Abstract
In eukaryotes, ubiquitination is an important posttranslational process achieved through a cascade of ubiquitin-activating (E1), conjugating (E2), and ligase (E3) enzymes. Many pathogenic bacteria deliver virulence factors into the host cell that function as E3 ligases. How these bacterial “Trojan horses” integrate into the eukaryotic ubiquitin system has remained a mystery. Here we report crystal structures of two bacterial E3s, Salmonella SopA and Escherichia coli NleL, both in complex with human E2 UbcH7. These structures represent two distinct conformational states of the bacterial E3s, supporting the necessary structural rearrangements associated with ubiquitin transfer. The E2-interacting surface of SopA and NleL has little similarity to those of eukaryotic E3s. However, both bacterial E3s bind to the canonical surface of E2 that normally interacts with eukaryotic E3s. Furthermore, we show that a glutamate residue on E3 is involved in catalyzing ubiquitin transfer from E3 to the substrate, but not from E2 to E3. Together, these results provide mechanistic insights into the ubiquitin pathway and a framework for understanding molecular mimicry in bacterial pathogenesis.
Keywords: crystallography, microbe-host interaction, NleL, SopA, ubiquitination
In order to coexist with eukaryotic hosts, bacterial pathogens have evolved mechanisms to alter the physiological and immune response of the host. For example, many gram-negative bacteria use the type III or type IV secretion system to deliver a large number of virulence factors into the host cell cytosol. These virulence factors, also known as effector proteins, play vital roles in bacterial attachment, entry, and survival. They modulate numerous host cellular functions such as cytoskeleton dynamics, gene expression, and posttranslational modification. Understanding how bacterial virulence factors exert their effects on host cell processes will offer new insights into eukaryotic host cell physiology and possibly lead to new approaches to antimicrobial therapy.
The ubiquitin (Ub) pathway is one of the host systems that is hijacked by bacterial pathogens (1, 2). In eukaryotes ubiquitination is central to many processes such as cell cycle, immune response, and DNA damage tolerance (3–6). The covalent attachment of Ub to a target protein requires the sequential actions of Ub-activating enzymes (E1), conjugating enzymes (E2), and ligases (E3). An Ub molecule is first attached to E1 through a thioester bond upon ATP hydrolysis and is subsequently transferred to the active site cysteine residue in E2. The E3 ligase is often needed to transfer Ub from E2 to a protein substrate. There are two major types of E3 ligases: the RING E3s that function as scaffold to bring E2 and substrate into proximity and HECT E3s that form a thioester intermediate with the Ub before transferring it to the substrate. Ubiquitination is absent in prokaryotes. However, some pathogenic bacteria deliver E3 ligases that interfere with the eukaryotic Ub pathway. These bacterial-encoded E3 ligases have little sequence identity to eukaryotic E3s and thus their mechanism of action has remained unknown.
Two homologous proteins, SopA from Salmonella and NleL from Enterohemorrhagic Escherichia coli, have been shown to exhibit E3 ligase activity (7–10). SopA regulates an intestinal inflammation response induced by Salmonella (8) while NleL is involved in pedestal formation in the host cell (10). Both SopA and NleL contain a cysteine residue near the C terminus that forms a transient thioester bond with Ub (7–10). Similar to eukaryotic HECT E3s, SopA and NleL function with a subgroup of E2 that contain a conserved phenylalanine residue (7, 8). The crystal structures of SopA and NleL show a common fold consisting of three domains: a β-helix domain that might serve as the substrate binding site, a central elongated domain that resembles the N-lobe of a eukaryotic HECT domain, and a C-terminal globular domain that is reminiscent of the C-lobe of HECT E3s (7, 9). Multiple structures of SopA and NleL differ in the orientation of their C-lobe relative to the other two domains, indicating that they possess the conformational flexibility characteristic of HECT E3 ligases (7, 9). However, the molecular surfaces of SopA and NleL bear no similarity to that of HECT E3 ligases. Therefore, it is unclear how these bacterial E3s interact with eukaryotic E2s to achieve ubiquitination. In this study we have determined crystal structures of SopA and NleL in complex with a human E2 to reveal the molecular details of this important microbe-host cell interaction.
Results
Crystal Structure of the UbcH7/SopA Complex.
