Abstract
Resistance to the antibiotic fusidic acid (FA) in the human pathogen Staphylococcus aureus usually results from expression of FusB-type proteins (FusB or FusC). These proteins bind to elongation factor G (EF-G), the target of FA, and rescue translation from FA-mediated inhibition by an unknown mechanism. Here we show that the FusB family are two-domain metalloproteins, the C-terminal domain of which contains a four-cysteine zinc finger with a unique structural fold. This domain mediates a high-affinity interaction with the C-terminal domains of EF-G. By binding to EF-G on the ribosome, FusB-type proteins promote the dissociation of stalled ribosome⋅EF-G⋅GDP complexes that form in the presence of FA, thereby allowing the ribosomes to resume translation. Ribosome clearance by these proteins represents a highly unusual antibiotic resistance mechanism, which appears to be fine-tuned by the relative abundance of FusB-type protein, ribosomes, and EF-G.
Keywords: antibacterial, protein synthesis, translocation, structure
The increasing prevalence of resistance to antibiotics among bacterial pathogens threatens effective treatment of the infections that they cause (1–3). In view of the medical importance of this phenomenon, substantial efforts have been directed toward understanding the molecular basis of antibiotic resistance. For the majority of antibiotic classes currently in clinical use, the primary resistance mechanisms have now been well defined in key pathogens, often characterized at the biochemical and even the structural level. However, some notable gaps in our understanding of resistance mechanisms remain. One of these concerns is the poorly defined mechanism underlying resistance to the clinically important antibiotic fusidic acid (FA) in the human pathogen Staphylococcus aureus.
FA has been in clinical use since the 1960s, predominantly in the treatment of superficial and systemic diseases caused by staphylococci and, in particular, S. aureus (4). Indeed, the continued rise of resistance to antibiotics in this species has rendered FA one of the few remaining oral agents available for treatment of infections caused by methicillin-resistant S. aureus (MRSA) (4). FA inhibits bacterial protein synthesis by binding to the ribosomal translocase, elongation factor G (EF-G), when the latter is associated with the ribosome (5, 6). FA does not interfere with the primary catalytic function of EF-G, which involves the translocation of mRNA and associated tRNAs on the ribosome in a reaction coupled to the hydrolysis of GTP (7). Rather, FA acts to prevent dissociation of the resulting EF-G⋅GDP complex from the ribosome once translocation has occurred (5, 8). Formation of ribosome⋅EF-G⋅GDP⋅FA complexes prevents further protein synthesis, with consequent cessation of bacterial growth. Recent studies indicate that FA also inhibits EF-G–mediated dissociation of ribosomes into subunits (“ribosome recycling”) following completion of polypeptide synthesis, thereby preventing subsequent rounds of protein translation (7).
Resistance to FA in clinical strains of S. aureus has increased dramatically in recent years (9). The most prevalent mechanism of resistance involves horizontal acquisition of the fusB-type FA resistance determinants (9, 10), a group named after the first discovered member, fusB (11, 12). At least two representatives of this group (fusB and fusC) are found in S. aureus (13), and studies to investigate the mechanism of fusB-type FA resistance have to date focused on fusB. The FusB protein has been shown to bind EF-G and protect the staphylococcal translation apparatus from the inhibitory effects of FA (11). The interaction between FusB and EF-G is central to FA resistance, because protection from FA is not observed when FusB fails to bind EF-G (11). How this protein–protein interaction gives rise to FA resistance is currently unknown: potentially, resistance might simply be the result of FusB-mediated steric hindrance of the interaction between FA and EF-G (11). Despite our limited understanding of FusB-type resistance, it is nonetheless evident that this antibiotic resistance mechanism is highly unusual. To our knowledge, there is only one other clinically relevant example of antibiotic resistance mediated through protein–protein interaction between a resistance protein and a drug target: the Qnr protein from Gram-negative bacteria, which protects DNA gyrase from fluoroquinolones (14).
An additional aspect of FusB-type proteins concerns their native role in the bacterial cell. Genes encoding FusB-type proteins are found in diverse Gram-positive bacterial genera, including organisms in the environment that are unlikely to face challenge with FA (11). This finding has led to the hypothesis that members of the FusB family of proteins represent previously uncharacterized accessory factors active in bacterial protein synthesis (11). Structural details of FusB-type proteins, their interaction with EF-G, their cellular role, and the mechanism by which resistance to FA is mediated have not been established. Here we describe the results of a series of biochemical and biophysical studies that together provide detailed insight into the structure and function of FusB-type proteins.
