Abstract
prdm1 is an important transcriptional regulator that plays diverse roles during development of a wide variety of vertebrate and invertebrate species. prdm1 is required for neural crest specification in zebrafish, but not in mouse embryos. The role of this gene in neural crest formation in other species has not been examined, and its regulation during embryonic development is poorly understood. Here, we investigate the expression pattern, function and the upstream regulatory inputs into prdm1 during lamprey neural crest development. prdm1 is strongly expressed in the lamprey neural plate border, suggesting a conserved ancestral role of this gene in the neural crest formation. We found that lamprey neural plate border expression of prdm1 is activated by Ap-2 and Msx, but is independent of Pax3/7 and Zic.
Introduction
prdm1 (PR domain containing 1, with ZNF domain) or Blimp-1 (B lymphocyte-induced maturation protein 1) is a zinc-finger-containing transcriptional regulator that plays critical developmental roles in a wide range of species including insects, worms, echinoderms, and all vertebrates (Bikoff et al., 2009; John and Garrett-Sinha, 2009). Several studies have demonstrated that prdm1 acts by recruiting a range of epigenetic histone modifiers (histone methylases and deacetylases) in a sequence-specific manner (Ancelin et al., 2006; Gyory et al., 2004; Su et al., 2009; Yu et al., 2000). prdm1 plays important roles in cell fate decisions, by repressing large sets of genes and dramatically altering a cell’s transcriptional profile. Loss-of-function prdm1 mutants and morphants exhibit complete loss of the cell type in which the gene is expressed, further underscoring the developmental importance of this gene (Brzezinski et al.; Robertson et al., 2007; Roy and Ng, 2004; Vincent et al., 2005; Wilm and Solnica-Krezel, 2005).
The neural crest is a vertebrate-specific multipotent migratory embryonic cell population, which forms most of the peripheral nervous system, cranium and pigment cells. Much progress has been made in the recent years in understanding the genetic mechanisms underlying neural crest formation, and this information has been assembled into a proposed neural crest gene regulatory network (Sauka-Spengler and Bronner-Fraser, 2008; Betancur et al., 2010). Extensive analysis of the neural crest gene regulatory network in the lamprey, the most basal extant vertebrate, demonstrated that there is a great deal of conservation in the genetic mechanisms responsible for neural crest formation to the base of vertebrate lineage, particularly at early stages (Sauka-Spengler et al., 2007).
Data from several zebrafish mutants show that prdm1 is essential for the specification of the common progenitor of the neural crest and Rohon-Beard (RB) sensory neurons. Two prdm1 fish mutants, narrowminded and U-boot, exhibit a decrease in the number of neural crest cells, and all neural crest derived structures are much smaller than in the wild-type fish (Artinger et al., 1999; Hernandez-Lagunas et al., 2005; Roy and Ng, 2004). The null prdm1 mutant, narrowminded, also exhibits a complete loss of RB neurons (Hernandez-Lagunas et al., 2005). Interestingly, however, there are no reports on a similar function for prdm1 or any other member of the prdm family in other species. prdm1 is not expressed in the neural plate border or neural crest of mouse embryos, and no defects in the early steps of neural crest formation are seen in any prdm1 mouse knockouts (Chang et al., 2002; Robertson et al., 2007; Vincent et al., 2005). It is possible, however, that prdm1 is expressed and functions in the neural crest component of the branchial arches in the mouse at later stages. Expression of prdm1 is seen at E10.5 in the mesenchyme of the mouse branchial arches (Chang and Calame, 2002; Robertson et al, 2007). prdm1-null mice exhibit a complete loss of all branchial arches posterior to the first arch, which could be due to a cell-autonomous neural crest defect or to a failure of the neural crest migration (Robertson et al, 2007; Vincent et al, 2005). No expression of prdm1 in neural crest of chick and lamprey embryos has been reported; however it is possible that the embryonic stages examined were too late to detect it (Ha and Riddle, 2003; Hammond et al., 2009). It is not clear at present whether prdm1’s role in zebrafish neural crest is specific to the fish lineage, or if this gene has an ancestral role in the neural crest development that had been lost in the mouse. In order to address this question, we isolated a full-length sequence for prdm1 from an embryonic cDNA library of the sea lamprey Petromyzon marinus, and examined prdm1 expression pattern at a range of embryonic stages. We found strong expression of this gene in the lamprey neural plate border and premigratory neural crest, suggesting an ancestral role in neural crest development. We next investigated prdm1 loss-of function phenotype and found that it plays a role in the specification of the mesoderm, which precedes and masks any possible function in the neural plate border. Finally, we examined the upstream inputs into prdm1 in the neural plate border, and found that Msx A and AP-2 function as upstream activators of prdm1 transcription.
