Abstract
Heterotrimeric G proteins are critical signal-transducing molecules controlled by a complex network of regulators. GIV (a.k.a. Girdin) is a unique component of this network and a nonreceptor guanine nucleotide exchange factor (GEF) that functions via a signature motif. GIV's GEF motif is involved in the regulation of critical biological processes such as phosphoinositide 3 kinase (PI3K)-Akt signaling, actin cytoskeleton remodeling, cell migration, and cancer metastasis. Here we investigated how the GEF function of GIV affects the wiring of its signaling pathway to shape different biological responses. Using a structure-guided approach, we designed a battery of GIV mutants with different Gαi-binding and -activating properties and used it to dissect the specific impact of changes in GIV's GEF activity on several cellular responses. In vivo signaling assays revealed a threshold effect of GEF activity for the activation of Akt by GIV in different cell lines and by different stimuli. Akt signaling is minimal at low GEF activity and is sharply increased to reach a maximum above a threshold of GEF activity, suggesting that GIV is a critical signal amplifier and that activation of Akt is ultrasensitive to changes in GIV's GEF activity. A similar threshold dependence was observed for other biological functions promoted by GIV such as remodeling of the actin cytoskeleton and cell migration. This functional characterization of GIV's GEF motif provides insights into the molecular interactions between nonreceptor GEFs and G proteins and the mechanisms that govern this signal transduction pathway.
Keywords: Hill coefficient, ultrasensitivity, switch-like, trimeric G proteins
Cellular responses to external stimuli are shaped by the intricate wiring of different components of signaling cascades. Much like enzymes with their substrates, signaling cascades and other complex biological systems are frequently characterized by hyperbolic responses that follow the Michaelis–Menten equation. However, in other cases the response is sigmoidal, resembling the properties of a cooperative enzyme that are defined by the Hill equation. (1). In these cases, signals that are low trigger little or no response, but once they go above a certain threshold the response increases sharply to reach a maximum, allowing for greater precision in biological control. Goldbeter and Koshland (2) coined the term “ultrasensitivity” to describe this phenomenon observed in complex systems, which is mathematically defined by Hill coefficients (nH) >1 (1, 2).
Heterotrimeric G-protein signaling is controlled by a complex regulatory network of proteins (3–5) of which nonreceptor guanine nucleotide exchange factors (GEFs) are by far the least characterized (6–9). There is virtually no information available about how nonreceptor GEFs influence the wiring of signaling cascades to shape biological responses. We have recently reported that the multidomain protein GIV (a.k.a. Gα-interacting, vesicle-associated protein, Girdin) is a nonreceptor GEF that activates G proteins via an evolutionarily conserved, well-defined motif (8). A major function of GIV's GEF motif is to enhance the activation of the phosphoinositide 3 kinase (PI3K)-Akt pathway in response to different stimuli (8, 10–13). GIV has been shown to be essential for cell migration during a variety of physiologic and pathologic processes, i.e., wound healing (14), macrophage chemotaxis (14), tumor cell migration (8, 10), neuronal development (12), and endothelial cell migration (15), as well as for inhibiting autophagy (16). Importantly, expression of GIV is dysregulated during cancer progression; in poorly metastatic carcinomas, full-length GIV is replaced by a GEF-deficient isoform via alternative splicing whereas, in highly metastatic carcinomas, the full-length isoform of GIV containing the GEF motif is up-regulated, and its expression correlates with decreased patient survival (10, 17). Thus, GIV's GEF function has emerged as a critical element of signaling cascades whose dysregulation is associated with the pathogenesis of human disease. Here we investigated how perturbation of the levels of GIV's GEF activity controls and shapes cell behavior in response to different stimuli by rationally designing GIV mutants that specifically increase or decrease its coupling to G proteins.
Results
Rational Design of Mutants That Either Decrease or Increase Gαi3 Binding and GEF Activity of GIV.
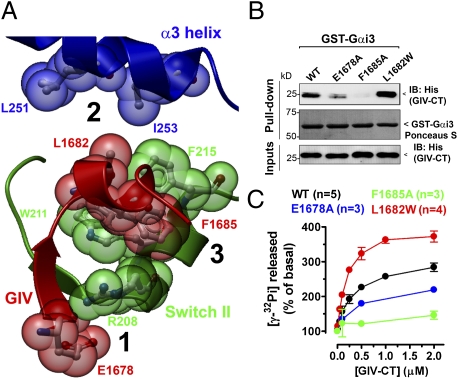
We set out to design GIV mutants with decreased or increased coupling to the G protein. For this, we used a previously described (8) homology model to identify critical structural features of the GIV–Gαi interface (Fig. 1A and Fig. S1). We predicted that (i) mutation of E1678 to A in GIV would negatively affect its binding to Gαi3, and (ii) mutation of L1682 in GIV to a bulkier hydrophobic residue such as W would improve Gαi3 binding. We chose W because this residue is found in the corresponding position of the GIV-like peptide KB-752 that binds to Gαi subunits (18). To test these predictions, we performed in vitro pulldown assays with purified GST-Gαi3 and hexahistidine (His)-GIV-CT (aa 1,660–1,870, which contains GIV's GEF motif). As an internal control, we used a previously described (8) GIV mutant, F1685A (Fig. 1A and Fig. S1), that is deficient in Gαi3 binding. As predicted, His-GIV-CT F1685A failed to bind to GST-Gαi3, and binding of His-GIV-CT E1678A was significantly reduced (50–60% by quantitative immunoblotting) compared with His-GIV-CT WT, whereas binding of His-GIV-CT L1682W was increased (approximately twofold) (Fig. 1B). Similar results were obtained when the same GIV mutants were expressed as full-length proteins in COS-7 cells and used in pulldown assays (Fig. S2).
