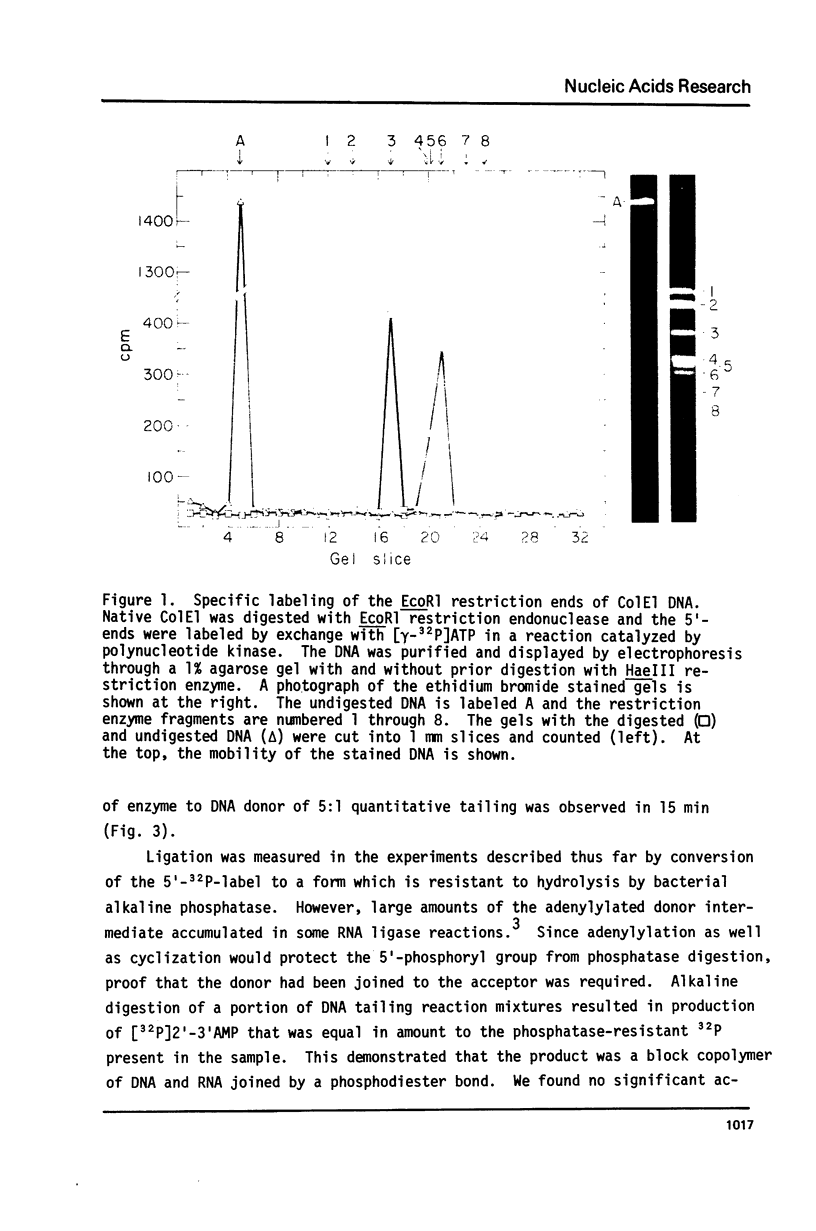
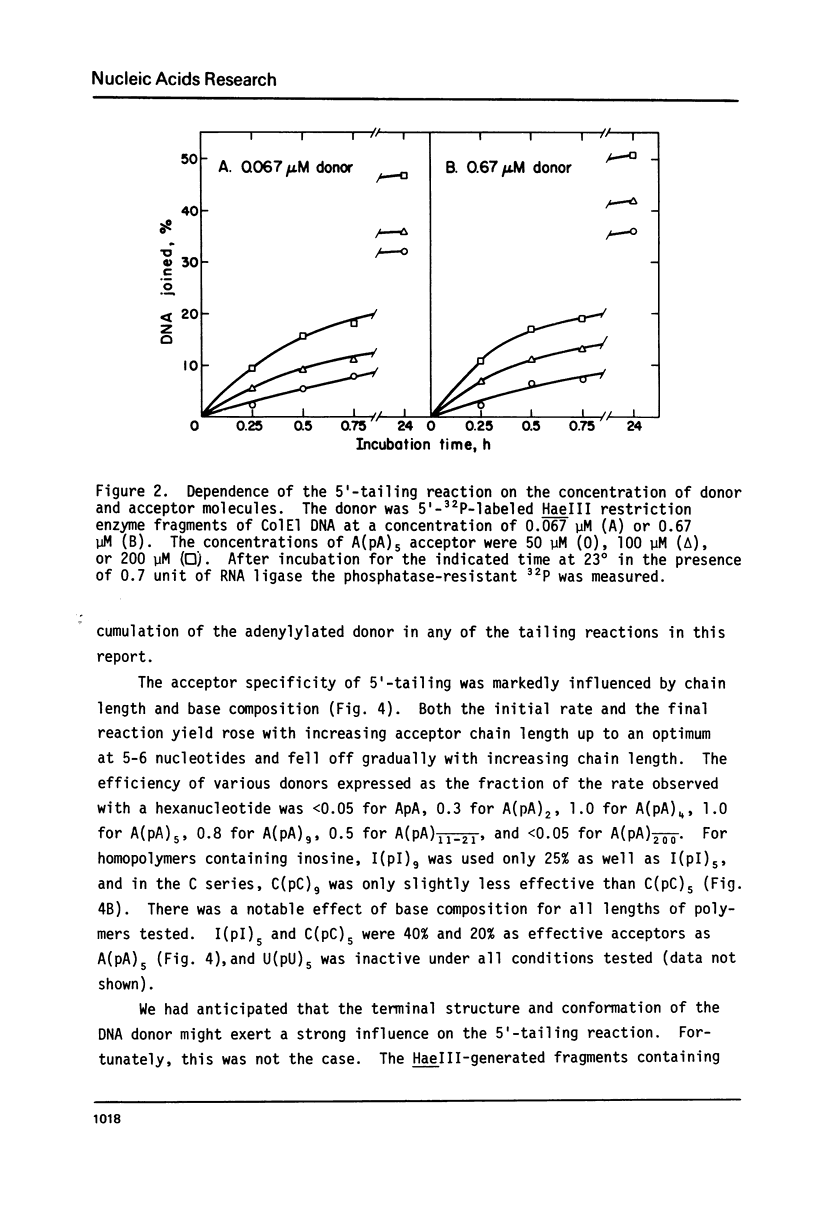
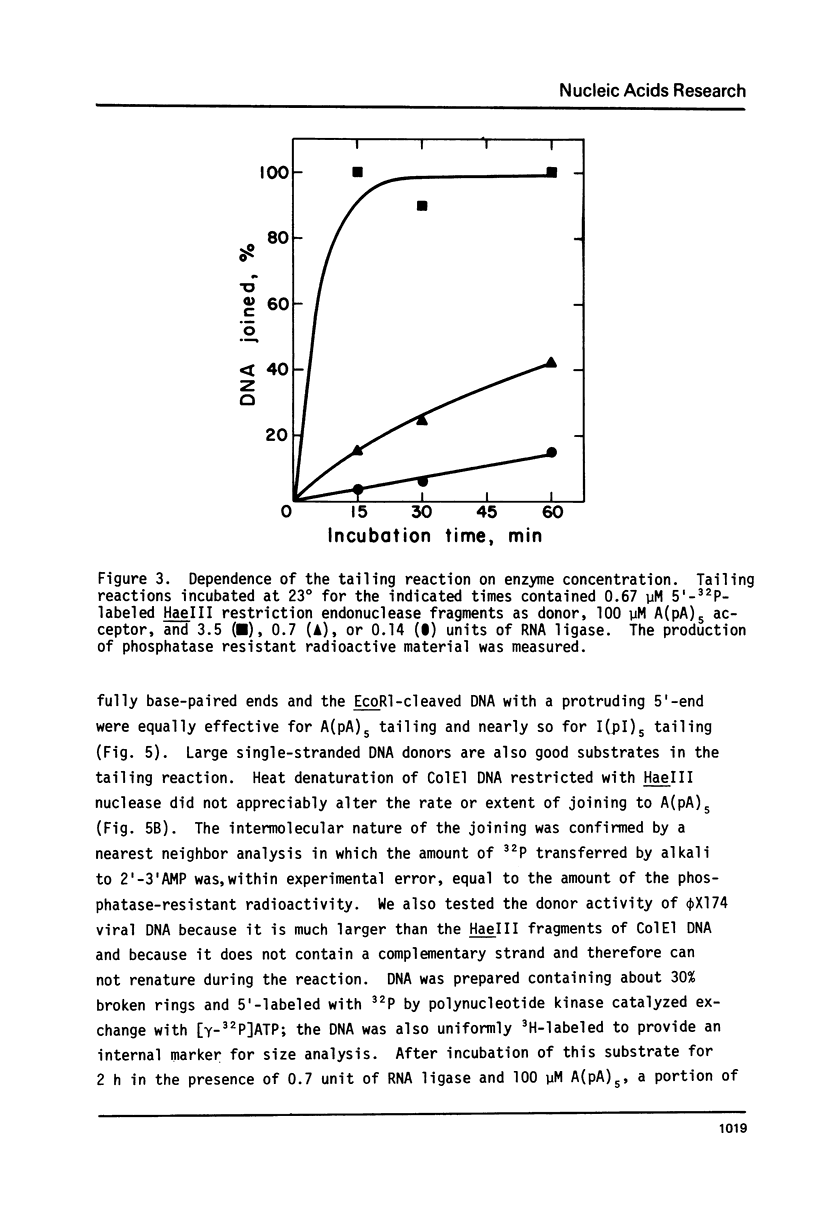
Abstract
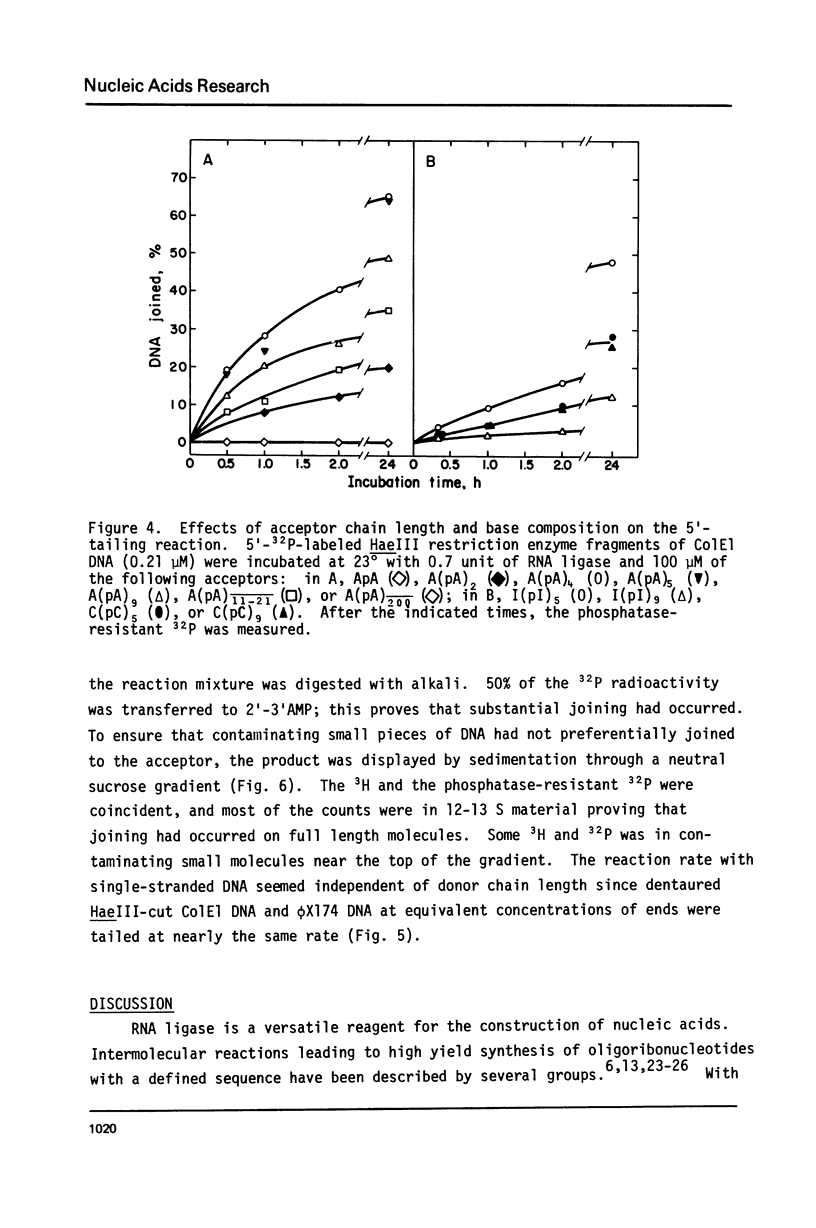
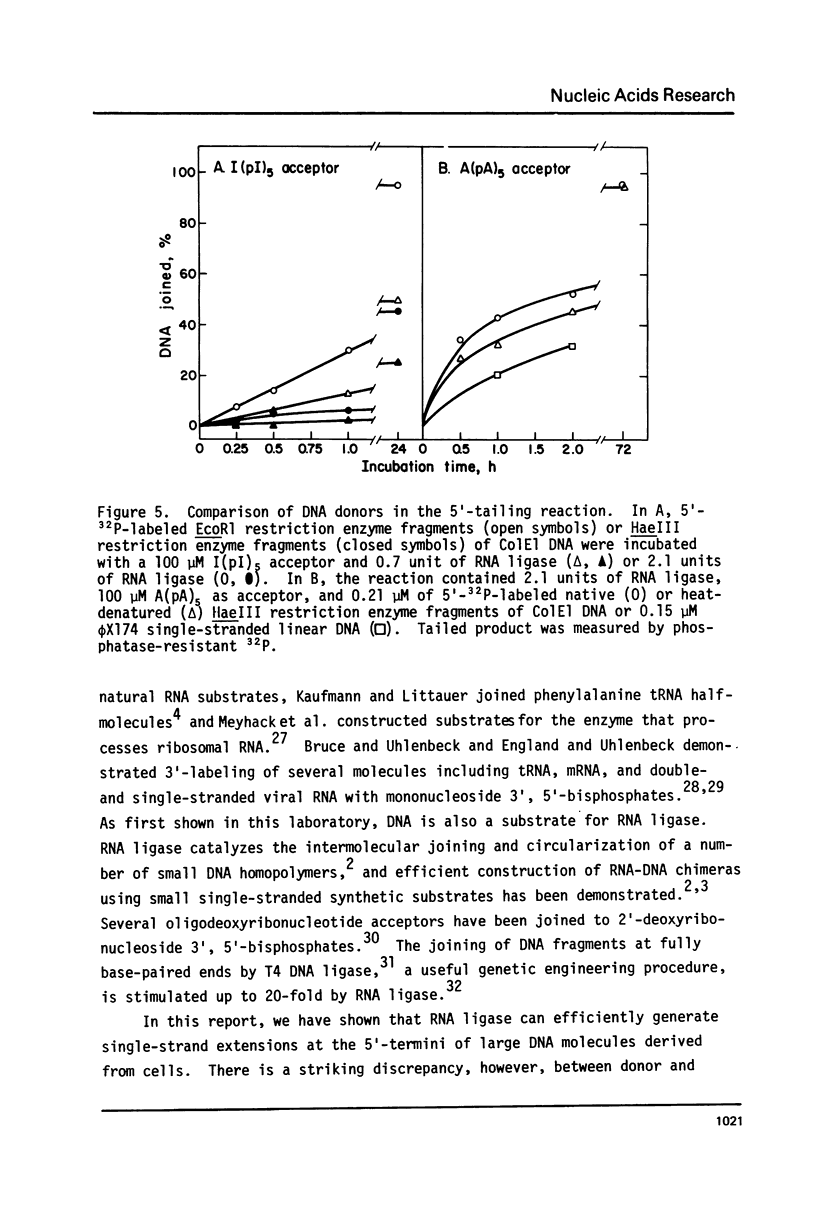
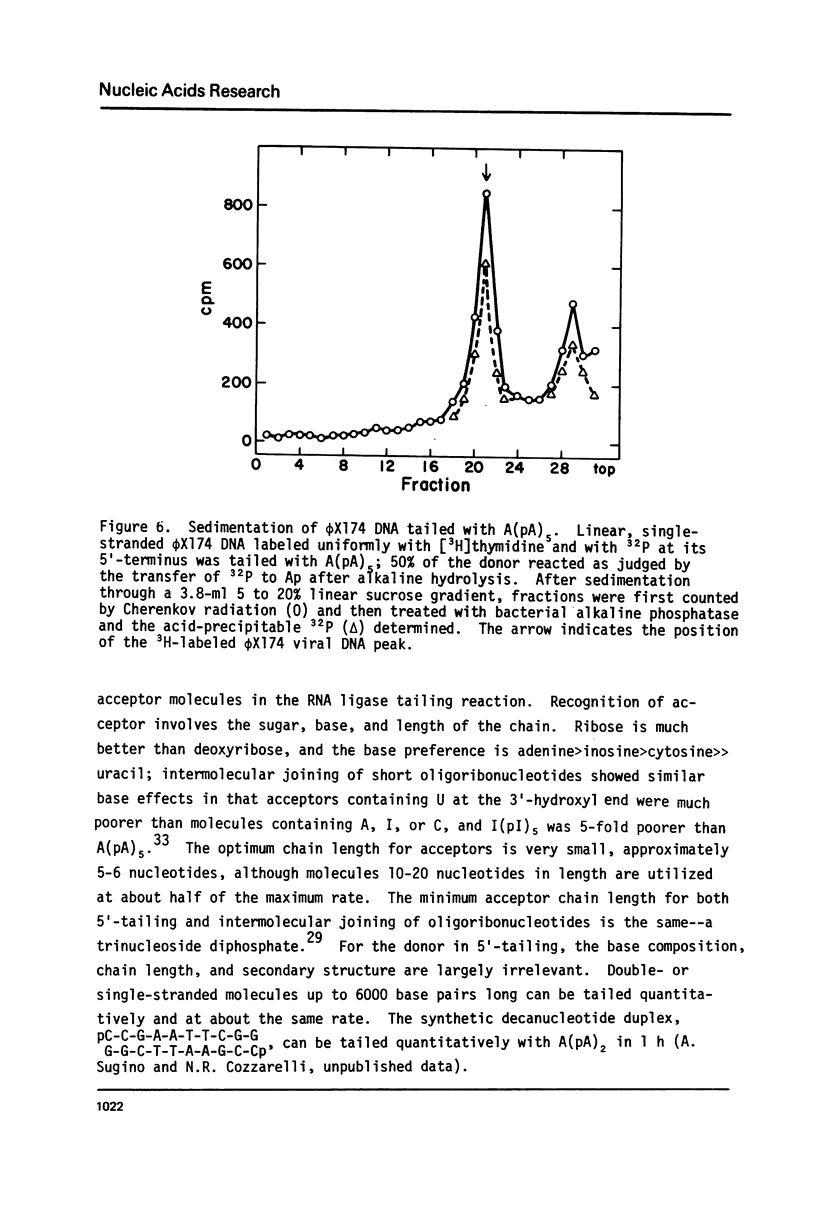
Bacteriophage T4-induced RNA ligase catalyzes the controlled template-independent addition of RNA to the 5'-phosphoryl end of large DNA molecules. Restriction enzyme-generated fragments of Co1E1 DNA with completely basepaired or cohesive ends and linear single-stranded øX174 viral DNA were all good substrates. DNA molecules from 10 to 6000 nucleotides long were quantitatively joined in an hour to a number of different RNA homopolymers. The most efficient of these was A(pA)5; I(pI)5 and C(pC)5 were also utilized while U(pU)5 was not. The optimum ribohomopolymer length was six nucleotides. Joining of ribohomopolymers between 10 and 20 nucleotides long occurred at approximately 1/2 the maximal rate and a trimer was the shortest substrate. Thus RNA ligase provides a method for generating extensions of predetermined