SopA functions with a number of E2s including UbcH5a, UbcH5c, and UbcH7 (8). We purified a complex consisting of UbcH7 and a stable fragment of SopA (residues 163–782) and obtained crystals that diffracted to 3.3 Å resolution. The structure was determined by molecular replacement and the final model was refined to an R-factor of 21.1% and Rfree of 27.5% (Table S1).
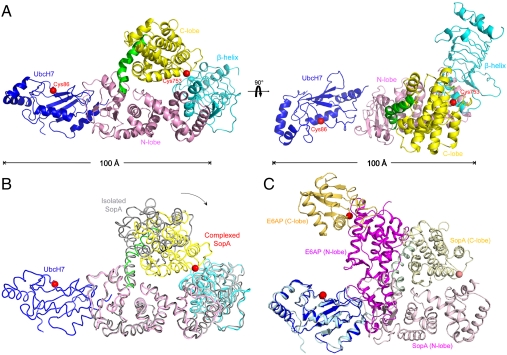
The crystal contains two E2/E3 complexes per asymmetric unit (AU), with a root-mean-square-deviation (rmsd) of 1.2 Å over 769 Cα positions, indicating no significant conformational variation between the two complexes. The overall structure of the complex has an elongated shape with UbcH7 and the N- and C-lobes of SopA lying in a plane; the β-helix domain protrudes out of plane by an angle of approximately 120° (Fig. 1A). Compared to the isolated SopA structure (9), the structures of each individual domain within the complex is essentially unchanged. Conformational differences occur in the relative orientations of the domains, in particular the C-lobe: In the E2/E3 complex, the C-lobe is displaced toward the β-helix domain (Fig. 1B).
Fig. 1.
Crystal structure of the UbcH7/SopA complex. (A) Ribbon representation of the UbcH7 (blue) in complex with SopA (β-helix domain, cyan; N-lobe, pink; C-lobe, yellow; hinge helix, green). The two views differ by a 90° rotation. (B) Superposition of the isolated SopA (gray, PDB code: 2QYU) and SopA in the complex via the N-lobes (PDB code: 3SY2). An approximately 37° rotation of the C-lobes was observed about the hinge helix. (C) Comparing UbcH7/SopA and UbcH7/E6AP by superposition of the E2 molecules (blue and cyan). SopA is rendered in pink (N-lobe), yellow (C-lobe), and green (hinge helix). E6AP is shown in magenta (N-lobe) and orange (C-lobe). The catalytic cysteine residues are represented as red spheres.
On the basis of eukaryotic HECT E3 structural studies one would predict that UbcH7 binds close to where the β-helix domain is attached to the N-lobe (9). However, the crystal structure shows that UbcH7 binds at the opposite end of the N-lobe, approximately 50 Å away from where E2 binds in eukaryotic HECT E3s, and has a different orientation with respect to the long axis of the N-lobe (Fig. 1C). These differences can be best appreciated by comparing the structures through superposition of the E2, UbcH7 (Fig. 1C). In this superposition, the bacterial E3 SopA and eukaryotic E3 E6AP are orthogonal to each other (Fig. 1C).
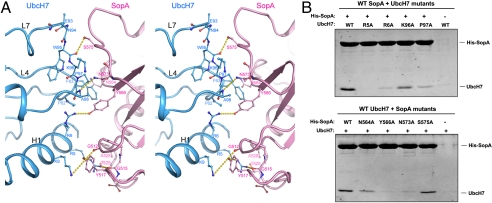
Although UbcH7 binds on the opposite sides of SopA and E6AP, it does so using the same surface in both interactions (Fig. 1C). This surface, comprising residues on the N-terminal helix (H1), loop 4, and loop 7 (L4 and L7), is the canonical E3 interaction surface through which UbcH7 interacts with both HECT and RING E3 ligases (11–23). A highly conserved phenylalanine residue in L4 (F63 in UbcH7) seems to be the determinant for HECT E3 recognition. Mutations at this position abolished the E2-HECT interaction, but had no effect on E2-RING complex formation (14, 24). In the UbcH7/SopA structure, three polar residues from the H1 helix of UbcH7 (R5, R6, and K9) form hydrogen bonds with SopA residues G515, Y566, and G512, respectively (Fig. 2A). F63 makes van der Waals interactions with SopA N564 and Y566. Additional contacts are made by residues on the L7 loop: K96 and P97 form van der Waals contacts to SopA N573 and Y566, and main chain atoms of W95 and A98 form hydrogen bonds with SopA S575 and N573, respectively. Previously we reported that replacing F63 with an alanine in UbcH7 severely reduces its affinity for SopA (9). Here we further tested the E2/E3 interface using mutagenesis and pulldown experiments (Fig. 2B and Fig. S1). Consistent with the crystal structure, UbcH7 mutants R5A did not bind to SopA in the pulldown experiment; and mutating R6, K96 and P97 to alanine lowered the affinity of the complex. Mutations on SopA showed that Y566, N573, and N564 play important roles in stabilizing the UbcH7/SopA complex. Thermal denaturation measurements indicate that these mutants are as stable as the wild-type proteins (Fig. S2).