Results
Interaction Between FusB-Type Proteins and EF-G.
In a previous study we demonstrated that purified FusB binds staphylococcal EF-G in whole-cell lysates (11). To characterize the interaction between these proteins, both FusB and EF-G from S. aureus were individually overexpressed in Escherichia coli and purified to >95% homogeneity. By mixing the proteins (with FusB in excess), followed by analytical gel filtration chromatography, it was established that they bind in vitro with a 1:1 stoichiometry in the absence of any other added biological component. This interaction was further examined by isothermal titration calorimetry (ITC; Fig. S1A). ITC confirmed the 1:1 stoichiometry of binding and established that FusB binds EF-G with high affinity (Kd = 63 ± 5 nM). The FusC protein is a homolog of FusB (∼45% amino acid identity) that also mediates resistance to FA in S. aureus (13). To confirm that FusC also binds EF-G, it was overexpressed and purified from E. coli, and binding to EF-G analyzed by ITC (Fig. S1B). FusC bound to EF-G with a slightly higher affinity than FusB (Kd = 25 ± 2 nM), and at 1:1 stoichiometry. Because FusB and FusC are homologous EF-G binding proteins, they likely mediate resistance to FA through the same mechanism.
To identify the region of EF-G with which FusB interacts, binding studies were performed with FusB and fragments of EF-G using analytical gel filtration chromatography and ITC (Fig. S1C). The EF-G fragments (Fig. S1C) were overexpressed in E. coli and purified, with the exception of the fragment encoded by construct 2, which did not prove amenable to soluble expression. FusB only bound to construct 3 (EF-GC3), a ∼35-kDa polypeptide spanning residues 401–691 and comprising domains 3, 4, and 5 of EF-G, implying that FusB does not interact with domains 1 and 2 of EF-G. ITC experiments established that FusB bound EF-GC3 with an affinity comparable to that for full-length EF-G (Fig. S1C), suggesting that the FusB-binding interface is completely contained within domains 3–5 of EF-G.
Crystal Structure of FusC.
To gain further insight into FusB-type proteins and their interaction with EF-G, we sought to determine their 3D structure by X-ray crystallography. Although FusB could be crystallized, we were unable to identify crystallization conditions generating well-diffracting crystals. By contrast, FusC produced crystals that diffracted to high resolution (∼2.1 Å). FusC crystals contained zinc, as detected by extended X-ray absorption fine structure (EXAFS) scanning; consequently, phasing of the FusC data were possible using single-wavelength anomalous data (SAD) collected at the zinc K-edge (1.28 Å). The FusC protein crystallized in space group P21 as a monomer with two molecules per asymmetric unit (see Table S1 for crystallographic data and refinement statistics). Anomalous data were collected from this crystal form, and the positions of the two zinc ions present in the asymmetric unit were defined for phasing.
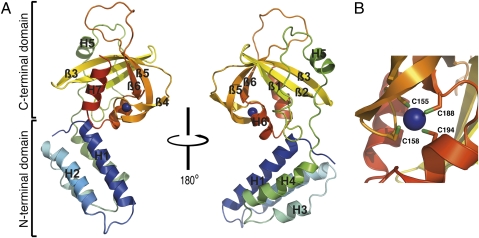
The structure of FusC reveals distinct N- and C-terminal domains, connected by a loop (Fig. 1A). The N-terminal domain forms a four-helix bundle, with helices designated H1–H4. Helices H1 and H2 span residues 6–25 and 32–49, respectively; H3 (residues 54–60) and H4 (residues 67–80) are shorter in comparison. Residues at the C terminus of H1 make contact with the C-terminal domain of the protein, fixing the angle between the N- and C-terminal domains and contributing to the overall shape of the molecule. The C-terminal domain consists of a combination of β-sheet and short helices. This domain comprises a C4 (four cysteine)-type zinc finger, with cysteine residues at positions 155, 158, 188, and 194 tetrahedrally coordinating a central zinc ion (Fig. 1B). Within this zinc-binding domain (ZBD) there are six antiparallel β-strands packed to form two β-sheets, and three helices (named H5–H7), of which H6 and H7 are situated within the cleft formed by the two β-sheets, with H5 flanking the outside of the first β-sheet. The C4 zinc finger is formed by residues residing in different secondary structural elements: C155 lies at the end of β4, C158 within the loop connecting β4 and β5, C188 at the end of β6, and C194 within H6. Both domains of FusC contain short disordered regions (residues 24–32 and 170–182) showing poor electron density, which probably correspond to unstructured loops.
