Abstract
Background
Regulatory T cells (Tregs) play an essential role in the control of the immune response. Treg cells represent important targets for therapeutic interventions of the immune system. Therefore, it will be very important to understand in more detail which genes are specifically activated in Treg cells versus T helper (Th) cells, and which gene regulatory circuits may be involved in specifying and maintaining Treg cell homeostasis.
Results
We isolated Treg and Th cells from a genetically diverse family of 31 BXD type recombinant inbred strains and the fully inbred parental strains of this family--C57BL/6J and DBA/2J. Subsequently genome-wide gene expression studies were performed from the isolated Treg and Th cells. A comparative analysis of the transcriptomes of these cell populations allowed us to identify many novel differentially expressed genes. Analysis of cis- and trans-expression Quantitative Trait Loci (eQTLs) highlighted common and unique regulatory mechanisms that are active in the two cell types. Trans-eQTL regions were found for the Treg functional genes Nrp1, Stat3 and Ikzf4. Analyses of the respective QTL intervals suggested several candidate genes that may be involved in regulating these genes in Treg cells. Similarly, possible candidate genes were found which may regulate the expression of F2rl1, Ctla4, Klrb1f. In addition, we identified a focused group of candidate genes that may be important for the maintenance of self-tolerance and the prevention of allergy.
Conclusions
Variation of expression across the strains allowed us to find many novel gene-interaction networks in both T cell subsets. In addition, these two data sets enabled us to identify many differentially expressed genes and to nominate candidate genes that may have important functions for the maintenance of self-tolerance and the prevention of allergy.
Background
Regulatory T cells (Tregs) are key modulators of immune responses in mice and humans and represent key candidates for therapeutic interventions of a broad variety of immunological diseases [1]. While reduction or functional inactivation of Tregs would be beneficial for restoration of anti-tumor immunity, selective expansion of Tregs is a promising approach for preventing autoimmunity, allergy and organ graft rejection in the transplantation setting. Initially being described as thymus-derived CD25+ subpopulation within the naïve CD4+ T-helper cell (Th) pool [2], during the last decade extensive gene expression studies based on the comparison of CD25+CD4+ Tregs and CD25-CD4+ T helper cells (Th) revealed a considerable number of additional genes critically involved in Treg development and function [3-9]. Among those, the transcription factor FOXP3 was identified as master-regulator of the Treg lineage [10-12].
Defects in the Foxp3 gene function in humans and mice result in fatal autoimmunity, and Foxp3 over-expression in previously naïve T cells converts them to Treg-like cells with in vivo and in vitro suppressive function. Despite increasing knowledge regarding the molecular signature of Tregs and mechanisms underlying their suppressive function, the extent to which Treg development and function are genetically controlled has not been studied to date.
To better understand gene variants that underlie disease predispositions related to Treg functions and to identify regulatory networks related to both Treg and Th cells, we undertook a systems genetics analysis of gene expression in these cell types using a genetic reference panel consisting of 31 members of the large BXD family of recombinant inbred strains [13,14]. Genetic reference panels (GRPs) such as the BXD family, are sets of strains that have a defined and fixed genetic architecture that can be used in classic linkage studies and complex trait analysis. The BXD family is one of the largest GRP, consisting of ~150 lines of which 80 are now fully inbred that all trace their descent from F2 progeny of crosses between C57BL/6J (B) and DBA/2J (D). Individuals within a single BXD strain are nearly isogenic (except for the sex chromosomes) and genotypes for the entire family of strains are known and stable [15]. The high level of genetic variation among BXD strains can be exploited to systematically study the genetic control of gene expression even at the level of single cell types [16] and even higher order genotype-to-phenotype relations, including for example global analysis of disease susceptibility [17-19].
Recently, whole-genome transcriptome data have been collected from GRPs. The expression level of a given transcript in a cell type or tissue may be then treated as a quantitative trait, and by employing standard linkage analyses so-called expression quantitative trait loci (eQTLs) can be identified. This links differences in expression to specific chromosomal intervals [16,20-22]. eQTL analysis can be used to identify regulatory interactions and to analyze specific effects of treatment or cell type on transcriptional control [23-27]. In addition, the analysis of eQTLs in chromosomal regions linked to disease susceptibility can help identify key candidate genes [23,28,29].
Here, we report the whole transcriptome analysis of steady-state Terg and Th cells that were isolated from 33 members of the BXD family, including the parental strains. Variation of expression across the strains allowed us to identify many novel gene-interaction networks in both T cell subsets. In addition, these two data sets enable us to nominate candidate genes that may have important functions for the maintenance of self-tolerance and the prevention of allergy.
Results
CD4+CD25+ T regulatory (Treg) cells and CD4+CD25-T helper (Th) cells were isolated from the spleens of 31 BXD strains and the parental strains DBA/2J and C57BL/6J by fluorescence activated cell sorting. Subsequently, whole genome transcriptome analysis was performed separately for each BXD strain and for Treg and Th cells.
The gene expression data has subsequently been used for two types of analysis. (1) A direct comparison of all genes that were expressed in Treg versus Th detected was performed from all samples together to identify differentially expressed genes in either cell type. Here, the variation in expression patterns between strains was not taken into account. These studies allowed us to identify a large number of novel, Treg-specific genes. (2) Differences in gene expression patterns between strains were used to determine eQTLs in Treg and Th cells, separately. This analysis allowed us to identify cis- and trans-eQTLs, potential gene regulatory interactions and candidate genes involved in autoimmune disease.
Analysis of differentially expressed genes reveals known and novel gene candidates important for Treg and Th functions
The large data set - for each cell type a total of 33 replicates has been generated - provided a high statistical power to identify genes that were differentially expressed in Treg versus Th cells. We identified differentially expressed genes by employing a statistical test on the difference between the two T cell populations, setting the threshold for the p-value to < 0.001 and using an expression difference of at least two-fold. In the following, probesets that exhibited a statistically significant and at least two-fold difference in expression between Treg and Th cells are referred to as 'DE-2fold genes'. In this way, a total of 14,117 probesets was found that exhibited a significantly higher expression level in Treg compared to Th cells (additional file 1, Table S1). 608 probesets were found from transcripts that were at least two-fold higher expressed in Treg cells (DE-2fold genes), including 413 annotated genes. On the other hand, using the same criteria, for Th expressed genes, a total of 11.944 probesets was found significantly higher expressed in Th compared to Treg cells, and 823 probesets, containing 481 annotated genes, showed a two-fold or higher expression level (DE-2fold genes) in Th cells (additional file 2, Table S2).
A total of 15 genes with known Treg functions (see additional file 3, Table S3) were found amongst the differentially expressed genes (additional file 3, Table S4), including well known Treg functional genes like Foxp3 (forkhead box P3), Nrp1 (neuropilin 1), Itgae/αEβ7 (integrin alpha E, epithelial-associated), Ctla4 (cytotoxic T-lymphocyte-associated protein 4), Ikzf4/Eos (IKAROS family zinc finger 4). Il2ra/CD25 (interleukin 2 receptor, alpha chain) was found to be highly expressed in Treg compared to Th cells (25-fold higher) which is expected, because the resting Treg subset was isolated on the basis of CD25 surface expression. Also, Foxp3, representing the most important Treg functional gene in mice, was found to be expressed 43-fold higher in Treg cells. GO term analyses for the DE genes revealed substantial differences between Tregs and Th. Whereas lymphocyte activation was indicated for Treg genes, antigen processing and presentation was strongly associated to Th genes (data not shown).
These results confirmed successful separation of highly pure Treg and Th subsets and corroborates the value of our data for subsequent analyses. Furthermore, the comparison of Treg and Th gene expression data from a large sample size allowed us to identify a large number of genes that are differentially expressed between Treg and Th cells and may play a role in the differentiation and maintenance or the function of the respective cell populations.
Global analyses of eQTL mapping reveals different eQTLs between Treg and Th cell populations
The analysis of gene expression patterns across 31 BXD and the two parental strains allowed determining eQTLs as quantitative traits by comparing expression values for each gene and associating them with markers along the genome in all strains.
In principle, two types of eQTLs can be distinguished. If an eQTL is located at the same genomic position as the gene itself (within an operationally defined 10 Mb interval of the gene), it is considered as a cis-eQTL. In this case, variations in the promoter sequence or in regions that determine the stability of the mRNA of the gene are the most likely causes for the observed differences. If the eQTL is at a distant location (operationally defined as a distance exceeding 10 Mb) from the regulated gene, it is referred to as a trans-eQTL. For example, a transcription factor may be located in the eQTL interval which regulates the expression of the target gene. In this way, many gene regulatory regions but also gene-gene interactions and networks may be identified.