Results and Discussion
prdm1 expression during sea lamprey embryonic development suggests ancestral roles in the development of neural crest and branchial arches
In order to determine if prdm1 is present in the lamprey embryo, we screened the Petromyzon marinus embryonic cDNA library (Sauka-Spengler et al., 2007) using a fragment of zebrafish prdm1 containing a conserved PR ( a type of SET protein-protein interaction) domain. We found a 5kb clone encoding a 1014-amino acid protein that corresponded to the full-length lamprey prdm1 (GenBank accession number JN242000). Analysis of the conserved domains using Prosite identified an SET domain and five C2Hs zinc fingers. Comparative sequence analysis revealed high levels of sequence conservation with prdm1 proteins from other vertebrates in the SET domain and the zinc finger domains, but low levels of similarity in the rest of the protein sequence (Figure 1). Interestingly, lamprey prdm1 does not show any conservation in the region corresponding to the Groucho binding site, which is highly conserved among other vertebrates.
Figure 1. Sequence alignment of vertebrate Prdm1 protein sequences to deduced P. marinus protein sequence.
Identical amino acids are highlighted in black, amino acids with similar properties in grey. The SET domain is boxed in blue, and the five zinc fingers in red. Sequences used for alignment: human (CAQ52608.1), mouse (AAI29802.1, chicken (ENSGALP00000024778), Xenopus laevis (AAH60348.1), zebrafish (AAR87139.1)
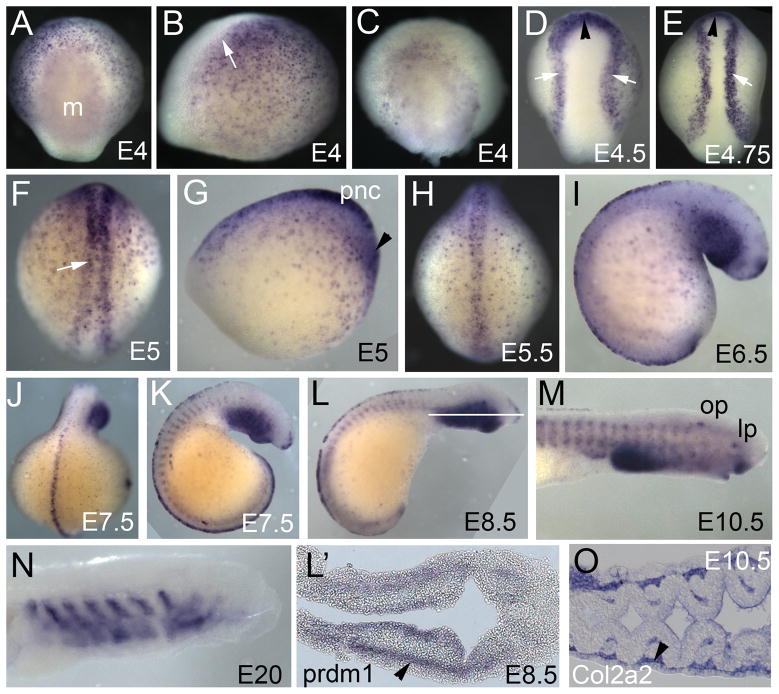
We analyzed the expression pattern of prdm1 during lamprey embryonic development starting at E4.0 to E20. At E4.0 (early neurula), prdm1 is expressed throughout the ectoderm with the exception of the neural plate, with particularly high expression at the neural plate border and anteriorly in the preplacodal domain (Figure 2A, B). prdm1 transcripts are also present at relatively low levels in the anterior endomesoderm and paraxial mesoderm (Figure 2C). As neurulation progresses, prdm1 expression in the ectoderm decreases, while that in the neural plate border and the preplacodal domain becomes enhanced (Figure 2D–H). Expression of prdm1 is particularly prominent in the premigratory neural crest (Figure 2G), but is turned off as soon as the neural crest starts migrating (compare Figure 2G to 2I). By E6.5, prdm1 expression remains only in the posterior dorsal neural tube. Additionally, it is expressed in the branchial arches (Figure 2I–M). This expression persists until E14 in the posterior arches. Comparison of the expression pattern of prdm1 to that of Col2a2 (which marks the neural crest, Figure 2O) suggests that prdm1 is expressed in the