009). In contrast, Scd6 family members from yeast, Arabidopsis and Xenopus can directly repress translation in vitro (Nissan et al., 2010, Xu and Chua, 2009; Tanaka et al., 2006), and at least for the yeast protein, this repression requires the C-terminal RGG domain and blocks translation initiation prior to formation of the 48S pre-initiation complex (Nissan et al., 2010). An unresolved issue is the mechanism by which Scd6 represses translation and leads to stable mRNA storage.
In this work, we show that Scd6 directly inhibits translation by binding the eIF4G subunit of eIF4F through its RGG domain, thereby forming an mRNP repressed for translation initiation. Such a complex also provides a possible mechanism by which Scd6 (and possibly its orthologs) can stably store translationally repressed mRNAs. In addition, we demonstrate that two other RGG domain containing proteins, Npl3 and Sbp1, also directly bind eIF4G and repress translation via their RGG-motifs. These observations identify the mechanism of Scd6 function and indicate that eIF4G also plays an important scaffolding protein for the recruitment of translation repressors with RGG-motifs.
RESULTS
Scd6 physically interacts with eIF4G
Since Scd6 blocks initiation upstream of 48S complex formation (Nissan et al., 2010), we hypothesized that Scd6 could either a) bind RNA and thereby limit the assembly of the translation machinery on the mRNA, b) bind to Pab1 and repress the stimulation of translation by poly(A) tail, or c) bind to the eIF4E/G complex and inhibit its binding to mRNA or its ability to recruit the MFC. To distinguish between these possibilities we tested the ability of Scd6 to bind RNA, eIF4E/G, or Pab1 using recombinant proteins. Moreover, we determined if any interaction was functionally relevant to Scd6 repression of translation by determining if it was affected by the deletion of the C-terminal RGG domain, which is required for growth inhibition in vivo and translation repression in vitro (Nissan et al., 2010).
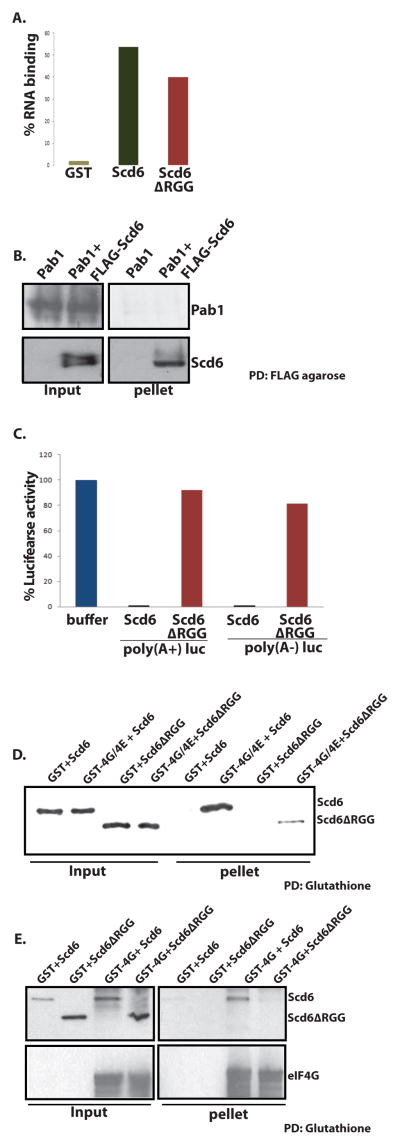
We observed that Scd6 bound to MFA2 mRNA using a column-binding assay (Figure 1A). However, the ability to bind RNA was not related to its ability to repress translation since Scd6Δrgg binds RNA almost as strongly as the wild-type protein (Figure 1A). This indicates that Scd6 can bind RNA but that RNA binding is not sufficient to repress translation.
Figure 1. Scd6 binds eIF4G.
A) 32P labeled MFA2 mRNA was incubated with GST, GST-Scd6 or GST-Scd6ΔRGG followed by pull down using glutathione sepharose. Radioactivity on beads after three washes was measured by Cerenkov counting. B) Purified His-Pab1 was incubated with FLAG-Scd6 followed by FLAG pull down. Pab1 and Scd6 were detected by using polyclonal anti-Pab1 and anti-Scd6 antibodies respectively. C) Purified Scd6 and RGG mutant were tested for their effects on translation of 100 ng of poly(A−) and poly(A+) luciferase message at 6uM concentration as described previously (Nissan et al., 2010). D) Glutathione sepharose pull downs performed to look at interaction of purified GST-4G/E with recombinant Scd6 and Scd6ΔRGG mutant protein. Scd6 protein was detected with anti-FLAG antibody following manufacturer’s instructions. E) Glutathione sepharose pull downs were performed to look at interaction of GST-4G in bacterial extracts with recombinant Scd6 and Scd6ΔRGG mutant protein. 6ug of each protein was used in 200ul reaction mixture. Scd6 protein was detected with anti-FLAG antibody following manufacturer’s instructions. Typically 10% of total reaction was loaded in the ‘input’ lanes and 100% of total pulled-down material was loaded in ‘pellet’ lanes.
Using a pull down assay we observed that Scd6 did not bind to Pab1 (Figure 1B). Moreover, we observed that at the tested concentration, Scd6 repressed translation of both A+ and A− luciferase mRNAs in cell extracts to a similar extent (Figure 1C). These observations argue that Scd6 does not repress translation directly through interaction with Pab1 or the poly(A) tail.
More importantly, we observed that Scd6 bound to eIF4E/G purified from E. coli as a complex, and this binding was strongly enhanced by the C-terminal RGG domain (Figure 1D). We also observed that Scd6 was pulled down by eIF4G directly in a manner dependent on its RGG domain (Figure 1E), although the pull down assay for eIF4G was performed in bacterial cell lysates since purified eIF4G precipitated in absence of eIF4E. Since the region of Scd6 required for strong translation repression (Nissan et al., 2010) is required for binding to eIF4G, these observations argue that Scd6 primarily represses translation by binding to eIF4G through its C-terminal RGG domain.