Fig. 1.
Crystal structure of FusC. (A) Ribbon diagrams of FusC, showing the protein in alternative orientations. The α-helices (H1–H7) and β-strands (β1–β6) are labeled, and the zinc ion is shown as a purple sphere. (B) Close-up view of the zinc finger, showing the four cysteine residues involved in coordination of the zinc ion.
The DALI server (15) was used to search the Protein Data Bank for structures and folds related to FusC. Searches using whole FusC identified the E. coli histidine kinase NarX (PDB ID code 3EZI) as similar, although structural similarity was only apparent for the N-terminal domain of FusC. Indeed, all significant matches were based on structural similarity of the N-terminal and not the C-terminal domain of FusC. Therefore, separate searches were performed with each domain. The N-terminal domain identified the same matches as whole FusC, whereas the C-terminal domain did not yield significant matches. No structural similarities were observed with known zinc fingers and, therefore, the ZBD of FusC appears to represent a unique zinc-binding fold. Sequence alignment of FusB-type proteins from different bacterial species revealed conservation of the ZBD (Fig. S2).
Mapping the EF-G Binding Interface on FusB.
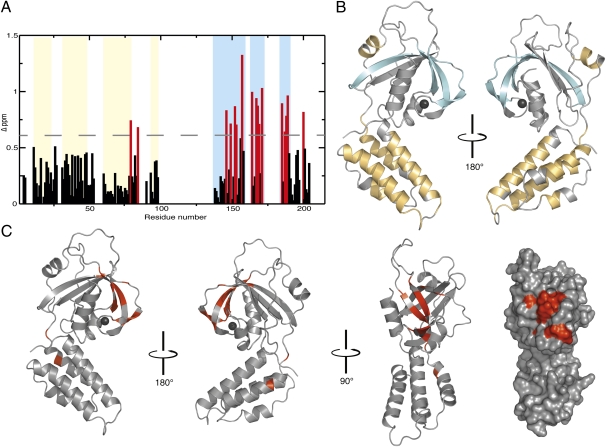
To define the regions of FusB involved in the interaction with EF-G, NMR chemical shift mapping was used. Initial experiments established that the 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of perdeuterated 15N-labeled FusB produced the expected number of backbone amide resonances (Fig. S3A). Analysis of 3D TROSY HN(CO)CA, HNCA, HNCO, HN(CA)CO, HN(CA)CB, and HN(COCA)CB FusB spectra allowed the assignment of ∼65% of the FusB backbone. Mapping of FusB 13C chemical shifts (16) yielded secondary structure information closely matching that determined crystallographically for FusC, with a clear division of α-helices in the N-terminal region and β-sheet in the C-terminal region (Fig. 2 A and B). Perdeuterated 15N-labeled FusB was saturated with unlabeled EF-GC3 and 1H-15N HSQC spectra of the purified complex collected (Fig. S3B). The residues of FusB involved in the interaction between FusB and EF-G were identified by chemical shift mapping of minimal chemical shift changes (17, 18), and were found to reside almost exclusively in the C-terminal portion of the protein (Fig. 2 A and C).
Fig. 2.
Identifying the EF-G binding site on FusB by 1H-5N HSQC minimal chemical shift mapping. (A) Plot of shift changes for assigned residues in FusB. Residues with Δ >0.6 ppm are colored red. Secondary structural elements, determined from the CSI of FusB, are represented as colored panels (α-helical regions in yellow, β-sheet in blue). (B) Ribbon diagram of FusC with secondary structure elements colored as in A. Regions shown in gray are areas of FusB that were not found to be in regular secondary structure according to the CSI or are residues that were not assigned. (C) Ribbon diagrams and surface representation of alternative orientations of FusC displaying the predicted EF-G binding site in red.
The residues of FusB predicted to interact with EF-G were subsequently mapped onto the crystal structure of FusC (Fig. 2C). The observed shift changes correspond to a region situated within the ZBD of FusC, with the majority mapping to exposed residues on the three antiparallel β-strands (β1, β5, and β6) that form the second β-sheet. Indeed, two of the cysteine residues (C155 and C188) involved in forming the C4 zinc finger showed significant chemical shift changes upon binding of FusB to EF-G.
Modeling the FusC⋅EF-G Complex.