Fig. 2.
The UbcH7/SopA interface. (A) Stereo view of the interactions between SopA N-lobe (pink) and UbcH7 (cyan). Residues making protein-protein contacts are shown in stick model. Hydrogen bonds are indicated by yellow dashed lines. (B) Testing the interface by mutagenesis. Top, His-tagged wt SopA was incubated with untagged wt or mutant UbcH7. Bottom, wt UbcH7 were incubated with His-tagged wt and mutant SopA and immobilized with Co2+ beads. Proteins immobilized by Co2+ beads were visualized by SDS-PAGE Coomassie staining.
Crystal Structure of the UbcH7/NleL Complex.
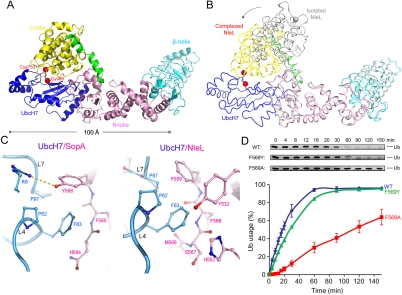
HECT E3 ligases are known to undergo large-scale conformational changes to transfer Ub from E2 to the target substrate (11, 21, 25, 26). Fortuitously, an alternative conformation of the E2/E3 complex appears to have occurred in the UbcH7/NleL structure (Fig. 3A). Superposition analysis shows that similar to SopA, individual domains of NleL maintained the same structure upon E2 binding. However, in the UbcH7/NleL complex the C-lobe of NleL has adopted an entirely different orientation than in the isolated NleL structures (Fig. 3B). Notably, the orientation of the C-lobe in the UbcH7/NleL complex is also different than the C-lobe in the UbcH7/SopA complex (Figs. 1 and 3). For this reason we think that the specific orientation taken by the C-lobe is not a direct result of UbcH7 binding, but rather a reflection of the range of conformations possible for the C-lobe because it is attached by a flexible stalk.
Fig. 3.
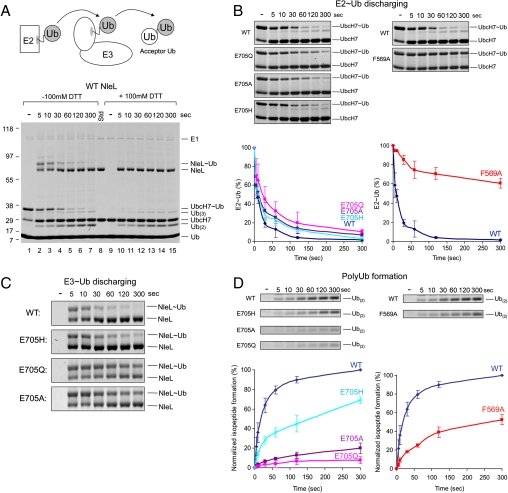
Crystal structure of UbcH7/NleL complex. (A) Ribbon representation of the UbcH7/NleL complex, same color theme as in Fig. 1. Catalytic cysteine residues are represented as red spheres. (B) Superposition of the isolated NleL (gray, PDB code: 3NB2 chain C) and NleL in the complex (colored as in panel A). An approximately 165° rotation of the C-lobes was observed about the hinge helix, and the catalytic cysteine residue C753 (gray and red spheres) is displaced by 45 Å. (C) F63 of UbcH7 interacts with different residues in SopA (left) and in NleL (right). (D) The ligase activities of wt and mutant NleL in multiturnover experiments, quantified by the percentages of mono-Ub usage. The error bars show the standard deviation from three independent experiments. Gel insets of the mono-Ub bands are also shown.