Fig. 1.
Effect of E1678A and L1682W mutations in GIV on Gαi3 binding and GEF activity. (A) Prediction of critical amino acid contacts between GIV and Gαi3 based on homology modeling. A homology model of Gαi3 in complex with GIV's aa 1,678–1,689 was generated (8) using the structure of the synthetic peptide KB-752 bound to Gαi1 (Protein Data Bank:1Y3A) as a template (18). Gαi3's α3 helix is shown in blue, the “switch II” region in green, and GIV's aa 1,678–1,689 in red. Three relevant structural features of the GIV–Gαi3 interface are displayed (black numbers): 1, positively charged R208 of Gαi3 (previously shown to reduce GIV binding when mutated to L (8) probably interacts with negatively charged E1678 of GIV to form an ionic pair; 2, the hydrophobic pocket formed by Gαi3's L251 and I253 is not filled by GIV's L1682, suggesting that a bulkier hydrophobic residue in this position might create a more favorable coupling with Gαi3; and 3, GIV's F1685 docks between Gαi3's W211 and F215, making an extensive hydrophobic contact previously shown to be critical for maintaining the GIV–Gαi3 interaction (8). A detailed view of the model and the predicted molecular contacts is shown in Fig. S1. (B) The F1685A mutation virtually abolishes His-GIV-CT binding to GST-Gαi3, and the E1678A mutation also impairs binding, whereas the L1682W mutation improves it. His-GIV-CT WT (1.2 μg) or the indicated mutants were incubated with 5 μg GST-Gαi3 preloaded with GDP immobilized on glutathione beads and analyzed by immunoblotting (IB) for His (Top). Equal loading of GST proteins was confirmed by Ponceau S staining (Middle), and equal loading of His-GIV-CT proteins by His immunoblotting of the inputs (Bottom). (C) Activation of His-Gαi3 by His-GIV-CT is dramatically decreased (∼85%) by the F1685A mutation (green) whereas it is significantly increased (∼75%) by the L1682W mutation (red) and moderately decreased (∼40%) by the E1678A mutation (blue) compared with His-GIV-CT WT (black). The steady-state GTPase activity of purified His-Gαi3 (50 nM) was determined in the presence of the indicated concentrations of purified His-GIV-CT WT or mutants by quantification of the amount of [γ-32P]GTP (0.5 μM, ∼50 cpm/fmol) hydrolyzed in 10 min. Data are expressed as percent of GTP hydrolyzed by the G protein alone (0.11 ± 0.02 mol GTP/mol Gαi3 in 10 min). Results are shown as mean ± SEM of the indicated number (n) of experiments.
Next we investigated if the ability of these mutants to activate the G protein correlates with the observed changes in Gαi3 binding. For this we measured the steady-state GTPase activity of purified His-Gαi3 [which directly depends on the rate of nucleotide exchange (19, 20)] in the presence of purified His-GIV-CT WT or mutants. Consistent with the in vitro binding results, the F1685A mutation dramatically reduced (∼85%) GIV's GEF activity whereas the E1678A mutation reduced it only partially (∼40%), and L1682W increased it (∼80%) (Fig. 1C). We conclude that the extent of G-protein activation by the GIV mutants tested correlates well with their binding properties.
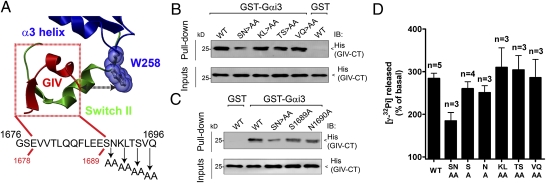
To further characterize GIV mutants with altered coupling to Gαi3, we investigated the role of residues located at the C terminus of GIV's GEF motif. On the basis of our homology model, we hypothesized that residues adjacent to the C terminus of GIV's GEF motif would interact with Gαi3's W258 (Fig. 2A), a residue required for GIV binding (19). To test this, we performed alanine-scanning mutagenesis of GIV's aa 1,689–1,696 by changing two residues at a time to alanine (Fig. 2A) and compared binding of the purified proteins to Gαi3. We found that among the mutants tested only the double mutant S1689A/N1690A (SN>AA) impaired His-GIV-CT binding to GST-Gαi3 (Fig. 2B; 60–70% reduction by quantitative immunoblotting). Single mutations S1689A and N1690A did not decrease binding significantly (Fig. 2C). GIV SN>AA expressed in COS7 cells also had decreased binding to Gαi3 as determined by pulldown (Fig. S3C) assays. Furthermore, we found that the SN>AA mutation did not affect GIV binding to Gαi3 W258F (Fig. S3D), suggesting that S1689/N1690 stabilizes GIV's interaction with Gαi3 only when W258 is intact. Taken together, these results indicate that residues S1689 and N1690 are required to interact efficiently with Gαi3 and suggest that they may make contact with Gαi3's W258.