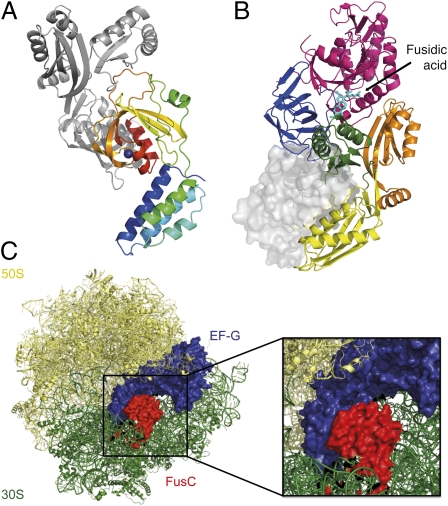
Using the recently solved 3D structure of S. aureus EF-G (PDB accession no. 2XEX) (19) in conjunction with the FusC structure, we performed computational docking of FusC and EF-GC3 via the ZDOCK server (20). ZDOCK performs rigid docking of target proteins, predicts the structure of multiple protein complexes, and uses pairwise shape complementarity to rank the results (21). Following docking of the proteins without reference to the NMR chemical shift mapping data, the highest-scoring prediction involved interactions between the ZBD of FusC and domains 3 and 4 of EF-GC3 (Fig. 3 A and B). Thus, the predictions for the FusC binding interface provided by chemical shift mapping and docking in silico are consistent.
Fig. 3.
Model of the FusC⋅EF-G complex, showing interactions between the ZBD of FusC and domains 3 and 4 of EF-G. (A) Ribbon diagram of the docked FusC⋅EF-GC3 complex. FusC is represented in rainbow colors, and EF-GC3 in gray. (B) Alternative representation of the two molecules, with a ribbon diagram of EF-G colored by domain (as per Fig. S1) and FusC surface representation in gray. The FA binding site on EF-G is also indicated. (C) Model of EF-G simultaneously bound to a FusB-type protein (FusC) and the ribosome, illustrating the multiple steric clashes that would occur between FusC and the 30S subunit. Coordinates for the EF-G–bound ribosome are from PDB ID code 2WRI.
The FusC⋅EF-G complex was subsequently modeled on the ribosome, using the crystal structure of FA-stalled EF-G on ribosomes from Thermus thermophilus (21) (Fig. 3C). From the crystal structure it is evident that the C-terminal region of EF-G, which has an important role in translocation (22), makes direct contact with the ribosome. It appears from visual inspection of the modeled FusC⋅EF-G complex and the conformation that EF-G adopts on the ribosome that, owing to its size and location of binding on EF-G, bound FusC would cause steric clashes with the 30S subunit of the ribosome, which would probably inhibit EF-G from making the normal ribosomal contacts.
Effect of FusB-Type Proteins on EF-G Function.
The effect of FusB on the function of EF-G on the ribosome was assessed using steady-state and transient kinetics measurements. These assays typically use 70S ribosomes and accessory proteins from E. coli; however, FusB-type proteins are not functional (i.e., do not mediate protection from FA) in an E. coli translation assay (11). We reasoned that the lack of functionality was due to the inability of FusB to bind to E. coli EF-G (11), and indeed substitution of E. coli EF-G with S. aureus EF-G yielded an experimental system in which protection from FA by FusB could be demonstrated.
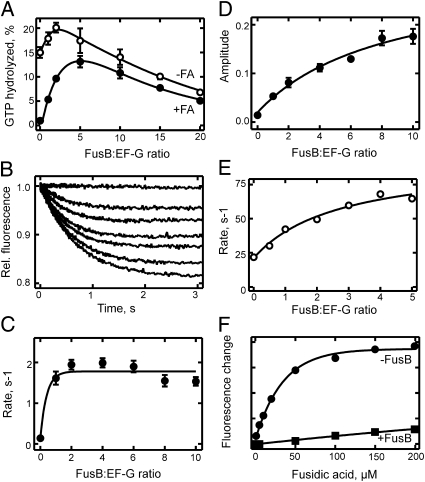
The residence time of EF-G on the ribosome is determined by a conformational rearrangement that limits EF-G turnover and is blocked by FA (23). When EF-G–dependent GTP hydrolysis was examined at conditions of EF-G turnover, the addition of FusB up to a FusB: EF-G ratio of ∼5 increased the rate of the turnover reaction, and the increase was particularly large (20-fold) in the presence of FA (Fig. 4A). This observation suggests that (i) FusB is capable of binding to EF-G on the ribosome, and (ii) FusB accelerates the conformational rearrangement of the EF-G⋅ribosome complex that ordinarily acts to limit the turnover rate. At higher FusB:EF-G ratios, the acceleration of turnover was reversed.