E2 binds to the N-lobe of NleL at a region equivalent to the E2-binding site in SopA. As the sequence conservation between SopA and NleL is low (26% identity), the atomic details of the E2/E3 interfaces are different. For example, in forming the UbcH7/SopA complex, the H-bond between UbcH7 R5 and SopA Y517 is essential; mutating R5 to alanine abolished binding (Fig. 2). But in the structure of UbcH7/NleL this interaction is absent. The residue corresponding to Y517 is a cysteine and the side chain of UbcH7 R5 is disordered, suggesting that it is not engaged in any specific contacts. In both structures, F63 makes critical intermolecular contacts, but with different sets of residues (Fig. 3C). In SopA, Y566 forms van der Waals contact with F63; in NleL, F569 occupies the same position. In an in vitro multiturnover experiment, NleL catalyzes formation of unanchored polyubiquitin (polyUb) chains (7). Compared with wt NleL, the F569A mutant showed strong defects in the rates of ubiquitination, determined by quantifying the amount of the free ubiquitin remaining at each time point (Fig. 3D). In contrast, mutating F569 to tyrosine as found in SopA had no effect in the ligase activity of NleL (Fig. 3D), suggesting that an aromatic side chain at this position is necessary to stabilize the E2/E3 interaction.
In addition to contacting the N-lobe, E2 also contacts the C-lobe of NleL, mainly through the active site cysteines. The Cα atoms of the two cysteines are approximately 7 Å apart, the side chains are orientated toward each other, and electron density indicates the formation of a disulfide bond (Fig. S3). Beyond this covalent interaction, no hydrogen bonding or van der Waals contacts are observed between E2 and the C-lobe of NleL. Indeed, residues of UbcH7 on this C-lobe-facing interface have high temperature factors and poor electron density for side chains. On the NleL C-lobe side of the interface electron density was extremely poor. These observations suggest that the interface between the C-lobe and UbcH7 is labile. It is likely that the disulfide bond observed in the crystal structure will not generally occur under the reducing conditions inside a cell. However, as we will discuss further below, the ability of these two cysteines to approach each other is undoubtedly essential for transthiolation.
Identifying Catalytic Residues.
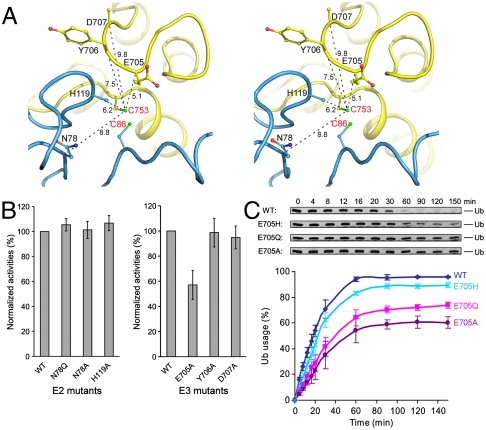
In the Ub transfer reaction one would expect catalytic residues to function either as a general base to deprotonate the attacking nucleophile or as a general acid to stabilize the negative charge on the tetrahedral intermediate. Previously, two residues of E2 were shown to be important in E2 catalyzed isopeptide bond synthesis, but not in HECT ligase catalyzed Ub conjugation (21, 27–29). The “catalytic asparagine” (N77 in UbcH5a) is important in stabilizing the oxyanion intermediate during RING E3 mediated conjugation (27); and D117 in UbcH5b (or D127 in UbcH9) facilitates efficient Ub and SUMO transfer from E2 to lysine residues of substrates (15, 17, 21, 28). In the UbcH7/NleL structure, the corresponding residues (N78 and H119) are located within 10 Å of the E3 catalytic cysteine C753 (Fig. 4A). Substitution of these residues by alanine or glutamine caused no defects in polyUb chain formation (Fig. 4B, Left), consistent with NleL functioning through a mechanism similar to that of eukaryotic HECT instead of RING E3s.
Fig. 4.
Identify catalytic residues. (A) Stereo view of the residues in the vicinity of NleL C753 (labeled in red), with the Cα distances indicated in Å. (B) The ligase activities of UbcH7 and NleL mutants assayed under multiturnover conditions, normalized with that of the wt proteins. (C) Activities of NleL mutants. The percentages of mono-Ub usage were calculated and plotted at indicated time points. The error bars show the standard deviation from three independent experiments.