Fig. 2.
Mutation of two amino acids in the C terminus of GIV's GEF motif impairs Gαi3 binding and activation. (A) Relative location of GIV's GEF motif and Gαi3's W258. Gαi3's α3 helix and the α3/β5 loop where W258 is located are shown in blue, the “switch II” region in green, and GIV (aa 1,678–1,689) in red. GIV's conserved GEF sequence (shown below the model) spans from aa 1,676 to aa 1,696 (8), but only aa 1,678–1,689 (indicated with red lines) are present in the homology model of GIV in complex with Gαi3. No direct contact between GIV's aa 1,678–1,689 and Gαi3's W258 is observed in this model. However, the orientation of GIV and Gαi3 suggests that residues adjacent to the C terminus of GIV's aa 1,678–1,689 might interact with W258 of Gαi3. The design of alanine-scanning mutants for the C-terminal segment (aa 1,689–1,696) of GIV's GEF motif is shown below. (B and C) His-GIV-CT S1689A/N1690A (SN>AA) shows a marked decrease (∼60–70%) in binding to GST-Gαi3 compared with His-GIV-CT WT (B), whereas binding of the His-GIV-CT S1689A or N1690A mutant to GST-Gαi3 is only marginally decreased (∼10%) (C). His-GIV-CT WT did not bind to the GST negative control. His-GIV-CT WT (1.2 μg) or the indicated mutants were incubated with 5 μg GST-Gαi3 or GST preloaded with GDP immobilized on glutathione beads. Bound proteins were analyzed by immunoblotting (IB) as in Fig. 1B. The confirmation of equal loading of GST proteins by Ponceau S staining is shown in Fig. S3. (D) The S1689A/N1690A (SN>AA) double mutation in His-GIV-CT markedly decreases the activation of His-Gαi3 by GIV (∼50%), and the S1689A or N1690A single mutation decreases it only marginally (∼10–15%). The steady-state GTPase activity of purified His-Gαi3 in the presence of 2 μM purified His-GIV-CT WT or the indicated mutants was determined as described in Fig. 1C and expressed as a percentage of GTP hydrolyzed by the G protein alone (0.11 ± 0.02 mol GTP/mol Gαi3 in 10 min).
Next we investigated how the alanine mutants described above affect GIV's GEF activity. We found that activation of Gαi3 by SN>AA was reduced ∼50% (Fig. 2D and Fig. S3E) whereas the remainder of the double-alanine mutants showed no significant differences (Fig. 2D), and the single mutants S1689A and N1690A had only a marginal effect (∼10–20% reduction) (Fig. 2D and Fig. S3E). These results indicate that the GEF activity of the alanine-scanning mutants correlates with their Gαi3 binding and that only GIV SN>AA has a significant binding defect.
Comparison of the results of the GEF assays with Gαi3 binding (Table 1) highlights the fact that the GEF activity of each mutant correlates qualitatively with their respective ability to bind to Gαi3. The GEF activity of the different mutants ranged from 0.17 ± 0.02 to 1.77 ± 0.14 (normalized to WT = 1). In summary, we have designed a battery of GIV mutants with a GEF activity significantly different from WT, which can be used to investigate the role of GIV's GEF function in controlling various biological functions.
Table 1.
Comparative analysis of GEF activity and Gαi3 binding for various GIV mutants
| Relative GEF activity† (mean ± SEM) | Gαi3 binding (range in %) | |
| WT | 1.00 ± 0.03 | 100 |
| F1685A‡ | 0.17 ± 0.01 (***) | 0–5 |
| S1689A/N1690A‡ | 0.51 ± 0.07 (**) | 30–40 |
| E1678A‡ | 0.63 ± 0.01 (***) | 40–50 |
| L1682W‡ | 1.77 ± 0.07 (***) | 180–200 |
| S1689A | 0.90 ± 0.04 (NS) | 85–100 |
| N1690A | 0.84 ± 0.07 (NS) | 85–100 |
| K1691A/L1692A | 1.14 ± 0.15 (NS) | 95–105 |
| T1693A/S1694A | 1.11 ± 0.10 (NS) | 95–105 |
| V1695A/Q1696A | 1.01 ± 0.13 (NS) | 95–105 |
Experiments were performed as described in Figs. 1 and 2 and Fig. S3. GEF activity of the different GIV-CT mutants relative to the activation observed for WT GIV-CT was calculated from the increases in steady-state GTPase activity as described in Experimental Procedures and expressed as mean ± SEM of n = 3–5 independent experiments. Gαi3 binding of the different GIV-CT mutants compared with WT is expressed as the percentage binding compared with WT determined by quantitative immunoblotting in three to four independent experiments as shown in Figs. 1 and 2. **P < 0.01; ***P < 0.001; NS, not significant.