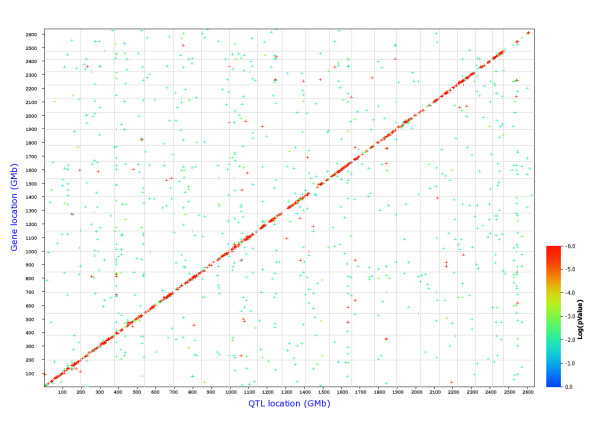
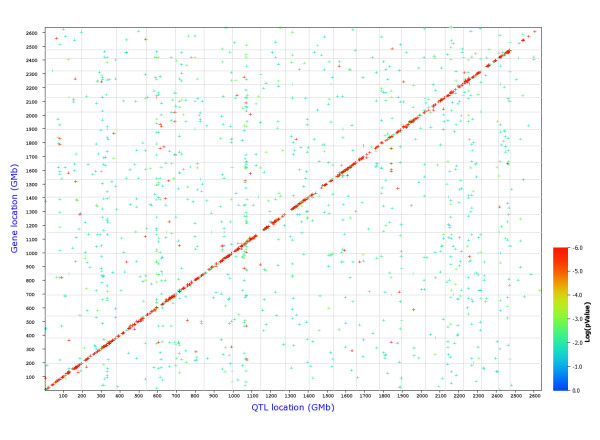
Therefore, we performed a search for e-QTLs at a global level for all probesets. Figures 1 and 2 show the position of all cis- and trans-eQTLs in Treg and Th cells, respectively. The analysis revealed that the overall distribution of trans-eQTLs between the two cell populations is different suggesting different regulatory circuits in the two cell types (see additional file 4, Figure S1 and S2 for direct comparisons of Treg and Th genome graphs).
Figure 1.
Genome-wide graph of cis- and trans-eQTLs in Treg cells. The positions of the eQTLs are plotted against the locations of the corresponding transcript along the genome. Cis-regulated genes are located along the diagonal, all other dots represent trans-regulated genes. The significance level of each QTL is indicated by the color. FDR = 0.3.
Figure 2.
Genome-wide graph of cis- and trans-eQTLs in Th cells. The positions of the eQTLs are plotted against the locations of the corresponding transcript along the genome. Cis-regulated genes are located at the diagonal, all other dots represent trans-regulated genes. The significance level of each QTL is indicated by the color. FDR = 0.3.
Many cis eQTL loci are shared between Treg and Th cells
We then identified all cis- and trans-eQTLs in both Treg and Th data sets with an LRS value of 18 or higher. An LRS of 18 for trans-eQTLs corresponds to a LOD score of 3.9 for trans-eQTLs and thus to a genome-wide p-value of about 0.05.
In the Treg data set, we identified 1,838 probesets with cis-eQTLs with LRS scores of 18 or greater. In comparison, we identified 2,153 probesets of this type in the Th data set. Of these 1,346 probesets (~62.5%) were common to both cell types.
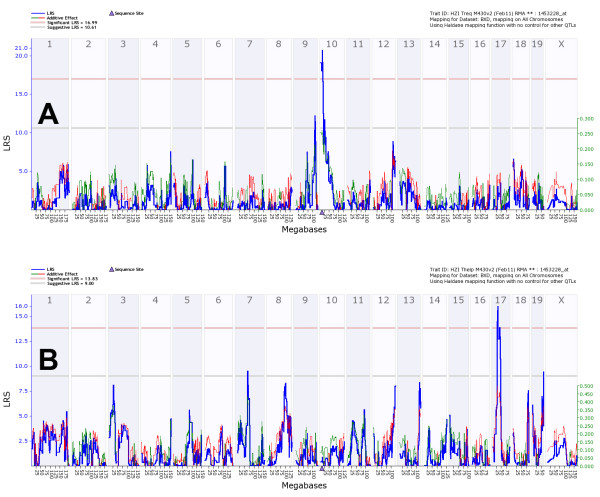
Sixty-seven of the cis-eQTL probesets in Tregs also represented DE-2fold genes for Tregs. Despite the large overlap of cisQTL in the Th and Treg samples, 35 probesets (52%) were only found in Treg samples and were not detected as cis-eQTLs in Th using the above criteria of a minimum LRS of 18 and DE-2fold for Treg (additional file 3, Table S5). However, it should be noted that some probesets, e.g. Lad1 (ladinin) Naip5/Birc1e (NLR family, apoptosis inhibitory protein 5) and Gbp4 (guanylate binding protein 4)) also exhibited significant cis-eQTLs in Th but the corresponding LRS did not reach the threshold level of 18 (data not shown). Figure 3 shows Stx11 (syntaxin 11) as an example for a gene that was expressed both in Treg and Th cells but exhibited a strong cis-eQTL only in Treg cells. Stx11, Gsta4, Ctram, Zfp467 exhibited a cis-eQTL in Treg and a trans-eQTL in Th cells.
Figure 3.
Genome wide eQTL mapping of Stx11 transcript in Treg and Th cells. (A) eQTL map for Stx11 (probeset 1453228_at) in Tregs and (B) in Th, respectively. The numbers at the top are chromosomes, and positions at the bottom are given in megabases along the chromosome. The blue line represents the significance level of the QTL expressed as LRS score (likelihood ratio statistic). A positive additive coefficient (green line) indicates that DBA/2J alleles increased trait values. A negative additive coefficient (red line) indicates that C57BL/6J alleles increased trait values. The two horizontal lines mark the genome-wide significance levels at p < 0.05 (red line) and p < 0.37 (gray line). A blue triangle marks the position of the gene.
In the Th data set, 70 of the identified cis-eQTLs in Th were also DE-2fold genes for Th cells. Of these, 35 probesets represented cis-eQTL genes that were only found in Th cells and were not detected as cis-eQTLs with a LRS of 18 or larger inTreg (additional file 3, Table S6). However, some probestes also exhibited strong cis-eQTLs in Treg but the corresponding LRS did not reach the threshold level of 18, e.g. Snx5 (sorting nexin 5, probeset 1417646_a_at) exhibited a significant cis-eQTL in Th and a strong trans-eQTL in Treg cells (data not shown). Similarly, Chn2, Cadm1 and Themis/E430004N04Rik showed a cis-eQTL in Th and a trans e-QTL in Treg cells.
Strong cis-eQTLs highlight genes that are differentially expressed from the two parental alleles. They can be employed in a forward genetic approach in subsequent studies to elucidate the biological function of the respective genes.
Trans-eQTL mapping reveals different mechanisms of gene regulation in Treg and Th cells
We then analyzed trans-eQTLs at a global level, using again a LRS threshold of 18 or higher. 1,019 probesets were identified which showed a trans-eQTLs in Treg cells. Of these, 65 probesets exhibited trans-eQTLs that were present both in Treg and Th cells. 25 probesets showed also a two-fold or higher level of expression (DE-2fold genes) in Treg cells (additional file 3, Table S7). Of these, all except BC010605 and Prdm5, represented Treg-specifc trans-eQTLs (see M&M for definition of Treg specific trans-eQTLs).
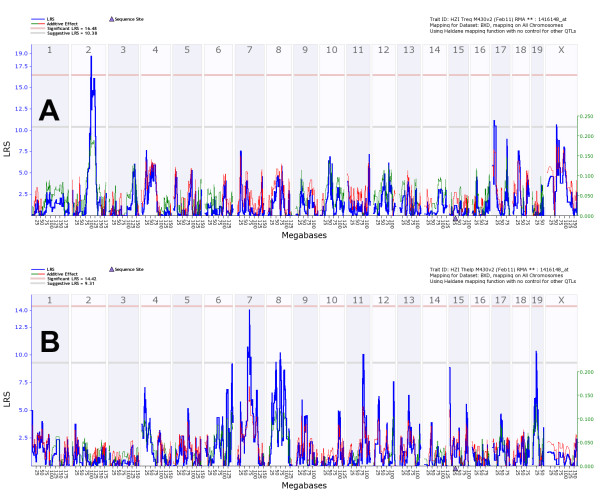
For Abcb1a (ATP-binding cassette, sub-family B (MDR/TAP), member 1A), Gata1 (GATA binding protein 1), Mapkbp1 (mitogen-activated protein kinase binding protein 1), Marco (macrophage receptor with collagenous structure), and Nr4a2/Nurr1 (nuclear receptor subfamily 4, group A, member 2) an immune-related function, in some cases with regulating activity, has been described (see discussion). Laptm4b (lysosomal-associated protein transmembrane 4B) and Lycat (lysocardiolipin acyltransferase 1) exhibited strong trans-eQTL signals in both cell types but at different chromosomal locations (Figures 4 and S3, respectively).