endodermal component and a subset of the neural-crest-derived component of the branchial arches (lateral neural crest). At later stages (E20) prdm1 expression is seen in the branchial arches, which by that time have become cartilaginous (Figure 2N) (Cerny et al, 2010). Expression in the somites is initiated at E7 and continues until E18 (Figure 2J, L, M), as described previously by Hammond et al (2009).
Figure 2. Expression pattern of prdm1 in sea lamprey embryos.
(A–N) whole mount in situ hybridization with the full-length lamprey prdm1 probe. Orientation: anterior is at the top in dorsal views, or in the top right-hand corner in side views. (L′, O) 12 micron sections through the stained embryos. (A) Dorsal view of an E4.0 embryo, showing prdm1 transcripts expressed uniformly throughout the ectoderm and the neural plate border, but absent from the neural plate. (B) Side view of the same embryo. (C) Dorsal view of the embryo in (A) with its ectoderm removed, demonstrating that prdm1 is also expressed in the paraxial mesoderm at this stage. (D and E) Starting at E4.5 (D), high levels of prdm1 are expressed in the neural plate border (white arrows) and in the preplacodal domain (black arrowhead), and lower levels in the ectoderm. (E) A similar pattern of prdm1 expression is seen 6 hours later at E4.75. (F) Dorsal and (G) side view of the same E5.0 embryo, showing strong expression in the premigratory neural crest (white arrow). (H) Embryo at E5.5. prdm1 is turned off in the neural crest as it starts to migrate. (I) Lateral view of an E6.5 embryo. prdm1 transcripts are no longer seen in the anterior dorsal neural tube. (J) Dorsal and (K) side view of an E7.5 embryo. prdm1 is expressed in the trunk dorsal neural tube, in the branchial basket and in somites. (L) E8.5 embryo showing prdm1 expression in the branchial arches. (M) Side view of the head of an E10.5 embryo, showing prdm1 expression in the lips and lens (lp) and otic placodes (op). (N) Side view of E20 embryo, showing prdm1 expression in the cartilage of the branchial arches. (L′) Transverse section through the head of the E8.5 embryo in (L) showing prdm1 expression in the endoderm and the lateral region of the neural crest (black arrowhead, compare to panel O). (O) Section through an E10.5 embryo stained for Col2a2, which at that stage is expressed in the neural crest components of the branchial arches. Black arrow marks the lateral component of the neural crest that corresponds to the prdm1 expressing cells in (L′).
Abbreviations: m-paraxial mesoderm, pnc – premigratory neural crest, op-otic placode, lp-lens placode. White arrows indicate neural plate border, black arrowheads – preplacodal domain.