†Concentration of His-GIV-CT wild type and mutants = 0.5 μM.
‡Mutants with significantly altered GEF activity in vitro, which were used in subsequent experiments in living cells.
Activation of Akt in Response to Different Stimuli Is Highly Sensitive to Changes in GIV's GEF Activity.
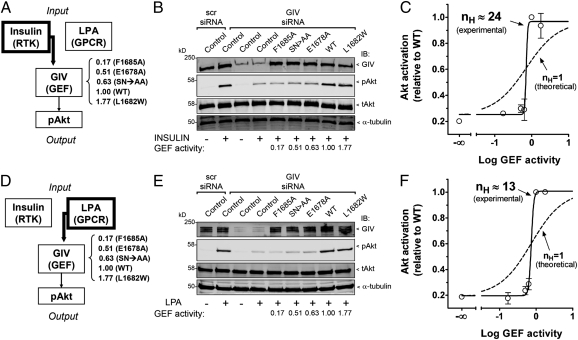
GIV has been previously reported by us (8, 14) and others (11, 12) to be an enhancer of PI3K-Akt signaling. We previously found that activation of Gαi3 by GIV is required for the efficient activation of Akt upon stimulation of receptor tyrosine kinases (RTKs) (8, 10, 14, 16, 19) as well as G-protein-coupled–receptors (GPCRs) (8, 19). To further characterize this function, we generated HeLa cell lines stably expressing similar levels of GIV WT or mutants with significantly different GEF activities (Table 1). These cell lines and control HeLa cells were depleted of endogenous GIV using siRNA, stimulated with either insulin (Fig. 3A) or lysophosphatidic acid (LPA) (Fig. 3D), and analyzed for Akt activation by immunoblotting for phospho-Akt (pAkt). Consistent with our previous reports (8, 16), Akt activation in response to either insulin (Fig. 3 B and C) or LPA (Fig. 3 E and F) was dramatically reduced (∼80%) after depletion of GIV from control HeLa cells. A similar reduction (70–80%, Fig. 3 B and E) was observed for the cells expressing the GEF-deficient GIV F1685A, SN>AA, or E1678A mutants, despite the fact that they display different levels of GEF deficiency in vitro (Table 1). By contrast, Akt activation after stimulation of GIV WT or L1682W cells was high and similar to control HeLa cells expressing endogenous GIV (Fig. 3 B and C). Identical results were obtained when COS7 cells overexpressing the same set of mutants were serum-stimulated (Fig. S4A). Taken together, these results indicate that Akt signaling remains at a minimal, basal level when the GEF activity is low and that it dramatically increases to reach a maximum when the GEF activity rises above a certain threshold. This suggests that Akt activation in response to different stimuli is highly sensitive to changes in the level of GIV's GEF activity.
Fig. 3.
Effect of different levels of GIV's GEF activity on Akt signaling after insulin and LPA stimulation. (A) Schematic representation of signaling pathway and experimental design for B and C. The signaling pathway is divided into three different modules: “input” consists of insulin stimulation of its target RTKs and is maintained constant (insulin = 100 nM); “output” is Akt activation that is monitored by quantification of the levels of pAkt; GIV transmits input signals from insulin-stimulated receptors to activation of Akt (8, 16), which is studied here in response to varying degrees of GEF activity associated with specific mutations. The in vitro GEF activity of the different mutants is indicated on the right. Akt activation was analyzed 5 min after stimulation with 100 nM insulin when activation is maximal and the level of pAkt can be quantified within a reliable range of detection for all mutants studied. (B) Depletion of endogenous GIV (∼80–95%) dramatically impairs insulin-stimulated activation of Akt (pAkt) in control HeLa cells or HeLa cells stably expressing siRNA-resistant GIV-3xFLAG F1685A, SN>AA, or E1678A, but it has virtually no effect in HeLa cells stably expressing siRNA-resistant GIV-3xFLAG WT or L1682W compared with HeLa cells treated with scramble (scr) siRNA. HeLa cell lines were treated with control (scr) or GIV siRNA, serum-starved (0.2% FBS, overnight), and stimulated with insulin (100 nM, 5 min) as indicated. Cell lysates were analyzed for GIV, pAkt, total Akt (tAkt), and α-tubulin by immunoblotting (IB). The in vitro GEF activity of each mutant (Table 1) is indicated below the corresponding lanes for reference. One experiment representative of three is shown. (C) Akt activation in response to insulin stimulation is highly sensitive to changes in GIV's GEF activity. Akt activation (y axis) for the mutants investigated in B was quantified by infrared immunoblotting as described in Experimental Procedures (○) and plotted as a function of GEF activity corresponding to each mutant (x axis). The data were fitted to the four-parameter logistic equation using Prism 4.0 (solid line), resulting in an nH value of 24. The dashed line represents a theoretical nonultrasensitive response (nH = 1). Results are average ± SEM of three independent experiments. (D) The signaling pathway studied and experimental design are identical to those in A except that the “input” component consists of LPA stimulation (10 μM, 10 min). (E) Depletion of endogenous GIV (∼80–95%) dramatically impairs LPA-stimulated activation of Akt (pAkt) in control HeLa cells or in HeLa cells stably expressing siRNA-resistant GIV-3xFLAG F1685A, SN>AA, or E1678A but has virtually no effect in HeLa cells stably expressing siRNA-resistant GIV-3xFLAG WT or L1682W. Cells were treated as in B except that stimulation was with LPA (10 μM, 10 min). One experiment of two with virtually identical results is shown. (F) Akt activation in response to LPA stimulation is highly sensitive to changes in GIV's GEF activity. Data from E was analyzed exactly as for C. nH was 13. The dashed line represents a theoretical non-ultrasensitive response (nH = 1). Results are shown as average ± SD of two independent experiments.