Scd6 forms a complex with eIF4E/G on mRNAs
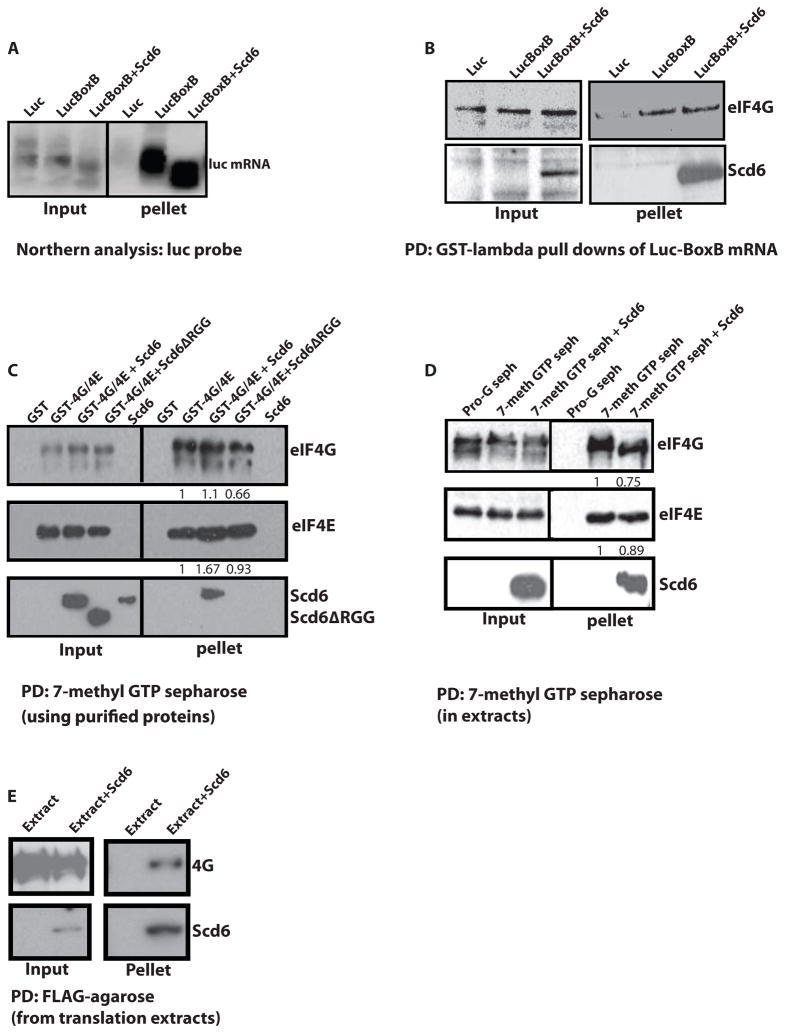
The interaction of Scd6 with eIF4G could inhibit translation either by binding eIF4E/G and preventing it from binding to the mRNA and/or initiation factors, or by binding eIF4E/G once bound to mRNAs and inhibiting eIF4F mediated recruitment of the 43S pre-initiation complex. To distinguish between these two possibilities, we first asked if Scd6 altered the interaction of eIF4E/G with mRNA in cell-free extracts under conditions where Scd6 inhibits translation. If Scd6 inhibits the interaction of eIF4E/G with mRNA, then Scd6 should reduce the amount of eIF4E/G co-purifying with mRNA. Conversely, if Scd6 binds to eIF4E/G complexes on the mRNA, then Scd6 should not reduce the levels of eIF4E/G bound to the mRNA, and more importantly, Scd6 would also be associated with the mRNA under translation repression conditions. For this experiment, we programmed the extracts with a luciferase mRNA containing two copies of the binding site for the lambda-N protein (referred to as BoxB sites), which allows the mRNA and any associated proteins to be purified from extracts (Czaplinski et al., 2005). The insertion of the BoxB sites into the luciferase mRNA did not affect its repression by exogenously added Scd6 (data not shown). Following incubation in the extracts, the luciferase-BoxB mRNA could be specifically purified using a GST-lambdaN fusion protein, while a control mRNA without the BoxB sites was not selected (Figure 2A).
Figure 2. Scd6 forms a tri-complex with eIF4E/G on cap.
A) Northern analysis of luciferase mRNA containing BoxB repeats pulled down from translation extracts with GST-lambda protein (see procedures) +/− purified His-tagged Scd6 at 6uM concentration. B) Western analysis of proteins coming along with luciferase mRNA containing BoxB using antibodies indicated. C) 7-methyl-GTP sepharose pull downs were performed using recombinant purified eIF4G/4E and Scd6 proteins (see procedures). D) 7-methyl-GTP sepharose pull downs performed from yeast translation extracts in presence of Scd6 or Scd6ΔRGG. E) FLAG-pull downs from translation reactions to pull down recombinant Scd6. 20 ul translation reactions were diluted to 200ul followed by addition of FLAG-agarose beads. Binding was performed for an hour with end to end rotation followed by three washes of beads for 10′ each. The beads were then boiled in SDS-loading buffer and analyzed by SDS-PAGE followed by western blotting. Typically 10% of total reaction was loaded in the ‘input’ lanes and 100% of total pulled-down material was loaded in ‘pellet’ lanes. Bands in C and D were quantified by using Image J program.
By western analysis, we observed that eIF4G co-purified with the BoxB tagged mRNA and the amount of eIF4G bound to the mRNA was not affected by the addition of exogenous Scd6 (Figure 2B). Moreover, we observed that the exogenously added Scd6 co-purified with the Box B tagged mRNA. These observations indicate that Scd6 does not block eIF4E/G recruitment to the mRNA and instead Scd6 can form a complex with mRNA and eIF4E/G.