Figure 4.
Genome wide eQTL mapping of Laptm4b transcript in Treg and Th cells. (A) eQTL map for Laptm4b (probeset 1416148_at in Tregs and (B) in Th, respectively. See figure 3 for explanation of labels.
In the Th data set, 1,099 probesets were identified exhibiting a trans-eQTLs of 18 or higher. Of these, 30 probesets showed a two-fold or higher level of expression (DE-2fold genes) in Th cells (additional file 3, Table S8) and all, except BC018101 and Prpf3, represented Th-specifc trans-eQTLs.
The expression of Prpf3 (PRP3 pre-mRNA processing factor 3 homolog (yeast)) showed strong and shared trans-eQTLs in Th and Treg cells (additional file 4, Figure S4), whereas St8sia1 (ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1), Crtam (cytotoxic and regulatory T cell molecule), Igfbp4 (insulin-like growth factor binding protein 4) and Sell (selectin, lymphocyte) exhibited strong trans-eQTL signals in both cell types but at different chromosomal locations (data not shown).
In summary, the identification of cell-specific trans-eQTL regions allowed the subsequent analyses of candidates in the respective regions and to propose novel gene-gene regulatory interactions (see below).
QTLs analysis for known functional Treg genes shows trans-regulation for Nrp1 in Treg
In order to identify gene interaction networks for genes that may play an important role for Treg function, we studied known Treg functional genes (see additional file 3, Table S3) that were differentially expressed in Treg cells and that showed a high expression signal in Tregs (expression signal > 8). This analysis revealed four genes with eQTLs of an LRS equal or larger than 14 (additional file 3, Table S9): Tnfrsf1b (tumor necrosis factor receptor superfamily, member 1b), Nrp1 (neuropilin 1), Stat3 (signal transducer and activator of transcription 3) and Ikzf4 (IKAROS family zinc finger 4). Nrp1 (additional file 4, Figure S5) and Tnfrsf1b (data not shown) exhibited a genome-wide significant trans-eQTL, whereas the eQTLs for Stat3 and Ikzf4 did not reach genome-wide significance levels. Also, Foxp3 and Ctla4 did not exhibit any significant cis- or trans-eQTLs in Treg cells.
These trans-eQTL regions were subsequently used to propose novel gene-gene regulatory interactions (see below).
Analysis of candidate genes in trans-eQTL intervals reveals novel gene-gene regulatory interactions and relation to classical phenotype traits
Next, we analyzed several trans-eQTLs that were identified in the above analyses for candidate genes that may be involved in the regulation of target gene expression (additional file 3, Table S10). Below, we present the analysis of one QTL interval as an example. The results for other QTL intervals can be found in additional file 5, 'Additional Results and Discussion.
The interval on chromosome 4 (28 - 35 Mb) was found to regulate expression of the F2rl1 gene (additional file 4, Figure S6). This region contains 37 probesets which were well expressed (> 8) in Treg cells (Table S11). Of these, Ccdc111 (coiled-coil domain containing 111), Zfp292 (zinc finger protein 292), 1810030N24Rik, AI448984 and Map3k7 (mitogen-activated protein kinase kinase kinase 7) exhibited strong cis-eQTLs (LRS > = 15) and thus are most likely candidates for regulating the expression of F2rl1 in Treg cells.
Furthermore, we analyzed the same eQTL intervals for the presence of a significant QTL in classical phenotypes that are contained in the GeneNetwork database. A total of 18 phenotypes could be found with an LRS of 18 or larger. Of these, eight were related to immune and infectious diseases traits (Table 1). Thus, the candidate genes may also be involved in regulating these traits. It should, however, be noted that our search algorithm only detects classical phenotypes that exhibit a peak QTL at this position. A more detailed analysis can be performed manually for any phenotype of interest.
Table 1.
QTLs from classical phenotypes overlapping with eQTLs
| Chromosome | Mb | Record | Phenotype | Max LRS | Max LRS Location (Chr: Mb) |
|---|---|---|---|---|---|
| 3 | 20-30 | 12958 | Blood chemistry, cardiovascular system: White blood cell count of 14-week old males | 19 | Chr3: 28.293133 |
| 4 | 28-35 | 10410 | Immune function: Antigenic activity of irradiated BXD spleen cells for Thy1+CD3+CD4+CD8- T-cell clone TGVH32 | 31 | Chr4: 31.651725 |
| 17 | 25-45 | 11025 | Infectious disease, immune function: Chlamydia psittaci (6BC) infection response (10^4 IFU ip), pathogen load in peritoneal cavity at 30 days among surviving 3-5 month males | 33.6 | Chr17: 31.769795 |
| 17 | 25-45 | 10466 | Immune function, gastrointestinal system: Intestinal intraepithelial gamma-delta type T lymphocytes (i-IELs) in 2-6 month old adults based on receptor expression (V-gamma-7 positive, Vgamma-4 negative T cells) | 33.1 | Chr17: 32.068297 |
| 17 | 25-45 | 10236 | Proliferation of JTL-G12 cells (T cell clone) with 50 ug/ml GAT (Glu60, Ala30, Tyr10) | 24.5 | Chr17: 32.068297 |
| 17 | 25-45 | 10238 | Proliferation of JTL-G12.8 (Tcell clone) with 50 ug/ml GAT (Glu60, Ala30, Tyr10) | 26.7 | Chr17: 32.068297 |
| 17 | 25-45 | 10441 | Anti-F antigen (liver protein) titer, experiment 1 | 19.1 | Chr17: 26.950946 |
| 17 | 25-45 | 12672 | Infectious disease, immune function: Ectromelia virus survival over two weeks after a 90 pfu intranasal inoculation, males and females between 40 and 150 days of age, residuals corrected for sex, age, and body weight | 19.5 | Chr17: 31.179359 |
Analysis of autoimmune traits
In a previous study for autoimmune disease traits, two QTLs were identifies that are associated with autoimmune traits in the BXD population. One QTL on chromosome 2 was associated with elevated serum titers of anti-DNA antibodies and a second locus on chromosome 4 with increased serum titers of rheumatoid factor (RF) levels [30]. Therefore, we investigated these intervals in our Treg and Th data sets.
The QTL interval (150 - 170 Mb) on chromosome 2 contains 286 probesets that were expressed in Treg cells (signal > 8) and 290 probesets were expressed in Th cells (signal > 8), respectively. We then identified the genes that exhibit a cis-eQTL in Treg cells because these may be involved in the regulation of autoimmunity to DNA. 19 probesets were identified that also exhibited a strong cis-eQTLs (LRS > = 15) in Treg cells (additional file 3, Table S12). The peak interval (160 - 165 Mb) contained the genes Ada (adenosine deaminase), Slpi (secretory leukocyte peptidase inhibitor), Sys1/2610042O14Rik (SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)), and Znf335 (zinc finger protein 335). The cis-eQTL signal for Ada is most likely caused by SNPs in the hybridization probe. Thus these genes, with the exception of Ada, represent likely candidates for regulating this trait in Treg cells. The results for the analysis of the chromosome 4 interval can be found in the supplement data.
Discussion
Here, we studied the expression patterns of genes in Treg and Th cells from a large number of individuals from the BXD recombinant inbred strain collection. The ultimate goal of these studies was the identification of expression quantitative traits (eQTLs) and of co-regulated genes that will give new insights into gene-gene interactions and regulatory pathways. In addition, the large data set of expression profiles from 33 mouse lines allowed us to validate known and to newly identify so far unknown genes that are expressed specifically in Treg but not Th cells.
Until now, several studies have aimed to identify genes that are specific for Treg cells (additional file 3, Table S3). Using our data set from 33 mouse stains allowed us to search for differences in expression levels in Treg versus Th cells with a so far unprecedented statistical power. Our analysis confirmed that many genes with a described Treg function are also differentially expressed. Of the 23 genes that were described in the literature as genes with an important function in Treg cells, 17 were also found to be Treg specific in our analysis. Of the remaining six genes, Galectin-10 does not have a homologue in humans. S1pr1a and Lrrc32 were not represented on the array. Also a Treg function for LRRC32/GARP was so far only described in humans. The remaining three genes, Gzmb, Lag3 and Il12rb2, were expressed at low levels in both Treg and Th but with no significant difference between the two cell types.