length and base composition at the 5'-end of large DNA molecules that complements the available procedures for extending the 3'-hydroxyl terminus of DNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranston J. W., Silber R., Malathi V. G., Hurwitz J. Studies on ribonucleic acid ligase. Characterization of an adenosine triphosphate-inorganic pyrophosphate exchange reaction and demonstration of an enzyme-adenylate complex with T4 bacteriophage-induced enzyme. J Biol Chem. 1974 Dec 10;249(23):7447–7456. [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978 May 30;17(11):2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- Gillam S., Waterman K., Smith M. Enzymatic synthesis of oligonucleotides of defined sequence. Addition of short blocks of nucleotide residues to oligonucleotide primers. Nucleic Acids Res. 1975 May;2(5):613–624. doi: 10.1093/nar/2.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Geballe A. P., Snopek T. J., Sugino A., Cozzarelli N. R. Bacteriophage T4 RNA ligase: preparation of a physically homogeneous, nuclease-free enzyme from hyperproducing infected cells. Nucleic Acids Res. 1977 Sep;4(9):3175–3186. doi: 10.1093/nar/4.9.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton D. M., Baez J. A., Gumport R. I. T4 RNA Ligase joins 2'-deoxyribonucleoside 3',5'-bisphosphates to oligodeoxyribonucleotides. Biochemistry. 1978 Nov 28;17(24):5091–5097. doi: 10.1021/bi00617a004. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Kallenbach N. R. Determination of recognition sites of T4 RNA ligase on the 3'-OH and 5' -P termini of polyribonucleotide chains. Nature. 1975 Apr 3;254(5499):452–454. doi: 10.1038/254452a0. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Littauer U. Z. Covalent joining of phenylalanine transfer ribonucleic acid half-molecules by T4 RNA ligase. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3741–3745. doi: 10.1073/pnas.71.9.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Hishinuma F., Sakaguchi K. Addition of mononucleotides to oligoribonucleotide acceptors with T4 RNA ligase. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1270–1273. doi: 10.1073/pnas.75.