Scd6 could be sufficient to form a complex with eIF4E/G on the cap structure or this complex might require additional factors in the extract and/or interactions of Scd6 with the mRNA body. To distinguish these possibilities, we determined if recombinant Scd6 and eIF4E/G were sufficient to assemble a complex on the cap structure using cap-affinity chromatography. We observed that Scd6 by itself did not bind to a cap column but was retained on the cap column in the presence of recombinant eIF4E/G (Figure 2C). Moreover, Scd6Δrgg failed to be retained on the cap column by eIF4E/G (Figure 2C). The levels of 4E/G bound to the cap resin were comparable in the presence and absence of Scd6 providing additional evidence that Scd6 does not affect the interaction of eIF4E/G with the cap. Similar results were obtained when the cap-binding assay was done from cell free extracts (Figure 2D). We conclude that Scd6 binding to eIF4E/G does not reduce eIF4E/G binding the cap structure, but instead leads to the formation of an Scd6-4E/G complex bound to the cap. This complex could then block 43S recruitment on the mRNA to limit translation. Note that such a complex could also protect mRNAs from decapping and thereby stabilize some mRNAs (see discussion).
If Scd6 represses translation by binding to eIF4G then we should be able to document this interaction in a reciprocal manner, where we pull down Scd6 from extracts and observe co-purification of eIF4G. In order to test this, we performed translation reactions in presence of Scd6 and then pulled out Scd6 from translation reactions. We observed that eIF4G co-purified with Scd6 providing additional evidence for a Scd6-eIF4G interaction (Figure 2E).
We also observed that the addition of Scd6 to the extracts decreased the size of the luciferase mRNA in a manner consistent with shortening of the 3′ poly(A) tail (Figure 2A). This suggests that Scd6 might also enhance deadenylation in extracts. However, deadenylation per se is not the mechanism by which Scd6 represses translation since Scd6 efficiently represses translation of poly(A)- mRNAs (Figure 1C). Whether Scd6 also affects deadenylation in vivo remains to be determined.
Scd6 does not perturb Pab1 and 4A interaction with 4G1
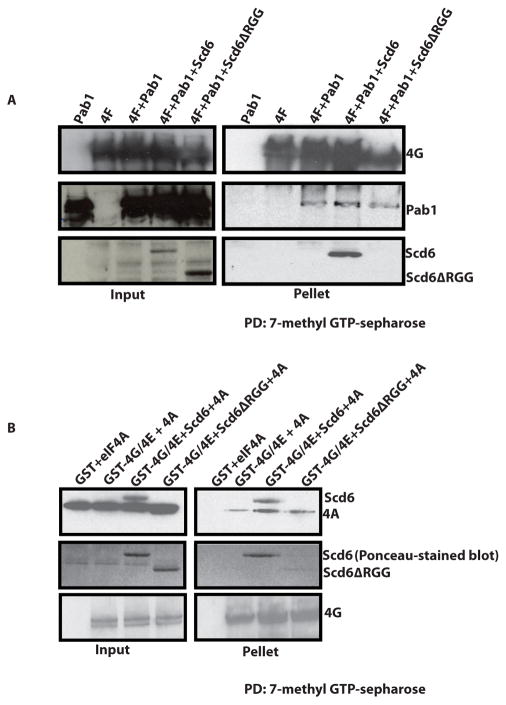
The eIF4G component of eIF4F is a scaffold protein that interacts with eIF4E, eIF4A and Pab1to promote translation initiation (Sonenberg and Hinnebusch, 2009). This raises the possibility that Scd6 might inhibit translation by blocking the interaction of Pab1 or eIF4A with eIF4G1. To test this possibility, we utilized recombinant proteins to examine how Scd6 affected the interaction of Pab1 with eIF4E/G using cap affinity chromatography.
We observed that the amount of Pab1 or eIF4A co-purifying with eIF4E/G on cap-sepharose did not change significantly in presence of Scd6 (Figure 3A & 3B). Again, Scd6 but not Scd6ΔRGG was detectable in these pull downs. This suggests that Scd6 binding to eIF4G does not strongly affect the Pab1-eIF4G or eIF4A-eIF4G interaction. The above results suggest that translation repression by Scd6 could involve the formation of a Scd6-eIF4E-4G-4A-Pab1 complex.
Figure 3. Scd6 does not destabilize Pab1-4G or 4A-4G interactions.
A) 7-methyl-GTP sepharose pull downs were performed to look at interaction of GST-eIF4G/eIF4E complex with recombinant Pab1 in presence of Scd6 or Scd6ΔRGG protein as described in materials and methods. 8ug of GST-eIF4G/eIF4E was incubated with wt or mutant Scd6 protein for 1h at 4°C with end-to-end rotation. Following this Pab1 was added to reaction mixture and incubated for 1h with end-to-end rotation. Finally, 7-methyl-GTP sepharose was added to reaction mix and incubated for 2h. Washing was performed as described in materials and methods. eIF4G and Pab1 were detected with polyclonal antibodies. B) Glutathione pull down assay to analyze effect of purified Scd6 on eIF4A-eIF4E/G interaction. Incubation and washing conditions were same as mentioned in A. eIF4G and eIF4A were detected with polyclonal antibodies. The anti-eIF4A antibody cross-reacts with purified Scd6 (contains both His and FLAG-tag). The middle panel depicts ponceau-stained blot with clearly visible wt and mutant Scd6 proteins. This was done since eIF4A and Scd6ΔRGG run at similar position (in top panel). Typically 10% of total reaction was loaded in the ‘input’ lanes and 50 or 100% of total pulled-down material was loaded in ‘pellet’ lanes.
Scd6 accumulates in and affects P-bodies and stress granules
The above observations argue that Scd6 can form a complex with translation initiation factors and thereby inhibit initiation. However, Scd6 also directly interacts with the decapping enzyme Dcp2 (Nissan et al., 2010). This suggests Scd6 might assemble into two different types of mRNPs and predicts that Scd6 might be a component of P-bodies, where Dcp2 is concentrated, and stress granules, where eIF4F and Pab1 accumulate when translation initiation is compromised (reviewed in Anderson and Kedersha, 2009; Buchan and Parker, 2009).
Previous results have shown that Scd6 accumulates in cytoplasmic foci during glucose deprivation, which overlap at least partially with P-bodies (Barbee et al., 2006). Since yeast stress granules often overlap or are adjacent to P-bodies (Brengues and Parker, 2007; Holmes et al., 2007; Buchan et al., 2008; Grousl et al., 2009), we determined whether Scd6 accumulated in yeast stress granules and/or P-bodies by examining the localization of a Scd6-GFP fusion protein during glucose deprivation as compared to mCherry-tagged markers of P-bodies (Edc3) or stress granules (Pbp1) (Swisher and Parker, 2010).