The genes Foxp3, Nrp1, Clta4, Tnfrsf18 /Gitr, Il2ra /Cd25, Itgae /CD103, Ikzf4 /Eos, Gpr83 have been reported to be hallmark genes for Treg cells [31,32]. Indeed, all of them appear as DE-2fold genes specifically higher expressed in Treg cells. Since we discovered Nrp1 as surface marker of murine regulatory T cells [3] comprehensive data have been obtained regarding its implications in Treg function [33-37]. Whereas expression of other Treg-markers such as Cd25 and Ctla4 are induced upon activation of Th cells, activated murine Th cells lack Nrp1 expression, suggesting fundamental differences in the transcriptional regulation of this molecule in Tregs when compared with other well described Treg markers.
In total, 608 probesets were found in our analysis as Treg-specific and with an expression difference of at least two-fold. Although a difference of two-fold in expression levels is often used in gene expression array studies in many studies, it may be considered as arbitrary. Our study allowed to find even more genes with statistical significant differences in expression levels and extended the known list to 14,117 probesets that are specific for differentiated Treg versus Th cells. Zheng et al. [38] performed chromatin immunoprecipitation and genome tiling array profiling to identify direct targets of the Foxp3 transcription factor. About 270 genes were identified were also up-regulated in Foxp3+ T cells. Of these, 162 genes were also found as differentially expressed genes in our analysis.
Recently, a genome wide association study in a large cohort of patients suffering from alopecia areata, which is among the most highly prevalent human autoimmune diseases, leading to disfiguring hair loss due to the collapse of immune privilege of the hair follicle and subsequent autoimmune attack revealed an association with genomic regions containing several genes controlling the activation and proliferation of Tregs including Ctla4, CD25, and Eos/Ikzf4 [39].
In conclusion, almost all genes described previously as Treg functional genes or in other screening assays were also found in our analysis. These findings confirm that our study is well suitable to identify genes that are important for Treg function, differentiation and/or maintenance of their differentiation status.
Cis-eQTL genes most likely carry a mutation in the promoter region or contain a structural variation in the transcribed region which results in low expression levels or unstable RNA of one but not the other allele. In our study, several cis-eQTL genes were found that were active both in Treg and Th cells and have a known function in the immune response. For example, Stx11 (syntaxin 11) has been associated with familial hemophagocytic lymphohistiocytosis 4 in humans [40,41].
BXD strains that carry a very low expressing allele inherited from one of the parental strains are similar to genetic knock-down experiments or hypomorphic mutants [42]. The function of such genes can be studied in a reverse genetics approach by comparing the mean phenotypes of BXD strains that have inherited either the low or high allele--a method referred to as reverse complex trait analysis. Prominent examples of genes that are nature "knock-downs" in the BXD strains, include Hc, Ahr, Gpnmb, Tyrp1, Sae1, Apoa2, and several CLEC and KLRA gene family members [23,42].
341 probesets represented cis-eQTLs shared by several tissues, in Treg and Th cells but also in brain, kidney and lung.
In Treg cells, 25 probeset were trans-regulated and represented genes that were differently expressed between Treg and Th cells and that showed a two-fold or higher level of expression (DE-2fold genes) in Treg cells. Several of them represented genes with a known immune-related function. For example, Abcb1a (ATP-binding cassette, sub-family B (MDR/TAP), member 1A) is involved in the down-modulation of dendritic cell functions through the regulation of pro-inflammatory cytokine secretion [43]. Gata1 (GATA binding protein 1) has been shown to be an important regulator of mast cell differentiation [44]. Mapkbp1 (mitogen-activated protein kinase binding protein 1) encodes a protein that enhances NF-kappaB activation induced by MAP kinase kinase kinase 7 and TNF Receptor-Associated Factor 2 [45]. Marco (macrophage receptor with collagenous structure) exhibits multiple functions in the innate immune response. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis [46]. MARCO-deficient mice exhibit lower IL-12 production in responses to stimulation [47]. A defect in Marco results in an impaired clearance of apoptotic cells and a generalized defect in both endocytosis and phagocytosis [48]. Expression of MARCO is required for TLR signaling [46]. But Marco also exhibits suppressive functions by decreasing inflammation in lungs after exposure to ozone [49]. Nr4a2/Nurr1 (nuclear receptor subfamily 4, group A, member 2) represents a transcriptional mediator of inflammatory signals [50,51]. Also, it plays an important role in modulating IL-8 expression [52].
Two genes, Laptm4b and Lycat exhibited strong trans-eQTL signals in both cell types but at different chromosomal locations. Thus, the same genes are expressed in both cell types, but are likely regulated by different mechanisms. Laptm4b (lysosomal-associated protein transmembrane 4B) is involved in cell proliferation and multidrug resistance [53], whereas no biological function has yet been described for Lycat (lysocardiolipin acyltransferase 1).
None of these genes represents a known Treg key gene. Our analysis thus allows expanding the list of genes with potentially important functions in Treg cells. We did not perform an extensive analysis for trans-eQTLs in Th cells but a similar result can be expected for Th cells.
We then analyzed several trans-eQTLs for potential regulator genes in order to propose possible novel gene-regulatory circuits in Treg and Th cells. These are by far not comprehensive but rather serve as examples to illustrate how our data set will allow searching for possible regulatory networks.
A QTL region was found on chromosome 4 that regulates the expression of the F2rl1 (coagulation factor II (thrombin) receptor-like 1) gene. F2rl1/Par2 gene expression has been associated with the activation and suppression of inflammatory responses. Overexpression of F2rl1 in allergic inflammation of the airway exacerbates eosinophil infiltration into the lumen and hyperreactivity of the airway, while F2rl1 deletion diminishes inflammatory cell infiltration and reduces hyperreactivity [54]. Also, F2rl1 plays a protective role during influenza virus type A infection through IFN-gamma production and decreased excessive recruitment of inflammatory cells to lung alveoli [55] and deletion of F2rl1 is associated with decreased clearance of P. aeruginosa [56]. Several candidate genes in the chromosome 4 QTL interval, which regulates F2rl1 expression, exhibited a cis-eQTL and thus represent potential regulators of F2rl1 (Table S11). These include the Map3k7/Tak1 (mitogen-activated protein kinase kinase kinase 7) protein kinase gene which exhibits many immune modulator functions. It mediates activation of IKK (inhibitor of kappaB kinase) and silencing of Map3k7 suppressed T cell receptor-dependent IKK activation and interleukin-2 production in T cells [57,58]. The Zfp292 (zinc finger protein 292) gene expression has been shown tocorrelate with growth hormone expression and may thus be involved in mediating proliferation signals [59]. The analysis of candidate genes in the other QTL regions is discussed in the supplement data.
Our analysis of the chromosome 2 regions autoimmune trait [30] identified several loci that exhibited a cis-eQTL in Treg cells and may thus be involved in regulating this trait: Slpi, Sys1/2610042O14Rik, and Znf335. Slpi (secretory leukocyte peptidase inhibitor) exhibits many biological functions, including anti-bacterial, anti-fungal, anti-viral, anti-inflammatory, wound healing and immuno-modulatory activities [60-68]. The Slpi protein represents a ligand for PLSCR1 and PLSCR4, which interact directly with the CD4 receptor at the cell surface of T lymphocytes [69]. Slpi is a prominent innate immune protein of the respiratory tract with serine protease inhibitor activity [70]. It attenuates excessive inflammatory responses resulting in a balanced innate immunity [71,72]. Constitutive expression of Slpi reduced the inflammatory response and improved lung function in an acute model of allergic asthma in Slpi transgenic and knockout mice [73]. The attenuation of the inflammatory response by Slpi is mediated through macrophages that secrete an increased amount of Slpi when encountering apoptotic cells [74,75]. Znf335 (zinc finger protein 335, also NIF1 in human) is a cotransducer that regulates the activity of the nuclear hormone receptor coactivator NRC [76] which may mediate the function of the CCR4 signal [77]. No biological functions were identified so far for Sys1 (SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)). The discussion of candidate genes in the chromosome 4 auto-immune QTL interval can be found in the supplemental data.
Conclusions
Our study revealed many novel Treg specific genes and also points to genes that may regulate important gene-gene interactions in Treg cells. These interactions, once validated, should allow proposing new targets for therapeutic interventions in cancer, auto-immunity, and allergies.
Methods
Mouse strains and sample preparation
Parental and BXD lines were received from Jackson Laboratory, or from The Oak Ridge National Laboratory (BXD43, BXD51, BXD61, BXD62, BXD65, BXD68, BXD69, BXD73, BXD75, BXD87, BXD90), and were bred in the facility of the Neuro-BSIK consortium (VU University Amsterdam). Mice were housed on sawdust in standard Makrolon type II cages with food (Harlan Teklad 2018) and water ad libitum under specific pathogen free conditions. For the analysis, mice were transferred to the animal facility in Braunschweig and adapted for at least two weeks to the new environment before preparing the spleen cells. All protocols involving mice were approved by national animal welfare committees.