Fig. 4.
Effect of FusB on the dissociation of S. aureus EF-G from the ribosome. (A) GTP hydrolysis at conditions of EF-G turnover. Ribosomes (0.5 μM) and EF-G (0.5 μM) were reacted with [γ-32P]GTP (1 mM) without FA (○) or in the presence of FA (50 μM; ●) for 5 min at 25 °C. The extent of GTP hydrolysis was determined by TLC (30). (B) FusB-induced dissociation of ribosome⋅EF-G⋅mant-GDP⋅FA complexes. Ribosome⋅EF-G⋅mant-GDP complexes were formed in the presence of FA (200 μM) and rapidly mixed with FusB at increasing concentrations (0–5 μM, from top to bottom) at 37 °C. Complex dissociation was monitored by mant fluorescence. (C) Rates from B. Stopped-flow traces were evaluated by single-exponential fitting. (D) Amplitudes from B. (E) FusB-induced dissociation of ribosome⋅EF-G⋅BODIPY FL-GDP complexes. Complex dissociation was followed at 37 °C monitoring BODIPY fluorescence. Rates were determined by single-exponential fitting of stopped-flow traces. (F) Effect of FusB on ribosome⋅EF-G⋅BODIPY FL-GDP⋅FA complex formation. Complex formation was examined without FusB (●) or in the presence of FusB (2.5 μM; ■) at 37 °C, monitoring BODIPY fluorescence. For experimental details, see Materials and Methods.
The rescue of EF-G from FA due to destabilization of the ribosome⋅EF-G⋅GDP⋅FA complex could also be shown directly by monitoring the fluorescence of a fluorescent GDP derivative, mant-GDP (Fig. 4B). FusB binding to ribosome-bound EF-G⋅GDP⋅FA increased the dissociation rate of the complex from ∼0.1 s−1 to 1.7 s−1 (Fig. 4C), consistent with the effect observed in the steady-state GTPase experiment (Fig. 4A). Thus, the rescue effect of FusB is due to the acceleration of the dissociation of the FA-stalled complex. From the amplitude increase of the FusB-induced dissociation of the ribosome⋅EF-G⋅GDP⋅FA complex (Fig. 4D), the KM of FusB binding to the complex was estimated at ∼4 μM, which is much higher than the Kd of the EF-G⋅FusB complex in the absence of ribosomes, suggesting that the binding of the two proteins is impaired on the ribosome. This finding is consistent with the results of modeling (Fig. 3), suggesting that ribosome-bound EF-G stalled by FA is less readily accessible for FusB binding compared with free EF-G. In keeping with the destabilizing effect of FusB on ribosome⋅EF-G complexes containing FA, FusB brought about a dose-dependent increase in the rate of dissociation of the EF-G⋅GDP complex in the absence of FA (Fig. 4E). This indicates that FusB also binds to the EF-G⋅ribosome complex in the conformation that is not stalled by FA, thereby accelerating EF-G turnover. The KM value of FusB binding to the complex was 1.4 μM, again much higher than the Kd of EF-G binding to FusB. Finally, we examined whether FusB is capable of preventing the formation of FA⋅EF-G⋅GDP⋅ribosome complexes in the presence of FA. Formation of a stable FA⋅EF-G⋅GDP⋅ribosome complex was monitored by an increase in fluorescence of a BODIPY group attached to GDP (24). The formation of the complex was strongly inhibited in the presence of excess FusB (Fig. 4F), suggesting that FusB protects EF-G from being stalled on the ribosome in the presence of FA.