Upon glucose deprivation, we observe that many Edc3 (P-bodies) and Pbp1 (stress granules) foci often overlap with each other but some of them are independent or in close proximity to each other (Figure 4A), consistent with previous report (Buchan et al., 2008). We observed that Scd6 behaves like a component of both P-bodies and stress granules. First, Scd6-GFP accumulates in foci that overlap 88% of time with Edc3-mCherry, but is also in separate or docked foci (Figure 4B). This indicates that Scd6 can accumulate in P-bodies, although the presence of some Scd6 foci that do not co-localize with Edc3 suggests that Scd6 can also accumulate in stress granules. Evidence that Scd6 can also be in stress granules is that Scd6-GFP foci co-localize 73.5% of the time with the stress granule Pbp1-mCherry (Figure 4C) (Swisher and Parker, 2010). The accumulation of Scd6 in both P-bodies and stress granules is a conserved feature of this protein family since the mammalian ortholog of Scd6, RAP55, also accumulates in both P-bodies and stress granules (Yang et al., 2006).
Figure 4. Scd6 localizes to and affects granules.
A) Strain bearing chromosomal GFP-Edc3 was transformed with plasmid expressing Pbp1-mCherry. Cells were grown to 0.3–0.4 OD600 in appropriate minimal media with 2% glucose and then stressed in minimal medium lacking glucose for 10 minutes before observing under the microscope. B) Strain bearing chromosomal GFP-Scd6 was transformed with plasmid expressing Edc3-mCherry. Cells were grown to 0.3–0.4 OD600 in appropriate minimal media with 2% glucose and then stressed in minimal medium lacking glucose for 10 minutes before observing under the microscope. C) Strain bearing chromosomal GFP-Scd6 was transformed with Pbp1-mCherry. Cells were grown to 0.3–0.4 OD600 in appropriate minimal media with 2% glucose and then stressed in minimal medium lacking glucose for 10 minutes before observing under the microscope. D) Wild type or scd6Δ strains were transformed with vector expressing Dcp2-mCherry. Cells were grown and stressed in absence of glucose as described in A and observed under the microscope. E) Wild type or scd6Δ strains were transformed with vector expressing Dcp2-mCherry and Pab1-GFP. Cells were grown up to 0.3–0.4 OD600 stressed by adding 0.5% sodium azide (or equal of water for control cells) to the medium, incubated for 30 minutes at 30°C before observing under microscope. F) Wild type strains expressing Edc3-mCherry and Pab1-GFP were transformed with either empty vector or vector driving over-expression of SCD6 or SCD6ΔRGG from the Gal-inducible promoter. These strains were grown to 0.3–0.4 OD600 in appropriate minimal media with 2% sucrose as carbon source followed by growth in medium containing 1.8% galactose-0.2% sucrose for 2 hours. Cells were then observed under the microscope. G) eIF4G-GFP, eIF4E-GFP and eIF3b-GFP strain were transformed with empty vector or vector driving over expressing SCD6 under Gal-inducible promoter. These strains were grown to 0.3–0.4 OD600 in appropriate minimal media with 2% sucrose as carbon source followed by growth in medium containing 1.8% galactose-0.2% sucrose for 2 hours. Cells were then observed under the microscope.
Several observations indicate that Scd6 can also affect the formation of stress granules and P bodies. First, during glucose deprivation or sodium azide treatment (Buchan et al., 2011), scd6Δ strains show reduced P-body and stress granule formation as judged by Dcp2-mCherry and Pab1-GFP, respectively (Figure 4D & E). Second, over-expression of Scd6 from a gal-inducible promoter led to the accumulation of both P bodies and stress granules as observed by Edc3-mCherry and Pab1-GFP foci respectively (Figure 4F). Strikingly, over-expression of the Scd6Δrgg variant induced P-bodies although to lesser extent, but not stress granules (Figure 4F), consistent with the induction of stress granules requiring the inhibition of translation by Scd6 interacting with eIF4G, while the induction of P-bodies would be due to different interactions of Scd6. We interpret these observations to indicate that Scd6 functions to increase the formation of stress granules and P-bodies during stress responses and that the induction of stress granules requires the RGG domain, presumably due to its interaction with eIF4G.
Stress granules contain translationally arrested mRNA with initiation factors (Buchan and Parker 2009). Our in vitro results suggest that Scd6 forms a complex on mRNAs minimally with Pab1, eIF4E, 4G and 4A and this mRNP could be blocked to subsequent recruitment of the 43S complex. This in vitro data predicts that over-expression of Scd6 in vivo should trap mRNAs in a similar biochemical complex, which would be revealed by the composition of stress granules formed in response to Scd6 over-expression. Thus, we utilized strains with GFP tagged versions of translation initiation factors to determine what proteins accumulate in stress granules when Scd6 is overexpressed. We observed the formation of eIF4E and eIF4G but not eIF3b foci (Figure 4G) upon Scd6 over expression. These observations argue that in vivo Scd6 also traps an mRNP containing Pab1, 4E and 4G, but lacking the MFC.
Multiple RGG-motif Containing Proteins interact with eIF4G
Our results indicated that Scd6 interacts with eIF4G through its RGG domain. By blast searching we identified the yeast proteins Sbp1, Psp2, Nop1, Npl3, Nsr1, Dbp2, Gar1, Gbp2, Nab2, and Ded1 as containing RGG domains similar to the Scd6 domain. Interestingly, from genomic analyses, eight of these ten proteins (all except Dbp2 and Nsr1) co-purify with eIF4E or one of the two eIF4G proteins in yeast (Krogan et al., 2004; 2006; Collins et al., 2007; Gavin et al., 2002 & 2006). Moreover, the Sbp1 and Npl3 proteins have been suggested to repress translation in vivo under certain conditions (Segal et al., 2006; Windgassen et al., 2004). This raised the possibility that other RGG domain containing proteins would also directly interact with eIF4G and modulate translation and/or mRNA degradation.