In the vicinity of the active site cysteine of NleL we focused our attention on three polar/charged residues (E705, Y706, and D707) in NleL because they are also present in SopA as E705, H706, and D707, respectively (Fig. 4A). We tested the catalytic functions of these residues by alanine substitution. Assayed under multiple-turnover conditions, only the E705A mutant altered the time course of the free ubiquitin band depletion (Fig. 4B, Right). At the 705 position mutation to glutamine (E705Q) also altered the time course of the reaction while the E705H mutant was very similar to the wild-type protein (Fig. 4C). From this assay we conclude that E705 is involved in ligase activity. However, because free ubiquitin depletion in this assay depends on multiple steps, in order to address whether the mutations of this site actually influence catalysis we had to develop an assay that focused only on reactions catalyzed by the E3 ligase.
HECT catalyzed ubiquitination involves two distinct Ub transferring steps: from E2 to E3 (transthiolation) and from E3 to the substrate (isopeptide bond synthesis) (Fig. 5A). To investigate the step in which E705 is involved, single turnover experiments were carried out to quantify the effects of mutations at each step (Fig. 5A). In this assay, UbcH7 was precharged with Ub by E1 before mixing with NleL (Fig. 5A, lane 1). E1 was then inactivated by EDTA to prevent recharging of E2. NleL was added to the UbcH7 ∼ Ub mixtures to initiate transthiolation. The reaction was quenched with SDS-PAGE sample buffer at indicated times and analyzed in nonreducing and reducing SDS-PAGE (Fig. 5A). The time courses of E2 ∼ Ub hydrolysis (Fig. 5B), E3 ∼ Ub discharging (Fig. 5C), and polyUb chain formation (Fig. 5D) showed that mutations at position 705 had a more pronounced effect in the isopeptide bond synthesis step than the transthiolation step. Furthermore, the E705H mutant was less defective compared with the E705Q and E705A mutants (Fig. 5D). Thus it seems that the ligase activity correlates with the relative nucleophilic strength at this position. The F569A mutation, on the other hand, had a much slower E2 ∼ Ub discharging and polyUb formation rates (Fig. 5B and D). Substituting F569 with alanine is likely to reduce the affinity of the E2/E3 complex, and thus hinder transthiolation between UbcH7 and NleL.
Fig. 5.
Single-turnover experiments. (A) The schematic presentation (up) and SDS-PAGE stained by Coomassie blue (bottom) of the overall reaction. Lanes: 1, Precharged E2 before addition of NleL; 2–7, time course of the reaction; 8, protein standards. Lanes 9–15, same samples as in lanes 1–7 but with the addition of 100 mM DTT in sample loading buffer to distinguish the presence of thioester bonds from isopeptide bonds. (B) Time-course of Ub discharging from E2. The intensities of the UbcH7 ∼ Ub bands were quantified and plotted over time normalized by the total amount of UbcH7 ∼ Ub. Gel insets for the plot are also shown. (C) Gel insets showing time-course of Ub transferring from NleL to acceptor Ub. (D) Time-course of polyUb chain formation. The amount of isopeptide bond formation was calculated based the intensities of Ub(2) + Ub(3) bands (Ub(2) + 2 × Ub(3)) and normalized by that of the wt protein at the 300 s time point. Gel insets of Ub(2) bands are shown. The error bars show the standard deviation from three independent experiments.
Discussion
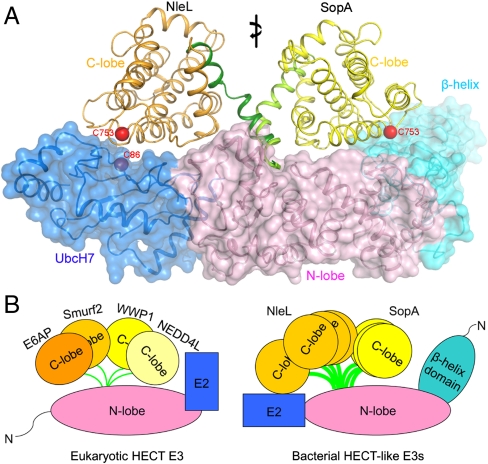
In this study we present two complex structures of bacterial HECT-like ligases binding to a human E2 enzyme. Superposition of these structures demonstrates that the C-lobe is able to move from one end to the other end of the E2/E3 complex (Fig. 6A), accomplished through conformational changes of the hinge helix (Fig. S4). The orientation of the C-lobe in the two structures differs by approximately 180° and the active cysteine is repositioned by 65 Å. For eukaryotic HECT E3s, the flexibility of the C-lobe is essential for catalytic activity, presumably allowing the C-lobe to rotate from E2 to the substrate to transfer Ub (25). The scale of the C-lobe displacement observed here is even larger than that observed in eukaryotic HECT structures. Despite the fact that E2 binds at a different region on the N-lobe (Fig. 6B), the ability of the C-lobe to move over a large distance would enable it to mediate Ub transfer through a mechanism similar to that proposed for eukaryotic HECT E3s (11, 21, 25, 26).