In the lamprey P. marinus prdm1 is expressed in the neural plate border as early as mid-gastrula (E4), and persists anteriorly until the start of neural crest migration (E5.5). It is maintained in the posterior dorsal neural tube until E9. This expression pattern is consistent with what has been reported for zebrafish (Hernandez-Lagunas et al., 2005; Roy and Ng, 2004), and suggests a likely ancestral role for this gene in the neural crest specification.
Expression pattern of prdm1 in other embryonic structures is consistent with what was reported in other vertebrates (Chang et al., 2002; Ha and Riddle, 2003).
Expression of prdm1 in the neural plate border is downstream of MsxA and AP-2
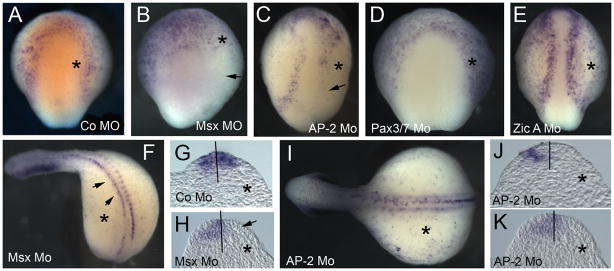
Very little is known about regulation of prdm1 expression during embryonic development, except that its expression in the neural plate border is downstream of Bmp signaling (Hernandez-Lagunas et al., 2005; Roy and Ng, 2004). To address interactions with potentially upstream genes, we investigated the effect of morpholino-mediated loss of function of four genes important for the neural plate border formation on prdm1 expression. Injection of MsxA morpholino in lamprey resulted in a complete loss of prdm1 transcripts from the ectoderm and the neural plate border on the injected side (73%, n=33) (Figure 3B). When these embryos were allowed to develop to later stages, we observed loss of prdm1 in the somites and dorsal neural tube on the injected side (Figure 3F, H) (47% at E8.5, n=17), suggesting that MsxA also regulates prdm1 in these tissues. Similarly, loss of AP-2 resulted in the absence of prdm1 expression from both the ectoderm and the neural plate on the injected side at E4.5 (74%, n=55) (Figure 3C). Similarly to the phenotype of MsxA morphants, loss of prdm1 expression in the dorsal neural tube was observed in AP-2-morpholino-injected embryos at E7 (72%, n=18) (Figure 3I–K). However, the expression of prdm1 in the somites was not affected (Figure 3I). In contrast to MsxA and AP-2, neither Pax3/7 (n=24) nor Zic morpholino (n=28) affected prdm1 expression (Figure 3D and E), suggesting that these two neural plate border specifiers are either downstream of prdm1 or act in a parallel pathway.
Figure 3. Expression of prdm1 in the neural plate border is regulated by AP-2 and Msx A.
prdm1 is detected by in situ hybridization in morpholino-injected embryos. Top row: E4.5 embryos injected with control (A) Msx A (B), AP-2 (C), Pax3/7 (D) and Zic A (E) morpholinos. All embryos are shown in dorsal view with anterior facing up. In all embryos, morpholino was incorporated into the right side (marked with an asterisk). Arrows in (B) and (C) indicate loss of prdm1 expression on the injected side. (F) MsxA-morpholino-injected embryo at E8.5, showing loss of prdm1 expression in somites and reduction of expression in half of the neural tube (arrowheads). (H) Section through the embryo in (F) showing loss of prdm1 expression in one half of the neural tube (arrow). (G) Section through an E8.5 embryo injected with the control morpholino. (I) E7.5 embryo injected with AP-2 morpholino, showing that prdm1 expression in the somites in not affected (asterisk marks the injected side). (J, K) Sections through the embryo in panel (I), showing loss of prdm1 expression on the injected side (asterisk).