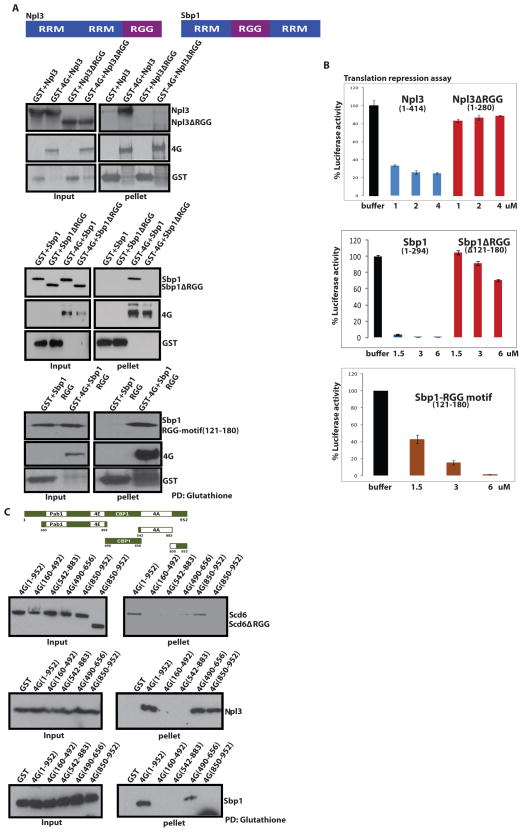
We tested if additional RGG containing proteins directly interact with eIF4G and if the RGG-motif was important for their interaction with eIF4G. We purified recombinant Sbp1, Npl3 and their respective RGG-motif deleted variants. We observed that both Sbp1 and Npl3 proteins directly bound eIF4G (Figure 5A), and that binding was impaired for their RGG-motif deleted variants (Figure 5A). In addition, we were able to express and purify the Sbp1 RGG-motif (121–180 amino acids), which we observed specifically interacted GST-eIF4G and not GST alone, suggesting that the Sbp1 RGG-motif is sufficient to bind eIF4G. Based on these observations we conclude that at least a subset of RGG-motifs are important for eIF4G-binding, although the specific sequence features of these motifs that interact with eIF4G have not been determined.
Figure 5. Multiple RGG Domain Containing Proteins bind eIF4G and repress translation.
A) Schematic representation of Sbp1 and Npl3 proteins with their domain organization. A) Top- GST pull downs with recombinant eIF4G in bacterial extracts and either full length His-Npl3 or its RGG-deletion mutants. Npl3 was detected using antisera against Npl3. A) Middle- GST pull downs with recombinant eIF4G in bacterial extracts and either full length His-Sbp1 or its RGG-deletion mutants. Sbp1 was detected using anti-His antibody. A) Bottom- GST pull downs with recombinant eIF4G in bacterial extracts and His-Sbp1 RGG-motif (121–180). RGG motif was detected using anti-His antibody B) In vitro translation assay was performed as described in Nissan et al., 2010 with the addition of recombinant full length or RGG-deleted variant of Npl3 (top panel), Sbp1 (middle panel) and Sbp1 RGG-motif (121–180) (bottom panel). Error bars represent standard error of three experiments. C) Top- Schematic representation of the 4G-deletion constructs used for mapping binding sites of Scd6, Npl3 and Sbp1. Bottom- GST pull downs of full length and subdomains of GST- tagged eIF4G with Scd6, Npl3, and Sbp1. Scd6, Npl3 and Sbp1 were detected in pull downs by antibodies against the His tag on these recombinant proteins. We observed approximately 60–70% pull downs for various 4G-fragments. Typically 10% of total reaction was loaded in the ‘input’ lanes and 100% of total pulled-down material was loaded in ‘pellet’ lanes. See also Fig. S1 which is connected to this figure.
The binding of Npl3 and Sbp1 to eIF4G suggests that these proteins might also affect translation. We tested this possibility by examining the affect of recombinant Sbp1, Npl3 and their RGG-mutants on translation of luciferase mRNA in yeast extracts. We observed that Sbp1 and Npl3 proteins strongly repressed translation and the repression activity was dependent on their RGG-motifs (Figure 5B). Consistent with its ability to bind eIF4G, the Sbp1 RGG-motif (amino acids 121–180) was sufficient to repress translation of luciferase mRNA (Figure 5B). This argues that the interaction of these proteins with eIF4G through their RGG-motifs contributes towards negatively regulating translation and identifies eIF4G as a major target of these types of translation repressors.
Both Npl3 and Sbp1 contain two RRM-domains (Figure 5A, top panel) and are RNA-binding proteins. However, and similar to Scd6, the ability of Npl3 to repress translation is not simply due to binding RNA since the Npl3Δrgg variant is compromised for its ability to repress translation but binds luc mRNA in comparable manner to wild type Npl3 (Supp. Figure 1A). The RGG-mutant of Sbp1 however binds RNA weakly in spite of having two intact RRM domains (Supp. Figure 1B) suggesting that the Sbp1 RGG-motif could play an important role in binding certain mRNAs. It is also possible that deletion of Sbp1 RGG-motif affects the structure of mutant polypeptide. However since the Sbp1 RGG-motif itself is sufficient to bind eIF4G and repress translation but binds luc mRNA poorly (Supp, Figure 1C) we conclude that the RGG-motif is important for Sbp1 repression activity independent of RNA binding.
To determine if Scd6, Sbp1 and Npl3 interacted with similar or different parts of eIF4G, we utilized a previously constructed set of eIF4G fragments (Figure 5C) expressed in bacteria and examined their ability to pull down Scd6, Sbp1, and Npl3 (Paquin et al., 2007). We observed that these three proteins showed different modes of interaction with eIF4G. Specifically, Scd6 bound to the very C-terminal domain of eIF4G (aa850–952), Sbp1 bound to an internal region (aa490–656), while Npl3 bound to both the aa850–952 and aa490–656 domains (Figure 5B). This observation suggests multiple regions of eIF4G interact with translation repressors and thereby might play a role in the inhibition of translation (see discussion).