Fig. 6.
Conformational flexibility of HECT E3 ligases. (A) Superposition of the two bacterial HECT/E2 structures. The β-helix domain, N-lobes, and E2 have been superimposed (shown as ribbon and transparent surface). The C-lobes are shown in different colors: yellow for SopA and orange for NleL. (B) Schematic comparison of eukaryotic HECT E3s and bacterial E3s, modeled based on structures of E6AP (11), Smurf2 (26), WWP1 (25), Nedd4L (21), isolated SopA (9), NleL (7), and the complex structures described in this study.
Both SopA and NleL bind to UbcH7 on the same region as eukaryotic HECT E3s. Therefore, the bacterial E3 ligases have utilized a different surface but nevertheless have managed to fulfill the molecular requirements for E2 binding. Comparing the structures of E2 in complex with HECT E3 ligases of eukaryotic and bacterial origins, the consistent theme is that hydrophobic interactions between the conserved phenylalanine in E2 and an aromatic residue in E3 are critical for molecular recognition. This finding contrasts with studies of SspH2, an E3 ligase from S. typhimurium, which has a structure that is distinct from either HECT or RING E3s but also forms a thioester bond with Ub (30, 31). NMR mapping experiments show that SspH2 only binds to charged E2 using a surface that has little overlap with the canonical E3 interaction site (30). Therefore it seems that different bacterial E3 ligases utilize different forms of molecular trickery to penetrate the host Ub pathway.
In the absence of substrate and Ub, it is difficult to specify which enzymatic stages the crystal structures of UbcH7/SopA and UbcH7/NleL each represent. Nevertheless, the structural and mutagenesis data presented here provide insights into the molecular mechanism of ubiquitination. In the UbcH7/SopA structure, the C-lobe makes no contact with E2. In the conformation observed in UbcH7/NleL complex, although the C-lobe is located close to E2, the interface seems to be labile. Instead of engaging in H-bonds or making close complementary interactions, residues located at the interface are somewhat disordered. Substituting polar/charged residues in the vicinity of the active cysteine residues had no effect on the thiotranslation rate between E2 and E3. It is therefore possible that transthiolation occurs spontaneously once the two active cysteines are in close proximity, as observed in the UbcH7/NleL structure. The dynamic nature of the C-lobe and the lack of strong interactions between E2 and the C-lobe suggest that once charged with Ub, C-lobe could move quickly away from E2 to prevent cycling Ub between E2 and E3.
In contrast, when the high-energy E3 ∼ Ub thioester bond is presented to the acceptor lysine in the substrate, a general base would be necessary to polarize the lysine ε-amino group for nucleophilic attack. Our data suggest that NleL E705 is relevant to the catalysis of isopeptide bond formation. The data presented in this work set the stage for future studies to test if E705 functions as the general base or it is only one of several residues that contribute to catalysis, as shown in the studies of SUMO conjugation (17).
Methods
Crystallization and Structure Determination.
Expression and purification of SopA163–782 and NleL170–782 were described previously (7, 9). Purified SopA163–782 and UbcH7 were mixed at 1∶5 molar ratio at 4 °C overnight and the complex was purified by gel filtration chromatography. Crystals were obtained by mixing protein sample with a reservoir solution containing 0.1 M MES, pH 6.5 and 1.6 M (NH4)2SO4 at 1∶1 ratio in sitting drops at 20 °C. Before data collection, crystals were stabilized in the reservoir solution with additional 20% ethylene glycol (v/v) and flash-frozen in liquid nitrogen. Purified NleL170–782 was mixed with UbcH7 at 1∶1 ratio in a buffer containing 5 mM DTT and 10 mM Tris-HCl pH 8.0. UbcH7/NleL complex (10 mg/mL) was crystallized using 1.5–1.7 M (NH4)2SO4 as precipitant at pH 5.6–6.8. UbcH7/NleL crystals were flash-frozen in reservoir solution plus 30% glycerol (v/v).