Sorting of Tregs and Th cells by fluorescent activated cell sorting
For sorting of Tregs and Th cells, splenocytes from 31 BXD recombinant inbred strains as well as from the parental mouse lines DBA/2J and C57BL/6J were isolated by flushing the spleens with erythrocyte-lysis-buffer. Cells were collected by centrifugation, re-suspended in cold FACS-buffer (PBS/2% FCS/0.5 mM EDTA). For each strain and sex, the cells from three individual mice were pooled. After passing the cells through a 100 μm cell strainer and an additional washing step with FACS-buffer, splenocytes were stained with anti-CD4-APC and anti-CD25-PE (BD Biosciences (Heidelberg, Germany)) for 10 minutes at 4°C, washed and re-suspended in FACS-buffer. CD4+ T cells were separated into CD4+CD25+ Treg and CD4+CD25- Th cells using a MoFlo cell sorter (Cytomation) and purity of the sorted T cell subsets reached 95-97%.
Microarray analysis
Quality and integrity of the total RNA isolated from 1 × 105 cells was controlled by running all samples on an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies; Waldbronn, Germany). RNA amplification and labelling was done according to manufactures protocol (Small Sample Target Labeling Assay Version II, Affymetrix; Santa Clara, CA). The concentration of biotin-labeled cRNA was determined by UV absorbance. In all cases, 10 μg of each biotinylated cRNA preparation were fragmented and placed in a hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre) as recommended by the manufacturer. Samples were hybridized to an identical lot of Affymetrix MOE430 2.0 for 16 hours at 46°C. After hybridisation the GeneChips were washed and stained using the Affymetrix's recommended EukGE-WS2v5 protocol for GeneChip® Fluidics FS400 station. Images were scanned using GeneChip® Scanner 3000 under the control of GCOS 1.3 software package (Affymetrix; Santa Clara, CA)
Data preprocessing and analysis
Microarray data was preprocessed using the RMA method [78] and log2 values were computed. The Z scores for each cell value was then calculated (including subtraction of means and division by standard deviations), multiplied by 2 and a value of 8 was added. The advantage of this variant of a Z transformation is that all values are positive and that 1 unit represents approximately a 2-fold difference in expression as determined using the spike-in control probesets (see [26] for details). The data was then batch corrected [79]. In this study, RNA was extracted at three different points in time for the Treg samples and also microarray processing was performed at three different points in time. Similarly, the Th samples were processed in two batches. Therefore, we performed a batch correction for both cell types using the following ANOVA model before further analysis of the data: yi = μ + Bi + ei where yi is the expression level of the ith microarray, μ is the overall mean, Bi is the batch to which the ith individual belongs and ei is the residual error. Finally, mean values were calculated if multiple samples from one BXD line were recorded (male and females or replicates) and the processed data were stored and are publically available in the GeneNetwork (GN) database [20]. Identification and QTL analyses were performed using the GeneNetwork web service [20] as previously described [23].
Differentially expressed probes between Treg and Th were calculated by applying the moderated F-statistic [80], an empirical Bayes method, Cut-offs were set to pFDR < = 0.001 and fold change > = 2×. Probes significantly higher expressed either in Treg or Th, respectively, were tested for statistically associated GO terms via the hypergeometric test. Data analyses were done in the R environment with Bioconductor [81] and packages limma [78] and GOstats [82].
The GeneRIF database http://www.ncbi.nlm.nih.gov/projects/GeneRIF/GeneRIFhelp.html was used as a primary source to search for known gene functions and to cite these in the discussion. We also tested whether a trans-eQTLs obtained in one data set is specific for this cell type. GeneNetwork does not have an automatic function for such an analysis. We therefore tested this feature manually in the following way. For trans-eQTLs with an LRS of 18 or higher in Treg cells, the LRS was determined at the same marker positions in the Th data set. If the LRS in Th was less that 6 (which corresponds to a p-value of less than 0.05 at this marker - not a genome-wide significance) the QTL in Treg was considered "Treg-specific". The analysis was performed likewise to determine Th-specifc trans-eQTLs.
Access to microarray data
The processed data and analysis tools are available at the GeneNetwork database http://www.genenetwork.org/webqtl/main.py, accession number for Treg data: GN122; accession number for Th data: GN319. The raw and processed data are accessible from the ArrayExpress database, ID: E-MTAB-836.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DB, HC and KS generated and analyzed the material. RA, KS and RW performed the analysis of the data. GS and SS provided the mice for the analysis. DB and KS wrote the manuscript. CP performed the identification of DE genes and GO term analyses. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Treg specific genes. The table lists genes that exhibited a differential expression in Treg compared to Th cells. Probeset: probeset ID of microarray hybridization probe, logFC: fold changes of expression level in Treg versus Th as difference of log2 values, pFDR: FDR-corrected p-value, Treg: expression level in Treg cells as log2 value, Th: expression level in Th cells as log2 value.
Table S2 Th specific genes. The table lists genes that exhibited a differential expression in Th compared to Treg cells. Probeset: probeset ID of microarray hybridization probe, logFC: fold changes of expression level in Treg versus Th as difference of log2 values (a negative value represents higher expression in Th compared to Treg cells), pFDR: FDR-corrected p-value, tr: expression level in Treg cells as log2 value, th: expression level in Th cells as log2 value.
Table S3 Genes with known functions in Treg cells. The table lists genes that have been described in the literature to exhibit a functional role in Treg cells. Table S4: Treg functional genes with a higher expression in Tregs. The table lists probesets of genes that were expressed at a higher level in Treg compared to Th cells and which exhibit known Treg functions. Table S5. Genes regulated by cis-eQTL in Treg cells. The table lists probesets from genes that exhibited a cis-eQTL larger or equal to an LRS of 18 in Treg but not in Th cells and which were expressed at least 2-fold higher in Treg cells. Table S6. Genes regulated by cis-eQTL (LRS > = 18) in Th. The table lists probesets from genes that exhibited a cis-eQTL larger or equal to an LRS of 18 in Th but not in Treg cells and which were expressed at least 2-fold higher in Th cells. Table S7. Treg specific genes regulated by trans-eQTL in Treg cells. The table lists probesets from genes that exhibited a trans-eQTL larger or equal to an LRS of 18 in Treg but not in Th cells and which were expressed at least 2-fold higher in Treg cells. Table S8. Th specific genes regulated by trans-eQTL in Th cells. The table lists probesets from genes that exhibited a trans-eQTL larger or equal to an LRS of 18 in Th but not in Treg cells and which were expressed at least 2-fold higher in Th cells. Table S9. Treg functional genes regulated and highly expressed in Treg cells. The table lists probesets of differentially expressed genes in Treg cells with a known Treg function, a high expression signal in Tregs (expression signal > 8) and which are regulated by a trans-eQTL with an LRS > = 14. Table S10: Intervals selected for further QTL analysis. The table lists the QTL intervals from selected Treg and Th cis- and trans-eQTLs that were further analyzed for the presence of candidate regulatory genes. Table S11 Candidate genes located in the F2rl1-QTL interval on chromosome 4. The table lists probesets of possible candidate genes that are located in the QTL interval on chromosome 4 which regulates the expression of F2rl1. Only genes with an expression signal larger than 8 were selected. Table S12 Candidate genes located in the QTL interval on chromosome 2 (autoimmunity to DNA). The table lists probesets of possible candidate genes that are located in the QTL interval on chromosome 2 which regulates autoimmunity to DNA. Only genes expressed in Treg cells (signal > 8 on log2 scale) and exhibiting a cis-eQTL of LRS > = 15 were selected.
Figure S1. Genome-wide graph of cis- and trans-eQTLs in Treg and Th cells. The file shows the genome-wide mapping graphs for Treg cells and the same graph for Th cells overlaid in grey. Figure S2. Genome-wide graph of cis- and trans-eQTLs in Th and Treg cells. The file shows the genome-wide mapping graphs for Th cells and the same graph for Treg cells overlaid in grey. Figure S3, Genome wide eQTL mapping of Lycat transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Lycat transcript in Treg and Th cells. Figure S4, Genome wide eQTL mapping of Prpf3 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Prpf3 transcript in Treg and Th cells. Figure S5, Genome wide eQTL mapping of Nrp1 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Nrp1 transcript in Treg and Th cells. Figure S6, Genome wide eQTL mapping of F2rl1 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the F2rl1 transcript in Treg and Th cells. Figure S7, Genome wide eQTL mapping of Ctla4 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Ctla4 transcript in Treg and Th cells.