In zebrafish double tfap2a/c morphant both prdm1 expression in the neural plate border and Rohon-Beard neuron formation was reduced, but not completely absent (Li and Cornell, 2007). This data suggests that, consistent with what we observed in the lamprey, prdm1 expression in at least a subset of the neural plate border-derived cells is downstream of the two zebrafish AP-2 homologues. Similarly, in triple MsxB, C, E morphants of zebrafish, a loss of a subset, but not all, Rohon-Beard neurons (a cell type dependent on prdm1 expression) is seen (Phillips et al., 2006). Taken together, these results suggest that there is a conservation of the upstream inputs into prdm1 between lamprey and zebrafish in a subset of the neural plate border cells. It is possible that those Rohon-Beard neurons that are still formed in zebrafish in the absence of tfap2a/c or Msx B, C, E (and the corresponding tfap2a/c and MsxB, C, E- independent prdm1 expression domain) is a teleost innovation. prdm1 regulation in these cells remain to be further explored.
In summary, our experiments demonstrate that MsxA and AP-2 are essential for prdm1 expression in the neural plate border. Two other neural plate border specifiers, Pax3/7 and ZicA, have no effect on prdm1 expression, suggesting that they act in a parallel pathway. Figure 5 shows a schematic representation of the current understanding of prdm1 regulation in the neural plate border.
Figure 5.
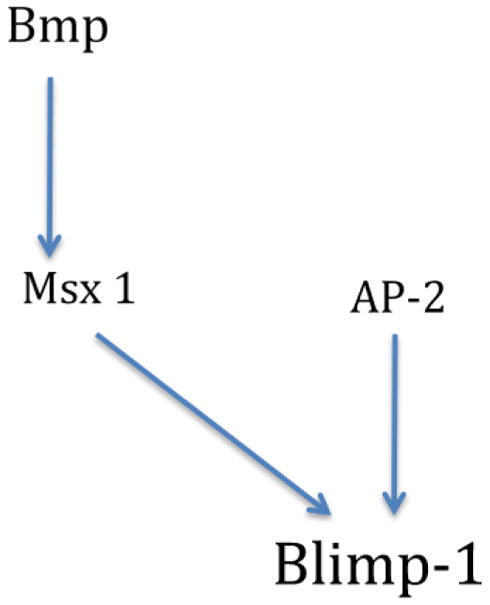
Schematic representation of prdm1 upstream regulatory inputs in the neural plate border.
Loss of prdm1 results in abnormal axis formation
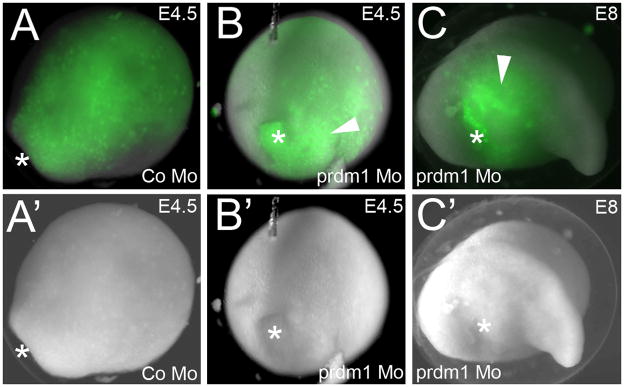
Strong and specific expression of prdm1 in the neural plate border and premigratory neural crest suggests that the function of this gene is important for the formation of the neural crest in lamprey. In order to examine the role this gene plays in the neural crest development, we designed a translation-blocking morpholino against prdm1 and injected it into lamprey embryos at the 4-cell stage. To confirm the specificity of prdm1 morpholino, we used both a standard control morpholino (Sauka-Spengler et al, 2007; Nikitina et al, 2008) and a prdm1 morpholino with 5 base pair mismatches. Neither of these morpholinos affected the phenotype of the injected embryos. Surprisingly, we found that none of about 300 embryos that received prdm1 morpholino incorporated it into the ectoderm, compared to ectodermal distribution in 25% of the control morpholino-injected embryos, as expected (compare Figure 4B and 4A). Since prdm1 is also expressed in the prospective anterior endomesoderm, we speculated that its loss might interfere with mesoderm formation. In an attempt to target the morpholino more specifically to the neural plate border, we injected it into one out of 16 cells at later cleavage stages. This resulted in progressively smaller regions of mesoderm and endoderm containing the morpholino, and never any ectodermal incorporation. A possible explanation for this result is that prdm1 plays a crucial role in the specification of embryonic layers during and before gastrulation. Continuous prdm1 expression may maintain ectodermal fate, while transient expression of low levels of prdm1 may instruct the cells to become paraxial mesoderm. Consistent with this possibility, embryos that received prdm1 morpholino also exhibited abnormal gastrulation and incorrect positioning of the anterior-posterior axis. Normally, the posterior end of the neural plate is formed just on top of the blastopore, and the head on the other end. In these embryos, we observed that the axis was perpendicular to its normal pattern: i.e. the head and the tail were formed on each side of the blastopore, which in some cases failed to close (asterisk in Figure 4C and 4C′).