The sensitivity of a biological response to changes in a certain parameter or stimulus can be mathematically determined by calculating the Hill coefficient (nH) (1, 2), such that nH values >1 indicate hypersensitivity. We calculated the apparent nH for Akt activation after insulin or LPA stimulation as a function of changes in GIV's GEF activity by analyzing the experimental data described in Fig. 3 B and E using the Hill equation. The curve fits of these analyses are shown in Fig. 3C (insulin) and Fig. 3F (LPA), which had nH values of 24 and 13, respectively, and revealed a threshold effect: When GIV's GEF activity is more than ∼0.6, there is a very sharp increase in Akt activation. Similar results were obtained when activation of Akt was analyzed in COS7 cells overexpressing similar levels of GIV mutants (Fig. S4B). Next we investigated Akt activation as a function of the number of GIV copies expressed instead of the specific GEF activity. Importantly, we found that expression of increasing amounts of GIV WT in COS7 cells promoted a graded increase in Akt activation rather than a threshold response (Fig. S4C), suggesting that the level of GIV expression does not promote an ultrasensitive response. Taken together, these results indicate that Akt signaling is ultrasensitive to GIV's GEF activity but not to the level of GIV expression, switching sharply from a minimum to a maximum above a certain threshold of GEF activity.
Threshold Response to GIV's GEF Activity Controls Stress Fiber Formation and Cell Migration.
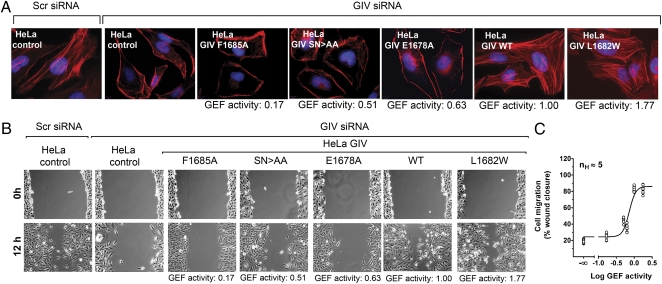
We previously reported that activation of Gαi3 by GIV is also required for remodeling of the actin cytoskeleton and efficient cell migration (8, 19) after stimulation by growth factors present in serum. We hypothesized that these responses might also be ultrasensitive to GIV's GEF activity and be promoted only above a certain threshold of activity. We found that this is likely the case because stress fibers are prominent in serum-stimulated control HeLa cells or in HeLa cells stably expressing GIV constructs with GEF activity ≥1 (Fig. 4A), whereas they are virtually absent in control HeLa cells depleted of endogenous GIV or in HeLa cells stably expressing GIV constructs with GEF activity ≤0.63 (Fig. 4A). A similar trend was observed when we assessed cell migration after scratch wounding (14, 21). Wound closure was greatly impaired for HeLa cells exclusively expressing GIV constructs with GEF activity ≤0.63 whereas, for cells expressing GIV constructs with GEF activity ≥1, it was as efficient as in control HeLa cells expressing endogenous GIV (Fig. 4B). When wound closure was quantified and plotted as a function of GEF activity, analysis of the curve fit revealed an ultrasensitive behavior with an nH of 5 (Fig. 4C). From these results we conclude that, like Akt activation, actin cytoskeleton remodeling and cell migration are highly sensitive to changes in GIV's GEF activity.
Fig. 4.