'Additional Results and Discussion': This file contains additional results on: Trans-eQTLs mapping in Treg and Th cells, the analysis of candidate genes in trans-eQTL intervals, and the analysis of autoimmune traits. Also, the file contains additional discussion points on Treg-specifc genes, the analysis of the interval on chromosome 2 found to regulate expression of Nrp1, analysis of an interval on chromosome × regulating expression of Klrb1f/A630024B12Rik (killer cell lectin-like receptor subfamily B member 1F) and Ctla4 (cytotoxic T-lymphocyte-associated protein 4), the analysis of known Treg functional genes for possible cis- or trans-eQTLs, and the analysis of the QTLs interval chromosome 4 associated with autoimmune traits.
Contributor Information
Rudi Alberts, Email: rudialberts@gmail.com.
Hairong Chen, Email: charis0224@gmail.com.
Claudia Pommerenke, Email: Claudia.Pommerenke@helmholtz-hzi.de.
August B Smit, Email: guus.smit@cncr.vu.nl.
Sabine Spijker, Email: sabine.spijker@falw.vu.nl.
Robert W Williams, Email: rwilliam@nb.uthsc.edu.
Robert Geffers, Email: robert.geffers@helmholtz-hzi.de.
Dunja Bruder, Email: dbr@helmholtz-hzi.de.
Klaus Schughart, Email: kls@helmholtz-hzi.de.
Acknowledgements
This work was supported by intra-mural grants from the Helmholtz-Association (Program "Infection and Immunity") and by a grant from the German Research Foundation (SFB 587 to D.B.). We like to thank Dr. Lothar Gröbe for his excellent support for the sorting of T cell populations. GeneNetwork and the production of BXD strains are supported by the UT Center for Integrative and Translational Genomics and NIH grants INIA U01 AA13499, AA016662.
References
- Sakaguchi S, Ono M, Setoguchi R, Yagi H, Hori S, Fehervari Z, Shimizu J, Takahashi T, Nomura T. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155(3):1151–1164. [PubMed] [Google Scholar]
- Bruder D, Probst-Kepper M, Westendorf AM, Geffers R, Beissert S, Loser K, von Boehmer H, Buer J, Hansen W. Neuropilin-1: a surface marker of regulatory T cells. Eur J Immunol. 2004;34(3):623–630. doi: 10.1002/eji.200324799. [DOI] [PubMed] [Google Scholar]
- Lechner O, Lauber J, Franzke A, Sarukhan A, von Boehmer H, Buer J. Fingerprints of anergic T cells. Curr Biol. 2001;11(8):587–595. doi: 10.1016/S0960-9822(01)00160-9. [DOI] [PubMed] [Google Scholar]
- Lehmann J, Huehn J, de la Rosa M, Maszyna F, Kretschmer U, Krenn V, Brunner M, Scheffold A, Hamann A. Expression of the integrin alpha Ebeta 7 identifies unique subsets of CD25+ as well as CD25- regulatory T cells. Proc Natl Acad Sci USA. 2002;99(20):13031–13036. doi: 10.1073/pnas.192162899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohning M, Hutloff A, Kallinich T, Mages HW, Bonhagen K, Radbruch A, Hamelmann E, Kroczek RA. Expression of ICOS in vivo defines CD4+ effector T cells with high inflammatory potential and a strong bias for secretion of interleukin 10. J Exp Med. 2003;197(2):181–193. doi: 10.1084/jem.20020632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh RS, Whitters MJ, Piccirillo CA, Young DA, Shevach EM, Collins M, Byrne MC. CD4(+)CD25(+) immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. 2002;16(2):311–323. doi: 10.1016/S1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30(5):636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Shimizu J, Yamazaki S, Takahashi T, Ishida Y, Sakaguchi S. Stimulation of CD25(+)CD4(+) regulatory T cells through GITR breaks immunological self-tolerance. Nat Immunol. 2002;3(2):135–142. doi: 10.1038/ni759. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8(5):457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. In: Genetic Variation in the Laboratory Mouse. Lyon ML, Searle AG, editor. Oxford: Oxford University Press; 1989. Recombinant inbred strains; pp. 773–796. [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2(11):RESEARCH0046. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF. et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using 'genetical genomics'. Nat Genet. 2005;37(3):225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- Boon AC, deBeauchamp J, Hollmann A, Luke J, Kotb M, Rowe S, Finkelstein D, Neale G, Lu L, Williams RW. et al. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J Virol. 2009;83(20):10417–10426. doi: 10.1128/JVI.00514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdeltawab NF, Aziz RK, Kansal R, Rowe SL, Su Y, Gardner L, Brannen C, Nooh MM, Attia RR, Abdelsamed HA. et al. An unbiased systems genetics approach to mapping genetic loci modulating susceptibility to severe streptococcal sepsis. PLoS Pathog. 2008;4(4):e1000042. doi: 10.1371/journal.ppat.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyairi I, Laxton JD, Wang X, Obert CA, Arva Tatireddigari VR, van Rooijen N, Hatch TP, Byrne GI. Chlamydia psittaci Genetic Variants Differ in Virulence by Modulation of Host Immunity. J Infect Dis. 2011;204(4):654–663. doi: 10.1093/infdis/jir333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7(5):485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17(7):388–391. doi: 10.1016/S0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Alberts R, Terpstra P, Bystrykh LV, de Haan G, Jansen RC. A statistical multiprobe model for analyzing cis and trans genes in genetical genomics experiments with short-oligonucleotide arrays. Genetics. 2005;171(3):1437–1439. doi: 10.1534/genetics.105.045930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts R, Lu L, Williams RW, Schughart K. Genome-wide analysis of the mouse lung transcriptome reveals novel molecular gene interaction networks and cell-specific expression signatures. Respir Res. 2011;12:61. doi: 10.1186/1465-9921-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peidis P, Giannakouros T, Burow ME, Williams RW, Scott RE. Systems genetics analyses predict a transcription role for P2P-R: molecular confirmation that P2P-R is a transcriptional co-repressor. BMC Syst Biol. 2010;4:14. doi: 10.1186/1752-0509-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Mulligan MK, Wang X, Miles MF, Lu L, Williams RW. A transposon in comt generates mRNA variants and causes widespread expression and behavioral differences among mice. PLoS ONE. 2010;5(8):e12181. doi: 10.1371/journal.pone.0012181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisert EE, Lu L, Freeman-Anderson NE, Templeton JP, Nassr M, Wang X, Gu W, Jiao Y, Williams RW. Gene expression in the mouse eye: an online resource for genetics using 103 strains of mice. Mol Vis. 2009;15:1730–1763. [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 2008;4(11):e1000260. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutnikova H, Laakso M, Lu L, Combe R, Paananen J, Kuulasmaa T, Kuusisto J, Haring HU, Hansen T, Pedersen O. et al. Identification of the UBP1 locus as a critical blood pressure determinant using a combination of mouse and human genetics. PLoS Genet. 2009;5(8):e1000591. doi: 10.1371/journal.pgen.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Badea A, Williams RW, Kas MJ. Identifying Human Disease Genes through Cross-Species Gene Mapping of Evolutionary Conserved Processes. PLoS ONE. 2011;6(5):e18612. doi: 10.1371/journal.pone.0018612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountz JD, Yang P, Wu Q, Zhou J, Tousson A, Fitzgerald A, Allen J, Wang X, Cartner S, Grizzle WE. et al. Genetic segregation of spontaneous erosive arthritis and generalized autoimmune disease in the BXD2 recombinant inbred strain of mice. Scand J Immunol. 2005;61(2):128–138. doi: 10.1111/j.0300-9475.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, Mak TW, Sakaguchi S. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192(2):303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen W, Loser K, Westendorf AM, Bruder D, Pfoertner S, Siewert C, Huehn J, Beissert S, Buer J. G protein-coupled receptor 83 overexpression in naive CD4+CD25- T cells leads to the induction of Foxp3+ regulatory T cells in vivo. J Immunol. 2006;177(1):209–215. doi: 10.4049/jimmunol.177.1.209. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Mueller C, Chae WJ, Alabanza LM, Bynoe MS. Neuropilin-1 attenuates autoreactivity in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(5):2040–2045. doi: 10.1073/pnas.1008721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka Y, Stoilova S, Mohammed N, Prud'homme GJ. Neuropilin-1 exerts co-receptor function for TGF-beta-1 on the membrane of cancer cells and enhances responses to both latent and active TGF-beta. Carcinogenesis. 2011;32(4):613–621. doi: 10.1093/carcin/bgq281. [DOI] [PubMed] [Google Scholar]