Diffraction data were collected at 100 K on beamline 23-ID at the Advanced Photon Source and processed with HKL2000 (32). The structure was determined by molecular replacement with Phaser (33). The structure was refined using CNS (34), Refmac (35), and Phenix (36).
Multiturnover Ubiquitination Assays.
Ubiquitination reactions were performed 25 °C in reaction mixtures containing 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 2 mM ATP, 5 mM MgCl2, 0.1 mM DTT, 100 ug/mL of Ub, 4 ug/mL of E1, 3 ug/mL of UbcH7, and 4 ug/mL of wild-type or mutant NleL170–782. Samples were removed from the reaction mixture at different time points, quenched with reducing SDS-PAGE sample buffer, and analyzed on NuPAGE Bis-Tris 4–12% gels (Invitrogen). The intensity of protein bands on SDS-PAGE were quantified using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Single-turnover Assay.
UbcH7 containing Cys17 to Ser and Cys137 to Ser mutations was used in the single-turnover experiments to prevent nonspecific cross-linking. To precharge UbcH7, a reaction mixture containing 20 ug/mL E1, 200 ug/mL UbcH7, 800 ug/mL of Ub, 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 2 mM ATP, 5 mM MgCl2, and 1 mM DTT was incubated at room temperature for 20 min and then quenched with equal amount of 500 mM EDTA, pH 7.3 on ice. The time course experiment was performed by mixing 20 uL UbcH7 ∼ Ub mixture with 1 uL of wild-type or mutants of NleL170–782 at 3.9 mg/mL on ice and quenched at indicated time points with nonreducing SDS-PAGE sample buffer. The samples were analyzed by SDS-PAGE using NuPAGE Bis-Tris 4–12% gels (Invitrogen). Gels were imaged and quantified with an Odyssey Infrared Imaging System (LI-COR Biosciences).
Supplementary Material
Acknowledgments.
We thank Dr. Daoguo Zhou for initiating this project; the staff at the Advanced Photon Source beam line 23-ID for assistance with data collection; Drs. Jon Huibregtse and Ning Zheng for comments on the manuscript; and Dr. Stanislav Zakharov for help with circular dichroism measurements. J.C. is an investigator of Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3SY2 and 3SQV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115025109/-/DCSupplemental.
References
- 1.Angot A, Vergunst A, Genin S, Peeters N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hicks SW, Galan JE. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol. 2010;13:41–46. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Malynn BA, Ma A. Ubiquitin makes its mark on immune regulation. Immunity. 2010;33:843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 7.Lin DY, Diao J, Zhou D, Chen J. Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. J Biol Chem. 2010;286:441–449. doi: 10.1074/jbc.M110.167643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 9.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol. 2008;15:65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- 10.Piscatelli H, et al. The EHEC type III effector NleL is an E3 ubiquitin ligase that modulates pedestal formation. PLoS One. 2011;6:e19331. doi: 10.1371/journal.pone.0019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L, et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 12.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez C, et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, et al. Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 17.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 18.Xu Z, et al. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mace PD, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 20.Das R, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, et al. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuber U, Scheffner M. Identification of determinants in E2 ubiquitin-conjugating enzymes required for hect E3 ubiquitin-protein ligase interaction. J Biol Chem. 1999;274:7576–7582. doi: 10.1074/jbc.274.11.7576. [DOI] [PubMed] [Google Scholar]
- 25.Verdecia MA, et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 26.Ogunjimi AA, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 27.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakata E, et al. Crystal structure of UbcH5b ∼ ubiquitin intermediate: insight into the formation of the self-assembled E2 ∼ Ub conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin I, et al. Identification of an unconventional E3 binding surface on the UbcH5 ∼ Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc Natl Acad Sci USA. 2010;107:2848–2853. doi: 10.1073/pnas.0914821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quezada CM, Hicks SW, Galan JE, Stebbins CE. A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci USA. 2009;106:4864–4869. doi: 10.1073/pnas.0811058106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Carter CW, Sweet RM, editors. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. Macromolecular Crystallography, part A. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.