Effect of GIV's GEF levels on actin stress fiber formation and cell migration. (A) Vector-transfected HeLa cells (HeLa control) treated with scramble (scr) siRNA show prominent stress fibers in the presence of serum, whereas those treated with GIV siRNA show a dramatic decrease in stress fibers. A similar reduction is observed in HeLa cells stably expressing siRNA-resistant GIV-3xFLAG F1685A, SN>AA, or E1678A, but not in those expressing GIV-3xFLAG WT or L1682W after siRNA depletion of GIV. F-actin (red) and DNA (blue) were visualized by fluorescence after costaining with phalliodin-Texas red and DAPI. (B) Control HeLa cells treated with scr siRNA migrate efficiently, and siRNA depletion of GIV dramatically impairs cell migration in control HeLa cells or in those stably expressing siRNA-resistant GIV-3xFLAG F1685A, SN>AA, or E1678A, but not in those expressing GIV-3xFLAG WT or L1682W. Monolayers of HeLa cells were monitored after scratch wounding (0 h) or 12 h later. (C) Cell migration is highly sensitive to changes in GIV's GEF activity. Cell migration (y axis) from B was quantified as a percentage of wound closure as described in Experimental Procedures, plotted as a function of the GEF activity (x axis) and analyzed as described in Fig. 3C. The nH was 5. Each data point represents the percentage wound closure for one field of 9–12 from three independent experiments.
Discussion
Our data reveal a threshold effect and ultrasensitive behavior of biological functions including Akt signaling, actin cytoskeleton remodeling, and cell migration in response to changes in the level of GIV's GEF activity when cells are exposed to different stimuli. This represents an example of ultrasentitive behavior in heterotrimeric G-protein signaling determined by the GEF activity of a nonreceptor protein and provides insights into how this class of G-protein regulators are assembled within signaling cascades to shape biological responses. We have exploited our previous findings on GIV's conserved GEF sequence (8) and its binding sites on the G protein (8, 19) to rationally design and characterize specific GIV mutants that impair or enhance its functional coupling to Gαi3 in vitro and in living cells. From the general standpoint, these findings provide insights into how nonreceptor GEFs work, as well as into the role of GIV's GEF function in controlling cell signaling and migration (8, 10, 13), which is particularly relevant in the context of cancer metastasis (10, 17).
A major advance provided by this work is the characterization of structural elements of a nonreceptor GEF for heterotrimeric G proteins that determine its biological functions. Although this type of study has been performed before for other G-protein regulators such as RGS and GoLoco/GPR proteins (22–24), nonreceptor GEFs have remained elusive due to the lack of knowledge about their specific functional domains or motifs. Our in vitro characterization of mutants in GIV's GEF motif shows a direct correlation between the GEF activity of these mutants and their ability to interact with Gαi3. Notably, none of the mutants studied was capable of affecting GEF activity without altering G-protein binding. We propose that GIV works as a GEF by binding to Gαi3 and stabilizing a conformation of the G protein that favors nucleotide exchange. This is supported by previous observations reported in the structural analysis of the complex between Gαi1 and the KB-752 peptide (18), a synthetic GEF peptide that shares significant sequence similarity with the GEF motif of GIV (8). In that work, the authors showed that KB-752 binding induces a conformational change in the G protein that repositions certain structural elements believed to secure the nucleotide in its binding pocket and thereby favors nucleotide exchange. With regard to how GIV docks on Gαi3 to stabilize this conformation that favors nucleotide exchange, our current work suggests that, in addition to making several contacts with residues in the α3 helix and the switch II of Gαi3 as predicted by our previously described homology model (8), GIV uses residues S1689 and N1690 in the C-terminal region of its GEF motif to extend the interaction surface to the adjacent α3/β5 loop and make contact with W258. To precisely define the nature of this interaction and identify other possible contacts established between the GEF and G protein will require analysis of the structure of the GIV/G-protein complex at atomic resolution.
Our work reveals how changes in GIV's GEF activity within a narrow range around a certain threshold sharply promote or inhibit signal transduction. Since Goldbeter and Koshland (2) coined the term “ultrasensitivity” (also known as “switch-like behavior” or “all-or-none responses”) to define this type of behavior, multiple examples have been described. These include the MAPK cascade in Xenopus oocyte maturation (1) or growth factor signaling in mammalian cells (25), SREBP-2 translocation upon changes in cholesterol levels in the endoplasmic reticulum (26), activation of the GTPase Rap1 in response to cannabinoid stimulation (27), and JNK-signaling cascades in Xenopus oocytes and mammalian cells (28), to name a few. Although some responses mediated by heterotrimeric G proteins have been shown to be ultrasensitive to the dose of certain GPCR ligands, e.g., cannabinoids (27), our findings represent an example of how this type of behavior can be controlled by a nonreceptor GEF for heterotrimeric G proteins. It is important to note that the nH values that we report are very high (>12), indicating that this response is highly ultrasensitive and approaches the behavior of an all-or-none decision switch. It remains to be investigated whether GIV's other known biological functions [e.g., in autophagy (16)] are ultrasensitive to its GEF activity.