Discussion
The interaction between FusB-type proteins and EF-G underlies the ability of the former to protect the bacterial translation apparatus from inhibition by FA (11). According to the present crystallographic analysis, FusC is a two-domain zinc-binding protein, the C-terminal portion of which comprises a unique fold of helices and β-sheets that together form a zinc-binding domain. Coordination of the zinc ion appears to be essential for stabilizing the tertiary structure of the C-terminal domain, although this does not exclude the possibility that the ZBD and the coordinated zinc ion also play a functional role. The NMR data indicate that the ZBD is directly responsible for mediating the interaction with EF-G. This interaction involves a high-affinity 1:1 binding of FusB-type proteins to the C-terminal part of EF-G. The FusB-binding interface lies within domains 3–5 of EF-G, with in silico modeling suggesting that the interaction between the two proteins may specifically involve domains 3 and 4 of EF-G. In view of the fact that FusB has the same secondary structure as FusC, and given the high degree of sequence conservation of the zinc finger among members of the FusB family, it is likely that all FusB-type proteins share the same structural architecture, at least within the C-terminal domain, and interact with EF-G in the same way. FusB-type proteins interact with a part of EF-G that is spatially distinct from the FA binding site, as the latter lies toward the N terminus of the protein, formed by domains 1–3 (21). Indeed, in our model of the FusC⋅EF-G complex, FusC lies >25 Å away from the FA binding site. Thus, the possibility that FusB-type proteins might directly sterically hinder binding of FA to EF-G seems unlikely in light of our results.
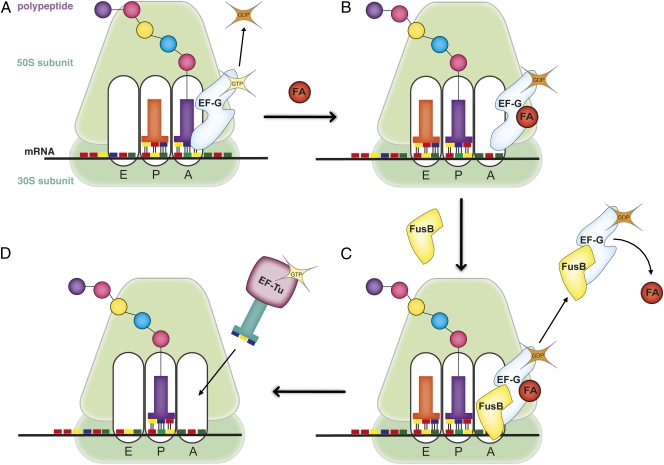
FusB-type proteins act to accelerate the release of EF-G from the ribosome. Our findings suggest that this effect is achieved through competition of FusB-type proteins with the ribosome for EF-G binding. Destabilization of EF-G binding to the ribosome in the absence of FA likely represents the cellular housekeeping function of FusB-type proteins (11)—i.e., to facilitate release of EF-G from the ribosome following translocation, thereby clearing stalled ribosomes and allowing translation to resume. In the presence of FA, the effect of FusB-type proteins on the dissociation of stalled ribosome⋅EF-G⋅GDP complexes is particularly striking, and provides an explanation for the mechanism of FA resistance mediated by this family of proteins (Fig. 5). As described previously, the mechanism of action of FA involves binding to EF-G, thereby interfering with the release of the latter from the ribosome. FA appears to function as a slow inhibitor, permitting several rounds of GTP turnover by EF-G before complete inhibition is achieved (7, 8), but it is not known whether the formation of FA⋅EF-G⋅GDP⋅ribosome complexes leads to shutdown of protein synthesis primarily by directly stalling the majority of ribosomes; by stalling a minority of ribosomes which then serve to blockade polysomes; or as a consequence of depleting the EF-G pool. The equilibrium of FA-mediated inhibition will be affected by conditions that either drive the formation of FA⋅EF-G⋅GDP⋅ribosome complexes or favor their dissociation (e.g., high or low concentrations of FA, respectively) (8). Because FusB-type proteins facilitate the release of EF-G from the ribosome, and inhibit the formation of the FA-stalled complex, they will shift the equilibrium toward complex dissociation. Consequently, a greater FA concentration will be required to bring about the same level of inhibition of translation in bacterial cells expressing FusB-type proteins, and such cells will therefore exhibit an FA resistance phenotype. Though conditions where FusB is slightly in excess over EF-G accelerate turnover of the latter, at high concentrations FusB may interfere with EF-G function. Consequently, the balance of interactions between ribosomes, EF-G, FA, and FusB-type protein appears important, and the precise amount of FusB-type protein in the cell must be fine-tuned to the concentrations of ribosomes and EF-G. The mechanism of FA resistance mediated by the FusB-type proteins provides a unique example where the interaction between two proteins, fine-tuned through their binding affinities and cellular abundance, rescues translation by clearing ribosomes from a stalled state and returns them to the actively translating pool.
Fig. 5.