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last J. A., Anderson W. F. Purification and properties of bacteriophage T4-induced RNA ligase. Arch Biochem Biophys. 1976 May;174(1):167–176. doi: 10.1016/0003-9861(76)90335-0. [DOI] [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Linné T., Oberg B., Philipson L. RNA ligase activity in phage-infected bacteria and animal cells. Eur J Biochem. 1974 Feb 15;42(1):157–165. doi: 10.1111/j.1432-1033.1974.tb03325.x. [DOI] [PubMed] [Google Scholar]
- Meyhack B., Pace B., Uhlenbeck O. C., Pace N. R. Use of T4 RNA ligase to construct model substrates for a ribosomal RNA maturation endonuclease. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3045–3049. doi: 10.1073/pnas.75.7.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka E., Nishikawa S., Fukumoto R., Tanaka S., Markham A. F. Joining of synthetic ribotrinucleotides with defined sequences catalyzed by T4 RNA ligase. Eur J Biochem. 1977 Dec 1;81(2):285–291. doi: 10.1111/j.1432-1033.1977.tb11950.x. [DOI] [PubMed] [Google Scholar]
- Ohtsuka E., Nishikawa S., Markham A. F., Tanaka S., Miyake T., Wakabayashi T., Ikehara M., Sugiura M. Joining of 3'-modified oligonucleotides by T4 RNA ligase. Synthesis of a heptadecanucleotide corresponding to the bases 61--77 from Escherichia coli tRNAfMet. Biochemistry. 1978 Nov 14;17(23):4894–4899. doi: 10.1021/bi00616a006. [DOI] [PubMed] [Google Scholar]
- Ohtsuka E., Nishikawa S., Sugiura M., Ikehara M. Joining of ribooligonucleotides with T4 RNA ligase and identification of the oligonucleotide-adenylate intermediate. Nucleic Acids Res. 1976 Jun;3(6):1613–1623. doi: 10.1093/nar/3.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Takanami M. Cleavage map of colicin E1 plasmid. Nature. 1976 Nov 11;264(5582):193–196. doi: 10.1038/264193a0. [DOI] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RANDERATH E., RANDERATH K. RESOLUTION OF COMPLEX NUCLEOTIDE MIXTURES BY TWO-DIMENSIONAL ANION-EXCHANGE THIN-LAYER CHROMATOGRAPHY. J Chromatogr. 1964 Oct;16:126–129. doi: 10.1016/s0021-9673(01)82446-8. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Apr;3(4):863–877. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber R., Malathi V. G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snopek T. J., Sugino A., Agarwal K. L., Cozzarelli N. R. Catalysis of DNA joining by bacteriophage T4 RNA ligase. Biochem Biophys Res Commun. 1976 Jan 26;68(2):417–424. doi: 10.1016/0006-291x(76)91161-x. [DOI] [PubMed] [Google Scholar]
- Snopek T. J., Wood W. B., Conley M. P., Chen P., Cozzarelli N. R. Bacteriophage T4 RNA ligase is gene 63 product, the protein that promotes tail fiber attachment to the baseplate. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3355–3359. doi: 10.1073/pnas.74.8.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaramella V., Khorana H. G. Studies on polynucleotides. CXVI. A further study of the T4 ligase-catalyzed joining of DNA at base-paired ends. J Mol Biol. 1972 Dec 30;72(3):493–502. doi: 10.1016/0022-2836(72)90170-2. [DOI] [PubMed] [Google Scholar]
- Stafford D. W., Bieber D. Concentration of DNA solutions by extraction with 2-butanol. Biochim Biophys Acta. 1975 Jan 6;378(1):18–21. doi: 10.1016/0005-2787(75)90132-x. [DOI] [PubMed] [Google Scholar]
- Sugino A., Goodman H. M., Heyneker H. L., Shine J., Boyer H. W., Cozzarelli N. R. Interaction of bacteriophage T4 RNA and DNA ligases in joining of duplex DNA at base-paired ends. J Biol Chem. 1977 Jun 10;252(11):3987–3994. [PubMed] [Google Scholar]
- Sugino A., Snoper T. J., Cozzarelli N. R. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J Biol Chem. 1977 Mar 10;252(5):1732–1738. [PubMed] [Google Scholar]
- Uhlenbeck O. C., Cameron V. Equimolar addition of oligoribonucleotides with T4 RNA ligase. Nucleic Acids Res. 1977 Jan;4(1):85–98. doi: 10.1093/nar/4.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C., Uhlenbeck O. C., Bedows E., Gumport R. I. T4-induced RNA ligase joins single-stranded oligoribonucleotides. Proc Natl Acad Sci U S A. 1975 Jan;72(1):122–126. doi: 10.1073/pnas.72.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Kleppe K., Khorana H. G. Reversal of bacteriophage T4 induced polynucleotide kinase action. Biochemistry. 1973 Dec 4;12(25):5050–5055. doi: 10.1021/bi00749a004. [DOI] [PubMed] [Google Scholar]