DISCUSSION
Mechanism of Translation Repression by Scd6
Several observations demonstrate that Scd6 represses translation by forming an mRNP complex with eIF4E/G and inhibiting the recruitment of the 43S pre-initiation complex. First, Scd6 binds directly to eIF4G through its RGG domain (Figure 1D & E), which is required for translation repression in vitro and growth inhibition in vivo (Nissan et al., 2010). Additional experiments indicate that Scd6 binds to the C-terminal domain of eIF4G (Figure 5C). Second, Scd6 is sufficient to form a complex with eIF4E/G on a cap structure and this depends on its RGG-motif (Figure 2C). Third, under conditions of translation repression Scd6 assembles on mRNAs with eIF4E/G in cell extracts (Figure 2B and D). Fourth, Scd6 inhibits 48S complex formation in vitro (Nissan et al., 2010). Finally, overexpression of Scd6 in vivo leads to the accumulation of stress granules containing eIF4E/G, Pab1, but not components of the MFC such as eIF3b, and this effect is dependent on its RGG domain (Figure 4F & G). This defines a mechanism of Scd6 translation repression by direct interactions with eIF4G both in vivo and in vitro. This is likely to be a conserved mechanism of translation repression since Scd6 orthologs in plants (DCP5), C. elegans (CAR-1), humans (RAP55), and Drosophila (Trailer-Hitch) all share a conserved structure and function as translation repressors (Xu and Chua, 2009; Jeske et al., 2011; Tanaka et al., 2006). Moreover, and consistent with our observations in yeast, one of the two RGG domains of the RAP55 protein is required for its accumulation in stress granules (Yang et al., 2006).
The formation of an Scd6-eIF4F-Pab1 mRNP complex has two implications. First, by having a pre-assembled partial translation initiation complex, such Scd6 repressed mRNAs would be poised to re-enter translation upon inactivation of Scd6 function. Interestingly, Scd6 is a target of arginine methyltransferase and methylation is thought to occur in the RGG domain and might thereby influence the interaction with eIF4G. Second, by forming a translationally repressed mRNP in combination with eIF4E/G assembled on the cap complex, the mRNA would be protected from decapping since eIF4E/G is known to inhibit mRNA decapping when bound to the 5′ cap structure (Schwartz and Parker, 1999; 2000). This provides a possible mechanism by which Scd6 functions to stabilize mRNAs in plasmodium (Mair et al., 2010), and possibly in C. elegans through interactions with CGH-1 (Boag et al., 2008). This formation of a translationally repressed mRNP with either eIF4E/eIF4G or eIF4E bound to the cap to block decapping may be a general mechanism for stable storage of mRNAs and has been proposed for the function of CUP in Drosophila (Igreja and Izaurralde, 2011).
eIF4G as a general target of translation repressors
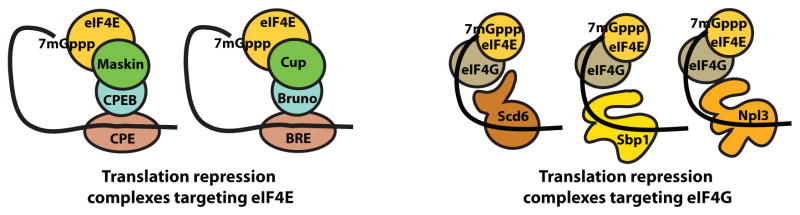
Several pieces of evidence indicate that, in addition to Scd6, several other RGG domain containing proteins will negatively regulate translation by interacting with eIF4F and the eIF4G subunit in particular. This possibility was first raised by the co-purification of numerous RGG containing proteins with eIF4E, eIF4G or both in yeast genomic analyses (Krogan et al., 2004; 2006; Collins et al., 2007; Gavin et al., 2002 & 2006). More directly, we show that Scd6, Sbp1 and Npl3 all bind eIF4G through their RGG-motifs and directly repress translation (Figure 5). This ability to repress translation for Sbp1 and Npl3 is related to their function in vivo since Sbp1 co-localizes with RNA granules in yeast and can contribute to translation repression in vivo (Segal et al., 2006), and Npl3 has been suggested to negatively control translation (Windgassen et al., 2004), and localizes to stress granules (Buchan and Parker, unpublished observation). The ability of Sbp1 RGG-motif alone to bind eIF4G and repress translation is striking (Figure 5A & B). It suggests that RGG-motifs from other repressors might also behave in same way. In addition, another RGG domain containing protein, Ded1, can repress translation through interactions with eIF4G in a manner dependent on a RGG containing domain, although this interaction can also stimulate translation following hydrolysis by Ded1 (Hilliker et al., 2011). This identifies at least a subset of yeast RGG domain containing proteins as binding eIF4G and thereby modulating translation. Such interactions are likely to be conserved in other eukaryotes since both the Scd6 and Ded1 orthologs contain conserved RGG motifs and numerous translation regulators in mammalian cells contain RGG domains. Moreover, the generality of eIF4G as a target of repressors is extended by the observations that the yeast Khd1 protein interacts with eIF4G to repress translation (Paquin et a., 2007), and the mammalian L13a protein inhibits translation in a manner dependent on eIF4G (Kapasi et al., 2007).
The interaction of several translation repressors with eIF4G identifies two general mechanisms for the repression of translation through components of the eIF4F complex (cartooned in Figure 6). Previous work defined a common mechanism for translational control where proteins bind eIF4E on the mRNA and inhibit the recruitment of eIF4G to the mRNA (reviewed in Richter and Sonenberg, 2005). We now suggest a related but distinct mechanism wherein multiple regulatory proteins bind eIF4G in association with the mRNA and thereby blocking recruitment of the 43S pre-initiation complex. Note that such a complex could give rise to mRNPs stalled in translation initiation after eIF4F assembly but before 43S joining as is seen during some stress responses in yeast (Buchan et al., 2008; Holmes et al., 2007). Moreover, by inhibiting translation after eIF4F has associated with the mRNA, the cap structure can be protected from decapping and the mRNA thereby stabilized.