- Catalano A. The neuroimmune semaphorin-3A reduces inflammation and progression of experimental autoimmune arthritis. J Immunol. 2010;185(10):6373–6383. doi: 10.4049/jimmunol.0903527. [DOI] [PubMed] [Google Scholar]
- Glinka Y, Prud'homme GJ. Neuropilin-1 is a receptor for transforming growth factor beta-1, activates its latent form, and promotes regulatory T cell activity. J Leukoc Biol. 2008;84(1):302–310. doi: 10.1189/jlb.0208090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris M, Andersen KG, Randow F, Mayr L, Betz AG. Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity. 2008;28(3):402–413. doi: 10.1016/j.immuni.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445(7130):936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, Kim H, Singh P, Lee A, Chen WV. et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson YT, Rudd E, Zheng C, Edner J, Ma D, Wood SM, Bechensteen AG, Boelens JJ, Celkan T, Farah RA. et al. Defective cytotoxic lymphocyte degranulation in syntaxin-11 deficient familial hemophagocytic lymphohistiocytosis 4 (FHL4) patients. Blood. 2007;110(6):1906–1915. doi: 10.1182/blood-2007-02-074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macartney CA, Weitzman S, Wood SM, Bansal D, Steele M, Meeths M, Abdelhaleem M, Bryceson YT. Unusual functional manifestations of a novel STX11 frameshift mutation in two infants with familial hemophagocytic lymphohistiocytosis type 4 (FHL4) Pediatr Blood Cancer. 2011;56(4):654–657. doi: 10.1002/pbc.22676. [DOI] [PubMed] [Google Scholar]
- Ciobanu DC, Lu L, Mozhui K, Wang X, Jagalur M, Morris JA, Taylor WL, Dietz K, Simon P, Williams RW. Detection, validation, and downstream analysis of allelic variation in gene expression. Genetics. 2010;184(1):119–128. doi: 10.1534/genetics.109.107474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooij G, Backer R, Koning JJ, Reijerkerk A, van Horssen J, van der Pol SM, Drexhage J, Schinkel A, Dijkstra CD, den Haan JM. et al. P-glycoprotein acts as an immunomodulator during neuroinflammation. PLoS ONE. 2009;4(12):e8212. doi: 10.1371/journal.pone.0008212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, Vannucchi AM, Migliaccio G, Orkin SH. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J Exp Med. 2003;197(3):281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Miyashita C, Koyano S, Kanda H, Yoshioka K, Shiba T, Takamatsu N, Ito M. JNK-binding protein 1 regulates NF-kappaB activation through TRAF2 and TAK1. Cell Biol Int. 2009;33(3):364–368. doi: 10.1016/j.cellbi.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Bowdish DM, Sakamoto K, Kim MJ, Kroos M, Mukhopadhyay S, Leifer CA, Tryggvason K, Gordon S, Russell DG. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog. 2009;5(6):e1000474. doi: 10.1371/journal.ppat.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefowski S, Arredouani M, Sulahian T, Kobzik L. Disparate regulation and function of the class A scavenger receptors SR-AI/II and MARCO. J Immunol. 2005;175(12):8032–8041. doi: 10.4049/jimmunol.175.12.8032. [DOI] [PubMed] [Google Scholar]
- Rogers NJ, Lees MJ, Gabriel L, Maniati E, Rose SJ, Potter PK, Morley BJ. A defect in Marco expression contributes to systemic lupus erythematosus development via failure to clear apoptotic cells. J Immunol. 2009;182(4):1982–1990. doi: 10.4049/jimmunol.0801320. [DOI] [PubMed] [Google Scholar]
- Dahl M, Bauer AK, Arredouani M, Soininen R, Tryggvason K, Kleeberger SR, Kobzik L. Protection against inhaled oxidants through scavenging of oxidized lipids by macrophage receptors MARCO and SR-AI/II. J Clin Invest. 2007;117(3):757–764. doi: 10.1172/JCI29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005;280(32):29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- McEvoy AN, Murphy EA, Ponnio T, Conneely OM, Bresnihan B, FitzGerald O, Murphy EP. Activation of nuclear orphan receptor NURR1 transcription by NF-kappa B and cyclic adenosine 5'-monophosphate response element-binding protein in rheumatoid arthritis synovial tissue. J Immunol. 2002;168(6):2979–2987. doi: 10.4049/jimmunol.168.6.2979. [DOI] [PubMed] [Google Scholar]
- Volakakis N, Joodmardi E, Perlmann T. NR4A orphan nuclear receptors influence retinoic acid and docosahexaenoic acid signaling via up-regulation of fatty acid binding protein 5. Biochem Biophys Res Commun. 2009;390(4):1186–1191. doi: 10.1016/j.bbrc.2009.10.116. [DOI] [PubMed] [Google Scholar]
- Kasper G, Vogel A, Klaman I, Grone J, Petersen I, Weber B, Castanos-Velez E, Staub E, Mennerich D. The human LAPTM4b transcript is upregulated in various types of solid tumours and seems to play a dual functional role during tumour progression. Cancer Lett. 2005;224(1):93–103. doi: 10.1016/j.canlet.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Schmidlin F, Amadesi S, Dabbagh K, Lewis DE, Knott P, Bunnett NW, Gater PR, Geppetti P, Bertrand C, Stevens ME. Protease-activated receptor 2 mediates eosinophil infiltration and hyperreactivity in allergic inflammation of the airway. J Immunol. 2002;169(9):5315–5321. doi: 10.4049/jimmunol.169.9.5315. [DOI] [PubMed] [Google Scholar]
- Khoufache K, LeBouder F, Morello E, Laurent F, Riffault S, Andrade-Gordon P, Boullier S, Rousset P, Vergnolle N, Riteau B. Protective role for protease-activated receptor-2 against influenza virus pathogenesis via an IFN-gamma-dependent pathway. J Immunol. 2009;182(12):7795–7802. doi: 10.4049/jimmunol.0803743. [DOI] [PubMed] [Google Scholar]
- Moraes TJ, Martin R, Plumb JD, Vachon E, Cameron CM, Danesh A, Kelvin DJ, Ruf W, Downey GP. Role of PAR2 in murine pulmonary pseudomonal infection. Am J Physiol Lung Cell Mol Physiol. 2008;294(2):L368–377. doi: 10.1152/ajplung.00036.2007. [DOI] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14(3):289–301. doi: 10.1016/S1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Singhirunnusorn P, Shimotabira E, Chino A, Suzuki S, Koizumi K, Saiki I. TAK1-mediated transcriptional activation of CD28-responsive element and AP-1-binding site within the IL-2 promoter in Jurkat T cells. FEBS Lett. 2005;579(29):6641–6646. doi: 10.1016/j.febslet.2005.10.059. [DOI] [PubMed] [Google Scholar]
- Wojtkiewicz PW, Phelps CJ, Hurley DL. Transcript abundance in mouse pituitaries with altered growth hormone expression quantified by reverse transcriptase polymerase chain reaction implicates transcription factor Zn-16 in gene regulation in vivo. Endocrine. 2002;18(1):67–74. doi: 10.1385/ENDO:18:1:67. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A, Gray LR, Patton LL, Caplan DJ, Slade GD, Tien HC, Shugars DC. Salivary secretory leukocyte protease inhibitor and oral candidiasis in human immunodeficiency virus type 1-infected persons. Infect Immun. 2004;72(4):1956–1963. doi: 10.1128/IAI.72.4.1956-1963.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau T, Baranger K, Dade S, Dallet-Choisy S, Guyot N, Zani ML. Multifaceted roles of human elafin and secretory leukocyte proteinase inhibitor (SLPI), two serine protease inhibitors of the chelonianin family. Biochimie. 2008;90(2):284–295. doi: 10.1016/j.biochi.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Nishimura J, Saiga H, Sato S, Okuyama M, Kayama H, Kuwata H, Matsumoto S, Nishida T, Sawa Y, Akira S. et al. Potent antimycobacterial activity of mouse secretory leukocyte protease inhibitor. J Immunol. 2008;180(6):4032–4039. doi: 10.4049/jimmunol.180.6.4032. [DOI] [PubMed] [Google Scholar]
- Sehnert B, Cavcic A, Bohm B, Kalden JR, Nandakumar KS, Holmdahl R, Burkhardt H. Antileukoproteinase: modulation of neutrophil function and therapeutic effects on anti-type II collagen antibody-induced arthritis. Arthritis Rheum. 2004;50(7):2347–2359. doi: 10.1002/art.20339. [DOI] [PubMed] [Google Scholar]
- Sehnert B, Gierer P, Ibrahim S, Kuhl A, Voll R, Nandakumar KS, Holmdahl R, Hallmann R, Vollmar B, Burkhardt H. Modulation of granulocyte-endothelium interactions by antileukoproteinase: inhibition of anti-type II collagen antibody-induced leukocyte attachment to the synovial endothelium. Arthritis Res Ther. 2006;8(4):R95. doi: 10.1186/ar1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PA, Lentsch AB. Endogenous regulation of the acute inflammatory response. Mol Cell Biochem. 2002;234-235(1-2):225–228. [PubMed] [Google Scholar]
- Weldon S, McGarry N, Taggart CC, McElvaney NG. The role of secretory leucoprotease inhibitor in the resolution of inflammatory responses. Biochem Soc Trans. 2007;35(Pt 2):273–276. doi: 10.1042/BST0350273. [DOI] [PubMed] [Google Scholar]