Figure 4. prdm1 Mo never incorporates in ectoderm.
(A–C) Fluorescent images showing incorporation of FITC-labeled morpholino (green). (A′–C′) Bright field images of the same embryos. (A, A′) E4.5 embryo injected with a control morpholino, showing incorporation of the morpholino throughout the dorsal ectoderm. The embryo is shown in dorsal view, with the anterior in the top right-hand corner. Asterisk denotes the position of the blastopore. (B, B′) E4.5 embryo injected with prdm1 morpholino. Morpholino is incorporated in the mesoderm and endoderm (white arrowhead). The embryo is shown with the dorsal side facing up, and the posterior (blastopore, marked with an asterisk) facing towards the viewer. (C, C′) prdm1 morpholino-injected embryo at E8. Orientation: the head is facing right, and the dorsal side is facing up. Asterisk denotes the abnormal position of the blastopore.
Thus, we observed prdm1 morpholino incorporation exclusively in endoderm and mesoderm, and never in the ectoderm, despite the large numbers of the embryos injected. Data from prdm1 overexpression experiments in Xenopus lends some support to the possibility that prdm1 expression might maintain ectodermal fate (de Souza et al., 1999), whereas transient low levels of prdm1 expression may promote mesodermal fate. Injection of prdm1 RNA into Xenopus eggs results in inhibition of mesoderm formation and repression of mesodermal markers. Also prdm1 overexpression in the animal cap explants induces endodermal markers (de Souza et al., 1999). In zebrafish, prdm1 overexpression results in the loss of anterior-dorsal structures, while MO-mediated loss of function causes an increase in dorsal extension movements and mild dorsalization (Wilm and Solnica-Krezel, 2005).
Experimental Procedures
Library screen
A directional high quality full-length cDNA library prepared from embryonic day 2–12 lampreys using Superscript Plasmid System from Invitrogen (Carlsbad, CA) (Sauka-Spengler et al., 2007) was screened at medium stringency using heterospecific probes per standard methods. The probes were synthesized using Danio rerio prdm1 (Gene Accession Number NM_199515.2) as a template in GC Rich PCR from Roche (Indianapolis, IN). Primers were synthesized by Integrated DNA Technologies (San Diego, CA) with sequences as follows:
-
A2
Forward: 5′-CTCAACTACCCTGCCTCTGG-3′
Reverse: 5′-GAGTAGTGCGGGTTTGGGTA-3′
-
B2
Forward: 5′-ATCGTGGCCTGAACCACTAC-3′
Reverse: 5′-CCTGTGGAACCTCGTTTCAT-3′
-
C2
Forward: 5′-GCAGAAAATGTCCCCAAAGA -3′
Reverse: 5′-GACTTCTCTCAGGGCTGCTG-3′
The radioactive probe was synthesized from the PCR product with P-32-labeled dATP using Amersham Rediprime II (GE Healthcare). The radioactive probe was hybridized at 52°C and washed at 42°C until radioactivity of the screen no longer decreased. Phosphor cassettes were exposed with the radioactive screen for 24 hours, then scanned and read using the BioArray program (The Biocomputation Group, Philadelphia, PA). Positive clones were selected, amplified, digested, and sequenced using SP6 and T7 promoter primers. Returned sequences were used in BLAST searches (more dissimilar sequences, discontinuous megablast) against the NCBI nucleotide collection. Sequences that returned with matches to prdm1 homologs in other species were sequenced to full length.