Like some other examples of ultrasensitive behavior, the precise molecular mechanisms by which ultrasensitivity to GIV's GEF activity arises remain unclear. It is possible that it is a consequence of the fact that GIV binding to Gαi not only promotes G-protein activation via its GEF function, but also simultaneously blocks the action of guanine nucleotide dissociation inhibitors (GDIs) such as Gβγ (8) or activator of G-protein signaling 3 (AGS3, a GoLoco/GPR motif protein) (16, 24) by molecular displacement. This is in keeping with a recent report by Lipshtat and coworkers indicating that a general mechanism by which GTPase-mediated signaling becomes ultrasensitive is when the increased action of GEFs is simultaneously accompanied by the decreased action of GDIs (27). Thus, it is tempting to speculate that the ultrasensitive response to GIV's GEF activity is a consequence of this dual effect of activating the G protein and displacing its GDIs; however, we cannot rule out other possibilities such as the existence of regulatory feedback loops and/or synergy with other components of this pathway. For example, it is possible that the ultrasensitive Akt response arises from the cooperative action of GIV's GEF activity and the direct activation of PI3K by tyrosine phosphorylated GIV (29).
What is the biological significance of the ultrasensitive response to GIV's GEF activity? Our previous findings (8, 10, 14, 19) indicate that GIV's GEF function is required to amplify signaling and initiate cell migration. The ultrasensitive behavior described in this work suggests that this amplification can be subject to precise regulation. When close to a certain threshold, minimal changes in GIV's GEF activity can make the cell switch sharply between two states. This suggests that altering GIV's GEF activity in vivo can potentially trigger or blunt cellular responses very efficiently and serve as a regulatory mechanism to control GIV's function in promoting cancer metastasis (10, 13, 17). We previously proposed (13) that GIV works as a rheostat that fine-tunes signaling during cancer progression in that increasing the number of GIV copies gradually increases the amplitude of prometastatic responses such as PI3K-Akt signaling and cell migration (10). This idea of a gradual, non-ultrasensitive response to the level of GIV expression is supported by our results presented here (Fig. S4C) and by previous observations (29) showing that Akt activation is gradually increased in cells of isogenic background from the 21T series that represent different stages of cancer progression and naturally express increasing levels of GIV. However, for a given number of GIV copies, Akt signaing is ultrasensitive to the level of GEF activity. Thus, we propose that the number of GIV copies determines the global amplitude of the response whereas GIV's GEF function serves as the critical regulatory element that shapes the response as an “all-or-none” switch. The molecular mechanisms by which GIV's GEF activity is regulated are still poorly understood but might involve its translocation to the proximity of its target G protein on membranes (10) and/or posttranslational modifications (e.g., phosphorylation) (21).
Another scenario in which the ultrasensitive response to GIV's GEF activity may be important is in the development of anti-metastatic agents. We have previously proposed that disruption of the interface between Gαi and GIV's GEF motif with small molecules might be therapeutically relevant for the treatment of cancer metastasis by abolishing enhanced PI3K-Akt signaling and cell migration (8, 10, 17, 19). It is conceivable that a drug capable of partially reducing GIV's GEF activity in vitro would still be very efficient in vivo if it is sufficient to shift GIV's GEF activity below the critical threshold and therefore completely blunt the GIV-dependent enhancement of PI3K-Akt signaling and cell migration associated with cancer metastasis.
Experimental Procedures
A detailed description of all of the experimental procedures can be found in the SI Experimental Procedures.
In Vitro Protein-Binding Assays and Quantitative Immunoblotting.
Protein binding was determined by pulldown assays as described previously (8, 14, 19), and immunoblot quantification was performed by infrared imaging following the manufacturer's protocols using an Odyssey imaging system (Li-Cor Biosciences).
Generation of Stable Cell Lines.
HeLa cell lines stably expressing p3xFLAG-CMV-14-GIV (GIV-3xFLAG) WT or mutants were selected after transfection in the presence of G418 (500 μg/mL) for 6 wk as previously described (8, 10). For each mutant, two different clones were investigated for each assay with similar results. GIV-3xFLAG expression was ∼2× endogenous levels.
Steady-State GTPase Assay.
This assay was performed as described previously (8, 16, 19), and a detailed description is available in SI Experimental Procedures. The relative GEF activity of the different GIV mutants compared with wild-type (Table 1) was calculated using the formula GEFmut − 100/GEFWT − 100, where GEFmut and GEFWT are the G-protein activities in the presence of different His-GIV-CT mutants or wild type, respectively, expressed as the percentage of the steady-state GTPase activity of Gαi3 alone. The His-Gαi3 protein used in this work was >95% functional on the basis of radiolabeled GTPγS binding and trypsin protection assays (19).
Data Analysis and Other Methods.