Figure 6. Two paradigms for translation repression complexes utilizing components of eIF4F.
It is notable that multiple different proteins interact directly with eIF4G and thereby modulate translation. One possibility is that each of these proteins regulates a subset of mRNAs through sequence specific RNA binding contacts. Consistent with that possibility, the Nsr1, Npl3, and Khd1 proteins have been shown to co-immunopurify with selected subsets of mRNAs, although they do show some overlap (Hogan et al., 2009). Another possibility is that different eIF4G interacting proteins bind eIF4G at different stages of the mRNAs life. For example, one set of proteins, such as Npl3, might bind nascent transcripts and play an important role in repressing translation initiation and or blocking decapping until the mRNP has been properly localized and remodeled in the cytoplasm. Consistent with that possibility, Npl3 is a nuclear-cytoplasmic shuttling protein that plays an important role in mRNA export (Zenklusen and Stutz, 2004). In contrast, other eIF4G binding proteins such as Scd6 might bind when mRNAs are exiting translation, either for degradation, or for mRNA storage in response to environmental cues. Thus, one future goal will be to determine the temporal and mRNA specificity of these eIF4G interacting proteins.
eIF4E/eIF4G as an integrator of mRNA Fate
Several observations now argue that the cap binding complex of eIF4E and eIF4G should be considered a general integrator of mRNA fate. First, it has long been appreciated that eIF4E/eIF4G functions to promote translation initiation, mainly based on its ability to act as scaffold for recruiting other initiation factors (Sonenberg and Hinnebusch, 2009). Now numerous studies, including this work, document translation repression complexes that form in conjunction with either eIF4E or with eIF4E/eIF4G (Richter and Sonenberg, 2005; Paquin et a., 2007; Kapasi et al., 2007; Igreja and Izaurralde, 2011) indicating a second role of this complex as a translation repressor. Finally, mammalian eIF4E can promote nuclear mRNP export through the formation of a specific mRNP (Topisirovic et al., 2009). Thus, the eIF4E and eIF4G proteins are general adaptors that interact with the capped mRNAs and then through interactions with other mRNP components modulate subcellular localization, translation, and mRNA degradation. Given this, a complete understanding of the proteins that interact with eIF4E or eIF4G, and their functional consequences, will be important to understanding the control of mRNA function in eukaryotic cells.
EXPERIMENTAL PROCEDURES
Yeast strains and plasmids
All plasmids used in this study are listed in Table S1. The genotypes of strains used in this study are listed in Table S2. Strains were grown on either standard yeast extract/peptone medium (YP) or synthetic medium (SC) supplemented with the appropriate amino acids and 2% glucose. Strains were grown at 30°C unless otherwise stated. eIF4E/G expression construct was a kind gift from Jon Lorsch. Pab1 expression construct was a kind gift from Alan Jacobson. pGLH2 was kindly provided by Kevin Czaplinski. pGLH2 is an improved version of previously reported vector (Czaplinski et al., 2005). It encodes for a mutant lambda peptide that has higher affinity for boxB sequence. The rare arginine codons in the previous version were changed to standard ones.
Protein purification, Cap-binding assays and Western blotting
Proteins were purified from E. coli according to standard protocols using glutathione sepharose beads (GE), Ni-NTA agarose (Qiagen) or FLAG-agarose (Sigma-Aldrich) according to standard protocols. To remove RNA that might provide bridging interactions, extracts after breaking open the cells during the time of protein purification were treated for 20 min with Qiagen RNase A (1mg/ml). Purified protein was concentrated and dialyzed into 20 mM Tris, 100 mM NaCl and 10% glycerol. Binding reactions for m7-GTP sepharose pull downs and GST-pull downs were performed at 4°C in binding buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 2 mM DTT, 2 mM MnCl2, 2 mM MgCl2, 1% Igepal CA-630 (USB), 10% glycerol, and 10 mg/ml BSA) containing 25 ng/μl of the target protein (without GST tag) and 25 ng/μl of the GST tagged bait protein. After 1h of binding the beads were washed with the binding buffer for 3× 10 minutes. Beads were then boiled in 1× SDS-loading buffer before SDS-PAGE analysis. Western analysis was performed using anti-GST (Abgent), anti-Flag M2 (Sigma), anti-His (Abcam), anti-4G and anti-4E (John McCarthy), anti-Pab1 (Alan Jacobson) antibodies or a polyclonal anti-Scd6 raised in rabbits to the purified protein (Cocalico Biologicals).
GRNA chromatography and RNA Analysis
Box B sequence was inserted at the 3′ UTR region of luciferase mRNA. GLH was purified using pGLH2 vector by a two step process, GST-purification followed by His purification and stored at −80 in storage buffer described above. Just before use GLH was freshly bound to glutathione sepharose beads for 1 hour followed by washing and blocking with BSA. 6 microgram of luciferase RNA was used per translation reaction. Translation was carried out for 10 minutes followed by binding to GLH-glutathione beads for 1h at 4C. Beads were washed 3x 10 minutes in binding buffer. 1/5th of the beads volume were used to recover RNA with MEGAclear (Ambion), followed by Northern blotting of agarose gels and probed for luciferase mRNA by oRP1435 (CAA TTT GGA CTT TCC GCC CTT). The rest of the beads were boiled in 1× SDS loading buffer to separate bound protein by SDS-PAGE analysis. Quantification of blots was performed using a Phosphorimager. Loading corrections were done using oRP100, an oligonucleotide directed against scR1 RNA, a stable RNA polymerase III transcript (Cao and Parker, 2001).
In vitro translation assays
Translation extract preparation and translation reactions were performed as described in Nissan et al., 2010.