- Wex T, Treiber G, Nilius M, Vieth M, Roessner A, Malfertheiner P. Helicobacter pylori-mediated gastritis induces local downregulation of secretory leukocyte protease inhibitor in the antrum. Infect Immun. 2004;72(4):2383–2385. doi: 10.1128/IAI.72.4.2383-2385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Nathan C, Jin W, Sim D, Ashcroft GS, Wahl SM, Lacomis L, Erdjument-Bromage H, Tempst P, Wright CD. et al. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111(6):867–878. doi: 10.1016/S0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- Py B, Basmaciogullari S, Bouchet J, Zarka M, Moura IC, Benhamou M, Monteiro RC, Hocini H, Madrid R, Benichou S. The phospholipid scramblases 1 and 4 are cellular receptors for the secretory leukocyte protease inhibitor and interact with CD4 at the plasma membrane. PLoS ONE. 2009;4(3):e5006. doi: 10.1371/journal.pone.0005006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon S, Taggart CC. Innate host defense functions of secretory leucoprotease inhibitor. Exp Lung Res. 2007;33(10):485–491. doi: 10.1080/01902140701756547. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Mori Y, Hagiwara K, Suzuki T, Sakakibara T, Kikuchi T, Igarashi T, Ebina M, Abe T, Miyazaki J. et al. Increased susceptibility to LPS-induced endotoxin shock in secretory leukoprotease inhibitor (SLPI)-deficient mice. J Exp Med. 2003;197(5):669–674. doi: 10.1084/jem.20021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen PA, Hitt M, Xing Z, Wang J, Haslett C, Riemersma RA, Webb DJ, Kotelevtsev YV, Sallenave JM. Adenoviral gene delivery of elafin and secretory leukocyte protease inhibitor attenuates NF-kappa B-dependent inflammatory responses of human endothelial cells and macrophages to atherogenic stimuli. J Immunol. 2004;172(7):4535–4544. doi: 10.4049/jimmunol.172.7.4535. [DOI] [PubMed] [Google Scholar]
- Marino R, Thuraisingam T, Camateros P, Kanagaratham C, Xu YZ, Henri J, Yang J, He G, Ding A, Radzioch D. Secretory leukocyte protease inhibitor plays an important role in the regulation of allergic asthma in mice. J Immunol. 2011;186(7):4433–4442. doi: 10.4049/jimmunol.1001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odaka C, Mizuochi T, Yang J, Ding A. Murine macrophages produce secretory leukocyte protease inhibitor during clearance of apoptotic cells: implications for resolution of the inflammatory response. J Immunol. 2003;171(3):1507–1514. doi: 10.4049/jimmunol.171.3.1507. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhu J, Sun D, Ding A. Suppression of macrophage responses to bacterial lipopolysaccharide (LPS) by secretory leukocyte protease inhibitor (SLPI) is independent of its anti-protease function. Biochim Biophys Acta. 2005;1745(3):310–317. doi: 10.1016/j.bbamcr.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mahajan MA, Murray A, Samuels HH. NRC-interacting factor 1 is a novel cotransducer that interacts with and regulates the activity of the nuclear hormone receptor coactivator NRC. Mol Cell Biol. 2002;22(19):6883–6894. doi: 10.1128/MCB.22.19.6883-6894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty S, Mahajan MA, Samuels HH. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting Factor NIF-1. J Biol Chem. 2008;283(11):6806–6816. doi: 10.1074/jbc.M706986200. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Alberts R, Fu J, Swertz MA, Lubbers LA, Albers CJ, Jansen RC. Combining microarrays and genetic analysis. Brief Bioinform. 2005;6(2):135–145. doi: 10.1093/bib/6.2.135. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Treg specific genes. The table lists genes that exhibited a differential expression in Treg compared to Th cells. Probeset: probeset ID of microarray hybridization probe, logFC: fold changes of expression level in Treg versus Th as difference of log2 values, pFDR: FDR-corrected p-value, Treg: expression level in Treg cells as log2 value, Th: expression level in Th cells as log2 value.
Table S2 Th specific genes. The table lists genes that exhibited a differential expression in Th compared to Treg cells. Probeset: probeset ID of microarray hybridization probe, logFC: fold changes of expression level in Treg versus Th as difference of log2 values (a negative value represents higher expression in Th compared to Treg cells), pFDR: FDR-corrected p-value, tr: expression level in Treg cells as log2 value, th: expression level in Th cells as log2 value.
Table S3 Genes with known functions in Treg cells. The table lists genes that have been described in the literature to exhibit a functional role in Treg cells. Table S4: Treg functional genes with a higher expression in Tregs. The table lists probesets of genes that were expressed at a higher level in Treg compared to Th cells and which exhibit known Treg functions. Table S5. Genes regulated by cis-eQTL in Treg cells. The table lists probesets from genes that exhibited a cis-eQTL larger or equal to an LRS of 18 in Treg but not in Th cells and which were expressed at least 2-fold higher in Treg cells. Table S6. Genes regulated by cis-eQTL (LRS > = 18) in Th. The table lists probesets from genes that exhibited a cis-eQTL larger or equal to an LRS of 18 in Th but not in Treg cells and which were expressed at least 2-fold higher in Th cells. Table S7. Treg specific genes regulated by trans-eQTL in Treg cells. The table lists probesets from genes that exhibited a trans-eQTL larger or equal to an LRS of 18 in Treg but not in Th cells and which were expressed at least 2-fold higher in Treg cells. Table S8. Th specific genes regulated by trans-eQTL in Th cells. The table lists probesets from genes that exhibited a trans-eQTL larger or equal to an LRS of 18 in Th but not in Treg cells and which were expressed at least 2-fold higher in Th cells. Table S9. Treg functional genes regulated and highly expressed in Treg cells. The table lists probesets of differentially expressed genes in Treg cells with a known Treg function, a high expression signal in Tregs (expression signal > 8) and which are regulated by a trans-eQTL with an LRS > = 14. Table S10: Intervals selected for further QTL analysis. The table lists the QTL intervals from selected Treg and Th cis- and trans-eQTLs that were further analyzed for the presence of candidate regulatory genes. Table S11 Candidate genes located in the F2rl1-QTL interval on chromosome 4. The table lists probesets of possible candidate genes that are located in the QTL interval on chromosome 4 which regulates the expression of F2rl1. Only genes with an expression signal larger than 8 were selected. Table S12 Candidate genes located in the QTL interval on chromosome 2 (autoimmunity to DNA). The table lists probesets of possible candidate genes that are located in the QTL interval on chromosome 2 which regulates autoimmunity to DNA. Only genes expressed in Treg cells (signal > 8 on log2 scale) and exhibiting a cis-eQTL of LRS > = 15 were selected.
Figure S1. Genome-wide graph of cis- and trans-eQTLs in Treg and Th cells. The file shows the genome-wide mapping graphs for Treg cells and the same graph for Th cells overlaid in grey. Figure S2. Genome-wide graph of cis- and trans-eQTLs in Th and Treg cells. The file shows the genome-wide mapping graphs for Th cells and the same graph for Treg cells overlaid in grey. Figure S3, Genome wide eQTL mapping of Lycat transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Lycat transcript in Treg and Th cells. Figure S4, Genome wide eQTL mapping of Prpf3 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Prpf3 transcript in Treg and Th cells. Figure S5, Genome wide eQTL mapping of Nrp1 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Nrp1 transcript in Treg and Th cells. Figure S6, Genome wide eQTL mapping of F2rl1 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the F2rl1 transcript in Treg and Th cells. Figure S7, Genome wide eQTL mapping of Ctla4 transcript in Treg and Th cells. The file contains a graph showing the result of a genome-wide mapping of eQTLs for the Ctla4 transcript in Treg and Th cells.
'Additional Results and Discussion': This file contains additional results on: Trans-eQTLs mapping in Treg and Th cells, the analysis of candidate genes in trans-eQTL intervals, and the analysis of autoimmune traits. Also, the file contains additional discussion points on Treg-specifc genes, the analysis of the interval on chromosome 2 found to regulate expression of Nrp1, analysis of an interval on chromosome × regulating expression of Klrb1f/A630024B12Rik (killer cell lectin-like receptor subfamily B member 1F) and Ctla4 (cytotoxic T-lymphocyte-associated protein 4), the analysis of known Treg functional genes for possible cis- or trans-eQTLs, and the analysis of the QTLs interval chromosome 4 associated with autoimmune traits.