Experiments were repeated at least three times, and results were presented either as one representative experiment or as average ± SEM. Statistical significance was assessed with the Student's t test. Data fitting the four-parameter logistic equation [Y = Min + (Max − Min)/(1 + 10exp((LogEC50 − X)*nH))] and the subsequent determination of nH (Hill slope) was performed using the sigmoidal dose–response (variable slope without constraints) of Prism 4.0 (GraphPad Software). In this equation, Y is either relative Akt activation (Fig. 3 and Fig. S4) or cell migration (Fig. 4), and X is the logarithm of GEF activity, which was considered negligible (log GIV's GEF → −∞) for control HeLa cells depleted of endogenous GIV. Protein structure analysis and visualization were performed using ICM Browser Pro software (Molsoft).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants CA100768 and DKI7780 (to M.G.F.). M.G.-M. was supported by Susan G. Komen Postdoctoral Fellowship KG080079. P.G. was supported by the Career Awards for Medical Scientists (CAMS) program of the Burroughs Wellcome Fund and a Research Scholar Award of the American Gastroenterology Association.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120538109/-/DCSupplemental.
References
- 1.Ferrell JE., Jr Tripping the switch fantastic: How a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 2.Goldbeter A, Koshland DE., Jr An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilman AG. G proteins: Transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 4.Sato M, Blumer JB, Simon V, Lanier SM. Accessory proteins for G proteins: Partners in signaling. Annu Rev Pharmacol Toxicol. 2006;46:151–187. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- 5.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 6.Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J Biol Chem. 2003;278:8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- 7.Lee MJ, Dohlman HG. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol. 2008;18:211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proc Natl Acad Sci USA. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cismowski MJ, et al. Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. J Biol Chem. 2000;275:23421–23424. doi: 10.1074/jbc.C000322200. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh P, et al. A Galphai-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol Biol Cell. 2010;21:2338–2354. doi: 10.1091/mbc.E10-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anai M, et al. A novel protein kinase B (PKB)/AKT-binding protein enhances PKB kinase activity and regulates DNA synthesis. J Biol Chem. 2005;280:18525–18535. doi: 10.1074/jbc.M500586200. [DOI] [PubMed] [Google Scholar]
- 12.Kim JY, et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh P, Garcia-Marcos M, Farquhar MG. GIV/Girdin is a rheostat that fine-tunes growth factor signals during tumor progression. Cell Adh Migr. 2011;5:237–248. doi: 10.4161/cam.5.3.15909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. J Cell Biol. 2008;182:381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura T, et al. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat Cell Biol. 2008;10:329–337. doi: 10.1038/ncb1695. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol Biol Cell. 2011;22:673–686. doi: 10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Marcos M, et al. Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. FASEB J. 2011;25:590–599. doi: 10.1096/fj.10-167304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston CA, et al. Structure of Galpha(i1) bound to a GDP-selective peptide provides insight into guanine nucleotide exchange. Structure. 2005;13:1069–1080. doi: 10.1016/j.str.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Marcos M, Ghosh P, Ear J, Farquhar MG. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. J Biol Chem. 2010;285:12765–12777. doi: 10.1074/jbc.M109.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay S, Ross EM. Quench-flow kinetic measurement of individual reactions of G-protein-catalyzed GTPase cycle. Methods Enzymol. 2002;344:350–369. doi: 10.1016/s0076-6879(02)44727-1. [DOI] [PubMed] [Google Scholar]
- 21.Enomoto A, et al. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev Cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Druey KM, Kehrl JH. Inhibition of regulator of G protein signaling function by two mutant RGS4 proteins. Proc Natl Acad Sci USA. 1997;94:12851–12856. doi: 10.1073/pnas.94.24.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- 24.Peterson YK, Hazard S, III, Graber SG, Lanier SM. Identification of structural features in the G-protein regulatory motif required for regulation of heterotrimeric G-proteins. J Biol Chem. 2002;277:6767–6770. doi: 10.1074/jbc.C100699200. [DOI] [PubMed] [Google Scholar]
- 25.Harding A, Tian T, Westbury E, Frische E, Hancock JF. Subcellular localization determines MAP kinase signal output. Curr Biol. 2005;15:869–873. doi: 10.1016/j.cub.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipshtat A, Jayaraman G, He JC, Iyengar R. Design of versatile biochemical switches that respond to amplitude, duration, and spatial cues. Proc Natl Acad Sci USA. 2010;107:1247–1252. doi: 10.1073/pnas.0908647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagowski CP, Besser J, Frey CR, Ferrell JE., Jr The JNK cascade as a biochemical switch in mammalian cells: Ultrasensitive and all-or-none responses. Curr Biol. 2003;13:315–320. doi: 10.1016/s0960-9822(03)00083-6. [DOI] [PubMed] [Google Scholar]
- 29.Lin C, et al. Tyrosine phosphorylation of the Gα-interacting protein GIV promotes activation of phosphoinositide 3-kinase during cell migration. Sci Signal. 2011;4:ra64. doi: 10.1126/scisignal.2002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.