Schematic of the proposed mechanism of FA resistance mediated by FusB-type proteins. (A) EF-G catalyzes translocation in a reaction driven by the hydrolysis of GTP. (B) In the presence of FA, the drug immobilizes EF-G⋅GDP complexes on the ribosome, sterically blocking the next stage of translation and stalling protein synthesis. (C) FusB-type proteins compete with the ribosome for binding to EF-G, thereby destabilizing the ribosome⋅EF-G⋅GDP⋅FA complex and prompting its dissociation; FA cannot bind to EF-G when the latter is not resident on the ribosome, and will spontaneously dissociate once the complex has been dislodged from the ribosome. (D) Dissociation of the EF-G⋅GDP⋅FA complex clears the ribosomal A site, allowing entry of the next incoming aminoacyl-tRNA molecule, and translation resumes.
Materials and Methods
Additional details are available in SI Materials and Methods.
Overexpression and Purification of Proteins.
The autoinduction method (25) was used for overexpression of proteins, with the exception of 15N- and 13C-labeled FusB, for which overexpression was induced by the addition of isopropyl-β-d thiogalactopyranoside. Cells were harvested by centrifugation at 6,000 × g for 20 min at 4 °C, resuspended in lysis buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, 250 KU rLysozyme (Novagen) and EDTA-free protease inhibitor tablets (Roche Diagnostics)], lysed by sonication, and lysates cleared by centrifugation at 16,000 × g for 30 min at 4 °C.
Nickel-affinity chromatography was performed using a 25-mL free-flow gravity column (GeneFlow) packed with 10 mL Ni-NTA agarose (Qiagen). Eluted fractions were dialyzed against running buffer [20 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, and 1 mM DTT]. Proteins were further purified by gel filtration chromatography using a Superdex 75 (S75) pg (26/60) prepacked column (GE Healthcare) in running buffer. For crystallization of FusC, protein preparations were digested with Tobacco etch virus protease at 4 °C for 24 h.
Isothermal Titration Calorimetry.
ITC experiments were undertaken using the VP-ITC calorimeter (MicroCal), with a cell volume of 1.409 mL. Titrations were performed in triplicate at 25 °C in degassed running buffer containing 0.5 mM Tris(hydroxypropyl)phosphine (Novagen) in place of DTT. Ligands (FusB or FusC) were used at concentrations of 500–600 μM, and EF-G at 50–60 μM. One 2-μL injection and 30× 8-μL injections were added at 5-min intervals. Heats of dilution were determined in the absence of EF-G or EF-GC3 and subtracted before curve fitting. MicroCal Origin v. 5.0 was used to fit curves using the one-site model (baseline correction and peak integration).
Crystallization of FusC and Data Collection.
Crystals of FusC were grown from hanging drops using the vapor diffusion method in 0.2 M ammonium acetate, 0.1 M Tris⋅HCl (pH 8.0), and 20% (wt/vol) PEG 3350. X-ray diffraction data were collected at Diamond Light Source beam line I02. The FusC structure was solved by SAD (26), and zinc sites determined using SHELX (27). The MolProbity Web service (28) was used to validate the refined FusC structure and produced a clash score within the 84th percentile and overall MolProbity score in the 90th percentile. Data collection and final refinement statistics are shown in Table S1.
NMR Spectroscopy.
NMR experiments were performed with perdeuterated 0.3 mM 15N- and 13C-labeled FusB. For analysis of the FusB⋅EF-GC3 complex, perdeuterated 15N-labeled FusB was saturated with unlabeled EF-GC3 and the complex purified by gel filtration chromatography. Spectra were recorded at 25 °C on a Varian Inova 600-MHz spectrometer with a room temperature probe, or a Varian Inova 750-MHz spectrometer with a cryogenic probe.
Backbone assignments of FusB were obtained from analysis of HNCA, HNCO, HN(CO)CA, HN(CA)CO, and HN(CA)CB spectra (all experiments used TROSY modifications, and where required, deuterium decoupling) (29). Chemical shift indexing (CSI) (16) was used to determine the secondary structure of FusB from the shifts of 1H and 13C nuclei. Conservative chemical shift differences between the 1H -15N spectra for FusB and FusB⋅EF-GC3 were calculated by finding the closest peak in the FusB⋅EF-GC3 spectrum to the assigned peaks in the FusB spectrum (18). Shift differences for which Δ >0.6 ppm were considered significant and indicated residues involved in forming the FusB⋅EF-GC3 interface.
Biochemical and Rapid Kinetic Experiments.