Lamprey husbandry and embryo culture
Adult lampreys (Petromyzon marinus) were obtained from Hammond Bay Biological Station, Millersburg, MI, USA. The animals were housed in a Caltech lamprey facility as previously described (Nikitina et al., 2009a). They were matured by expanding their daylight cycle and raising the temperature by 1°C per day until 18°C was reached. Eggs were harvested from gravid females, and fertilized in 100–200ml of 18°C spring water (Sparkletts). The eggs were washed after 15 minutes to prevent hyperspermy. After 6 hours when the embryos had undergone the first division, they were transferred to 0.1X MMR (Marc’s Modified Ringer’s solution) media, which was then replaced every day to prevent fungal or other infections. Embryo fixation was done with MEMFA (4% formaldehyde, 0.1M MOPS (pH 7.4), 1 mM MgSO4, 2 mM EGTA), dehydrated, and stored in MeOH at −20°C.
Morpholino injection
FITC-labeled translation blocking morpholinos against lamprey Pax3/7, ZicA, MsxA, and AP-2 were obtained from Gene Tools (Philomath, OR, USA). Their sequences were as follows (Sauka-Spengler et al, 2007; Nikitina et al, 2008):
Pax3/7 MO: 5′-TGTCCTGGTGCCGGGCGCATCATCC-3′; Msx MO: 5′-
GACCGCGAAGCGAAATGCGTTCATG-3′; ZicA Mo: 5′-
CGCCTCCAGAAGCATCGCGTGCGGT-3′; AP2 Mo: 5′-
CCTGTAATTTCAAAAGCATGACTCC-3′; control Mo: 5′-
CCTCTTACCTCAGTTACAATTTATA-3′
FITC-labeled translation blocking morpholino targeting lamprey Blimp-1 (5′-ATCACCATGCGCCCTCACCGCTGTT-3′) was injected into a single blastomere at 4-, 8-, 16- and 32-cell stage, and the embryos were cultured and collected as previously described (Nikitina et al., 2009; McCauley and Bronner-Fraser, 2006). A 5-nucleotide mismatch morpholino (5′-ATCtCCAcGCGCCCTCAtCGCaGTT-3′) was used as a negative control.
In situ hybridization
In situ hybridization was performed as described (Nikitina et al., 2009c). Following color development, the embryos were transferred to 7.5% gelatin/15% sucrose/PBS, then mounted in 20% gelatin/PBS and frozen in liquid nitrogen. Cryosections (10–12μm) were collected on Super Frost Plus slides (Fischer Scientific, Pittsburg, PA).
Conclusion
The expression pattern of prdm1 in the lamprey P marinus suggests that it may play an evolutionarily conserved role in early events of neural crest specification. We show that prdm1 expression in the neural plate border is turned on by AP-2 and Msx1, but is independent of Pax3/7 and Zic. In addition, loss-of-function experiments suggest an early role in anterior endomesoderm specification.