Microscopy
Cells were stressed either by glucose depletion or by 0.5% Sodium Azide treatment (Buchan et al., 2011). For all experiments yeast cultures were grown to OD600 of 0.3–0.4 in the appropriate SC media. For glucose depletion, cells were collected by brief centrifugation, washed in fresh SC medium with/without 2% glucose, resuspended in fresh SC medium with/without glucose, and incubated in a flask shaking 30°C water bath for 10 min. Cells were harvested and washed once more as described above, before spotting on slides and immediate microscopic examination at room temperature. For sodium azide stress, cells were grown as above followed by addition of 0.5% sodium azide or equal volume water (mock) to the medium. Cells were allowed to grow for another 30 minutes before observing under microscope. All images were acquired using a Deltavision RT microscope system running softWoRx 3.5.1 software (Applied Precision, LLC), using an Olympus 100x, oil-immersion 1.4 NA objective. They were collected as 512 × 512 pixel files with a CoolSnapHQ camera (Photometrics) using 1 × 1 binning for yeast. All yeast images were deconvolved using standard softWoRx deconvolution algorithms (enhanced ratio, low noise filtering). ImageJ (Abramoff et al., 2004) was used to adjust all images to equal contrast ranges according to the experiment conducted or protein examined.
Supplementary Material
Highlights.
Proteins with RGG-motifs bind eIF4G and repress translation through their RGG-motifs
Scd6 forms a tricomplex with eIF4E-4G and prevents the recruitment of 43S complex
eIF4G acts as a scaffold to recruit the RGG-motif containing translation repressors
eIF4G protein functions as an integrator of mRNP fate in an mRNA life cycle
Acknowledgments
We thank Allan Jacobson, John McCarthy, Alan Hinnebusch and Christine Guthrie for their generous gifts of antibodies; Jon Lorsch and Kevin Czaplinski for plasmids; Jon Lorsch for gift of purified eIF4A. We thank all the members of Parker lab for their suggestions and support. PR would like to thank Dr. Meenal Vyas for her constant support. This work was supported by a grant to RP (NIH R37GM45443) and by funds from the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramoff M, Magelhaes P, Ram S. Image processing with ImageJ. Biophotonics Intl. 2004;11:36–42. [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10(6):430–6. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Barbee S, Estes P, Cziko A, Hillebrand J, Luedeman R, Coller J, Johnson N, Howlett I, Geng C, Ueda R, Brand A, Newbury S, Wilhelm J, Levine R, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag P, Nakamura A, Blackwell K. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- Boag P, Atalay A, Robida S, Reinke V, Blackwell K. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol. 2008;182(3):543–57. doi: 10.1083/jcb.200801183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18(7):2592–602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan R, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan R, Parker R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol Cell. 2009;36(6):932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan R, Yoon J, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci. 2011;124(Pt 2):228–39. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;23:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S, Schuldiner M, Krogan N, Weissman J. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7(7):R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Köcher T, Schelder M, Segref A, Wilm M, Mattaj I. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-related mRNA during oogenesis. Dev Cell. 2005;8(4):505–15. doi: 10.1016/j.devcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle J, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci USA. 2008;105:5821–6. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P-bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang S, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;20:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–15. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N, Wilusz J, Wilusz C. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gavin A, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick J, Michon A, Cruciat C, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gavin A, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen L, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frýdlová I, Vasicová P, Janda F, Vojtová J, Malínská K, Malcová I, Nováková L, Janosková D, Valásek L, Hasek J. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–88. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Hilliker H, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4FmRNA complex. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D, Riordan D, Gerber A, Herschlag D, Brown P. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6(10):e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes L, Campbell S, De Long S, Sachs A, Ashe M. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol Cell Biol. 2004;24:2998–3010. doi: 10.1128/MCB.24.7.2998-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja C, Izaurralde E. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev. 2011;25(18):1955–67. doi: 10.1101/gad.17136311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske M, Moritz B, Anders A, Wahle E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J. 2011;30(1):90–103. doi: 10.1038/emboj.2010.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapasi P, Chaudhuri S, Vyas K, Baus D, Komar AA, Fox PL, Merrick WC, Mazumder B. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N, Peng W, Cagney G, Robinson M, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards D, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13(2):225–39. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Krogan N, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Iizuka N, Sarnow P. Translation-competent extracts from Saccharomyces cerevisiae: effects of L-A RNA, 5′ cap, and 3′ poly(A) tail on translational efficiency of mRNAs. Methods. 1997;11:353–360. doi: 10.1006/meth.1996.0433. [DOI] [PubMed] [Google Scholar]
- Mitchell S, Walker S, Algire M, Park E, Hinnebusch A, Lorsch J. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell. 2010 Sep 24;39(6):950–62. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker P. Decapping Activators in Saccharomyces cerevisiae Act by Multiple Mechanisms. Mol Cell. 2010;39(5):773–83. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Ménade M, Poirier G, Donato D, Drouet E, Chartrand P. Local Activation of Yeast ASH1 mRNA Translation through Phosphorylation of Khd1p by the Casein Kinase Yck1p. Mol Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Richter J, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433(7025):477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Schwartz D, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Parker R. mRNA decapping in yeast requires dissociation of the can binding protein, eukaryotic translation initiation factor eIF4E. Mol Cell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal S, Dunckley T, Parker R. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:5120–30. doi: 10.1128/MCB.01913-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch A. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher K, Parker R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS One. 2010;5(4):e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem. 2006;281:40096–40106. doi: 10.1074/jbc.M609059200. [DOI] [PubMed] [Google Scholar]
- Tarun S, Jr, Sachs A. Association of the yeast poly (A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;16:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes A, Lennertz M, Beggs J, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Siddiqui N, Lapointe L, Trost M, Thibault P, Bangeranye C, Piñol-Roma S, Borden L. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009;28(8):1087–98. doi: 10.1038/emboj.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun J, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Truffault V, Hartmann M, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol. 2007;27:8600–8611. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Stary J, Wilhelm J, Newmark P. A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev. 2010;24(18):2081–92. doi: 10.1101/gad.1951010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wilhelm J, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in. Drosophila Dev Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas I, González C, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004 Dec;24(23):10479–91. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua N. Arabidopsis Decapping 5 Is Required for mRNA decapping, P-Body formation, and translational repression during postembryonic development. Plant Cell. 2009 doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Yu J, Gulick T, Bloch K, Bloch D. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–54. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Stutz F. Nuclear export of mRNA. FEBS Letters. 2001;498:150–156. doi: 10.1016/s0014-5793(01)02482-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.