Experiments were performed at 37 °C in 50 mM Tris⋅HCl [pH 7.5], 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2 supplemented with 1 mM DTT. The effect of FusB on the rate of dissociation of EF-G⋅BODIPY-GDP⋅ribosome complexes was measured on a stopped-flow apparatus (Applied Photophysics) using an excitation of 520 nm and emission of >545 nm. Reactions consisted of E. coli 70S ribosomes (0.5 μM), EF-G (0.5 μM), BODIPY FL-GTP (50 nM; Molecular Probes), and a range of FusB concentrations. Analogous rapid kinetic experiments using mant-GTP (Molecular Probes) were performed to analyze the FusB-induced dissociation of preformed FA⋅EF-G⋅mant-GDP⋅ribosome complexes. The inhibition by FusB of the formation of FA⋅EF-G⋅BODIPY-GDP⋅ribosome complexes was examined in a PTI500 fluorimeter (Photon Technology International), using the same reaction components as above and a range of FA concentrations, in the absence and presence of FusB (2.5 μM).
Supplementary Material
Acknowledgments
We thank Dr. A. Kalverda for technical assistance with the NMR studies, R. Malham for technical advice regarding ITC, Dr. J. Haddon and Prof. S. Phillips for preliminary work on FusB crystallization, and staff at the Diamond Light Source for assistance with FusC data collection. This work was supported by a Biotechnology and Biological Sciences Research Council (United Kingdom) Doctoral Training Grant and Research Grant BB/H018433/1, International Human Frontier Science Program Organization Grant ST00163/2005-C, and a grant from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates of FusC reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2yb5) and the FusB chemical shift data reported in this paper have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 17503).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117275109/-/DCSupplemental.
References
- 1.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chopra I, et al. Treatment of health-care-associated infections caused by Gram-negative bacteria: A consensus statement. Lancet Infect Dis. 2008;8:133–139. doi: 10.1016/S1473-3099(08)70018-5. [DOI] [PubMed] [Google Scholar]
- 3.Spellberg B, et al. Infectious Diseases Society of America The epidemic of antibiotic-resistant infections: A call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 4.Howden BP, Grayson ML. Dumb and dumber—the potential waste of a useful antistaphylococcal agent: Emerging fusidic acid resistance in Staphylococcus aureus. Clin Infect Dis. 2006;42:394–400. doi: 10.1086/499365. [DOI] [PubMed] [Google Scholar]
- 5.Bodley JW, Zieve FJ, Lin L, Zieve ST. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem Biophys Res Commun. 1969;37:437–443. doi: 10.1016/0006-291x(69)90934-6. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka N, Kinoshita T, Masukawa H. Mechanism of protein synthesis inhibition by fusidic acid and related antibiotics. Biochem Biophys Res Commun. 1968;30:278–283. doi: 10.1016/0006-291x(68)90447-6. [DOI] [PubMed] [Google Scholar]
- 7.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo HS, et al. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 9.McLaws FB, Larsen AR, Skov RL, Chopra I, O'Neill AJ. Distribution of fusidic acid resistance determinants in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:1173–1176. doi: 10.1128/AAC.00817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castanheira M, Watters AA, Mendes RE, Farrell DJ, Jones RN. Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008) J Antimicrob Chemother. 2010;65:1353–1358. doi: 10.1093/jac/dkq094. [DOI] [PubMed] [Google Scholar]
- 11.O'Neill AJ, Chopra I. Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol Microbiol. 2006;59:664–676. doi: 10.1111/j.1365-2958.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 12.O'Neill AJ, Larsen AR, Henriksen AS, Chopra I. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob Agents Chemother. 2004;48:3594–3597. doi: 10.1128/AAC.48.9.3594-3597.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Neill AJ, McLaws F, Kahlmeter G, Henriksen AS, Chopra I. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob Agents Chemother. 2007;51:1731–1740. doi: 10.1128/AAC.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother. 2005;49:3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wishart DS, Sykes BD. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 17.Farmer, BT, 2nd, et al. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 18.Williamson RA, Carr MD, Frenkiel TA, Feeney J, Freedman RB. Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry. 1997;36:13882–13889. doi: 10.1021/bi9712091. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Koripella RK, Sanyal S, Selmer M. Staphylococcus aureus elongation factor G—structure and analysis of a target for fusidic acid. FEBS J. 2010;277:3789–3803. doi: 10.1111/j.1742-4658.2010.07780.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen R, Li L, Weng Z. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 21.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- 23.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 24.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci USA. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 27.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 28.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Sp. 1999;34:93–158. [Google Scholar]
- 30.Rodnina MV, et al. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc Natl Acad Sci USA. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.