References
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–30. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126:3969–79. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. AnnuRev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikoff EK, Morgan MA, Robertson EJ. An expanding job description for Prdm1/PRDM1. Curr Opin Genet Dev. 2009;19:379–85. doi: 10.1016/j.gde.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Brzezinski JAt, Lamba DA, Reh TA. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 137:619–29. doi: 10.1242/dev.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DH, Cattoretti G, Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Prdm1) during mouse embryonic development. Mech Dev. 2002;117:305–9. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Cerny R, Cattell M, Sauka-Spengler T, Bronner-Fraser M, Yu F, Medeiros DM. Evidence for the prepattern/cooption model of vertebrate jaw evolution. Proc NatlAcad Sci U S A. 2010;107(40):17262–7. doi: 10.1073/pnas.1009304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza FS, Gawantka V, Gomez AP, Delius H, Ang SL, Niehrs C. The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann’s organizer. EMBO J. 1999;18:6062–72. doi: 10.1093/emboj/18.21.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- Ha AS, Riddle RD. cPrdm1 expression in chick limb bud development. Gene Expr Patterns. 2003;3:297–300. doi: 10.1016/s1567-133x(03)00042-5. [DOI] [PubMed] [Google Scholar]
- Hammond KL, Baxendale S, McCauley DW, Ingham PW, Whitfield TT. Expression of patched, prdm1 and engrailed in the lamprey somite reveals conserved responses to Hedgehog signaling. Evol Dev. 2009;11:27–40. doi: 10.1111/j.1525-142X.2008.00300.x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Lagunas L, Choi IF, Kaji T, Simpson P, Hershey C, Zhou Y, Zon L, Mercola M, Artinger KB. Zebrafish narrowminded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev Biol. 2005;278:347–57. doi: 10.1016/j.ydbio.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John SA, Garrett-Sinha LA. Blimp1: a conserved transcriptional repressor critical for differentiation of many tissues. Exp Cell Res. 2009;315:1077–84. doi: 10.1016/j.yexcr.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Li W, Cornell RA. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivativesin zebrafish embryos. Dev Biol. 2007;304:338–54. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley DW, Bronner-Fraser M. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature. 2006;441(7094):750–2. doi: 10.1038/nature04691. [DOI] [PubMed] [Google Scholar]
- Nikitina N, Bronner-Fraser M, Sauka-Spengler T. Culturing lamprey embryos. CSH Protoc. 2009a;2009:pdb prot5122. doi: 10.1101/pdb.prot5122. [DOI] [PubMed] [Google Scholar]
- Nikitina N, Bronner-Fraser M, Sauka-Spengler T. Microinjection of RNA and morpholino oligos into lamprey embryos. CSH Protoc. 2009b;2009:pdb prot5123. doi: 10.1101/pdb.prot5123. [DOI] [PubMed] [Google Scholar]
- Nikitina N, Bronner-Fraser M, Sauka-Spengler T. Whole-mount in situ hybridization on lamprey embryos. CSH Protoc. 2009c;2009:pdb prot5125. doi: 10.1101/pdb.prot5125. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Kwon HJ, Melton C, Houghtaling P, Fritz A, Riley BB. Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border andregulate cranial placodes and neural crest development. Dev Biol. 2006;294:376–90. doi: 10.1016/j.ydbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Charatsi I, Joyner CJ, Koonce CH, Morgan M, Islam A, Paterson C, Lejsek E, Arnold SJ, Kallies A, Nutt SL, Bikoff EK. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development. 2007;134:4335–45. doi: 10.1242/dev.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ng T. Prdm1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr Biol. 2004;14:1772–7. doi: 10.1016/j.cub.2004.09.046. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Barembaum M. Gain-and loss-of-function approaches in the chick embryo. Methods Cell Biol. 2008;87:237–56. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–68. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–20. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Prdm1-mediated gene repression during plasma cell differentiation. Mol Cell Biol. 2009;29:1421–31. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–25. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Wilm TP, Solnica-Krezel L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development. 2005;132:393–404. doi: 10.1242/dev.01572. [DOI] [PubMed] [Google Scholar]
- Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by prdm1(PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20:2592–603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]