Abstract
A critical regulator of autophagy is the Class III PI3K Vps34 (also called PIK3C3). Although Vps34 is known to play an essential role in autophagy in yeast, its role in mammals remains elusive. To elucidate the physiological function of Vps34 and to determine its precise role in autophagy, we have generated Vps34f/f mice, in which expression of Cre recombinase results in a deletion of exon 4 of Vps34 and a frame shift causing a deletion of 755 of the 887 amino acids of Vps34. Acute ablation of Vps34 in MEFs upon adenoviral Cre infection results in a diminishment of localized generation of phosphatidylinositol 3-phosphate and blockade of both endocytic and autophagic degradation. Starvation-induced autophagosome formation is blocked in both Vps34-null MEFs and liver. Liver-specific Albumin-Cre;Vps34f/f mice developed hepatomegaly and hepatic steatosis, and impaired protein turnover. Ablation of Vps34 in the heart of muscle creatine kinase-Cre;Vps34f/f mice led to cardiomegaly and decreased contractility. In addition, while amino acid-stimulated mTOR activation was suppressed in the absence of Vps34, the steady-state level of mTOR signaling was not affected in Vps34-null MEFs, liver, or cardiomyocytes. Taken together, our results indicate that Vps34 plays an essential role in regulating functional autophagy and is indispensable for normal liver and heart function.
Keywords: LC3, SQSTM1/p62, 3-MA, epidermal growth factor receptor, transferrin
Macroautophagy (referred to as autophagy hereafter) is a dynamic membrane trafficking process that involves the delivery of intracellular content to lysosomes for degradation. A fully executed autophagy includes the formation of double-membraned autophagosomes, the fusion of autophagosomes to late endosomes/lysosomes, and the digestion of the enclosed content by lysosomal hydrolases. Autophagy is constantly maintained at the basal level and is up-regulated in response to stress conditions, such as nutrient and energy limitation, hypoxia, and DNA damage. Autophagy is necessary for cellular and tissue homeostasis, by eliminating damaged organelles and misfolded proteins, and its dysregulation is implicated in developmental defects and numerous diseases (1–5).
Vps34 is the Class III phosphoinositide 3-kinase (PI3K) that phosphorylates phosphatidylinositol to generate phosphatidylinositol 3-phosphate [PI(3)P], a phospholipid central for membrane trafficking processes. The localized production of PI(3)P acts to recruit effector proteins containing FYVE or PX domains that control membrane docking and fusion during the formation of internal vesicles (6). Vps34 is the only PI3K identified in yeast thus far, and its essential role in vacuolar protein delivery was initially described through yeast genetics studies (7). In metazoans, Vps34 has been found to play a critical role in endocytosis, autophagy, and TOR activation (8, 9). The essential role of Vps34 in autophagy has been established largely through the use of the pharmacological inhibitors wortmannin and 3-methyladenine (3-MA), which have been used to suppress autophagy in many studies in the literature. However, bevause of the lack of specificities of these PI3K inhibitors, the precise role of Vps34 in autophagy remains unclear. It was only recently that the physiological role of mammalian Vps34 has been investigated in mouse gene knockout studies (10, 11). Surprisingly, it was reported that although genetic deletion of Vps34 in sensory neurons leads to disruption of the endocytic pathway, the autophagic pathway is still intact in these cells, raising the question as to whether Vps34 is necessary for autophagy in mammals (10).
In the present study, we have generated conditional Vps34 gene knockout mice. Through tissue-specific expression of the Cre recombinase, Vps34 is deleted in the liver or heart, two organs where autophagy is known to play physiological roles. We have also generated Vps34f/f murine embryonic fibroblasts (MEFs), in which Vps34 can be acutely deleted through infection with adenoviral Cre. We report here that ablation of Vps34 leads to a blockade of autophagy in MEFs, liver, and heart, and that Vps34 plays an essential role in maintaining normal liver and heart functions.
Results
Generation of Vps34 Conditional Knockout Mice.
Vps34 (PIK3C3) conditional knockout mice (Vps34f/f) were generated with exon 4 of the Vps34 gene flanked by loxP sites (Fig. 1A and Fig. S1A). When exposed to Cre recombinase, exon 4 is excised, causing a frame-shift mutation leading to the deletion of 755 of the 887 amino acids of Vps34. The Vps34f/f mice were bred to muscle creatine kinase (Mck)-Cre or albumin (Alb)-Cre mice to generate tissue-specific knockouts in heart and liver, respectively, to study the effect of Vps34 loss in these organs.
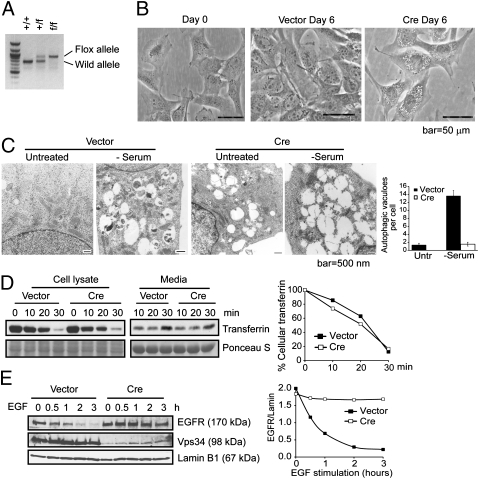
Fig. 1.
Ablation of Vps34 leads to defective autophagosome formation and endocytic protein turnover in MEFs. (A) PCR analysis with the Pik3c3-5′ and 3′ arm primers (Fig. S1) using genomic DNA isolated from MEFs generated from Vps34+/f breeding pair. (B) MEFs were observed by phase/contrast microscopy upon control or Cre infection for 6 d. Representative images are shown. (C) MEFs infected with vector control or Cre were left in full medium or serum starved for 6 h and observed by EM. Large-sized empty vacuoles are observed in Vps34-null MEFs representing single-membraned swollen endosomes/lysosomes. Serum starvation induced the appearance of autophagosomes in wild-type, but not Vps34-null MEFs. Quantification of the number of autophagosomes was obtained by counting 10–20 cells and the averages ± SEM are shown. (D and E) Endocytic EGFR degradation but not transferrin recycling is blocked in Vps34-null cells. (D) Vector control or Cre-infected MEFs were serum starved for 4 h and incubated with biotinylated-transferrin, then stripped and chased with unlabeled transferrin for the indicated times. Cells and culture media were collected and analyzed for biotinylated-transferrin levels. The relative amounts of intracellular transferrin at each time point was quantified by densitometry and normalized to that of time 0. (E) Vector control or Cre-infected MEFs were serum-starved overnight and stimulated with EGF (100 ng/mL) for the indicated times. EGFR protein level is quantified relative to Lamin B1.
Characterization of Vps34-Null MEFs.
Mouse embryonic fibroblasts (MEFs) were generated from day 13–16 embryos of Vps34+/f matings. MEFs with each of the possible genotypes were immortalized with SV40 large T antigen. Expression of Cre recombinase resulted in virtually complete loss of Vps34 protein as shown by immunoblotting (Fig. 2A). Vps34+/+ MEFs infected with adenoviral Cre or Vps34f/f MEFs infected with control adenovirus were used as controls for possible effects of flox or Cre expression. No obvious differences between the two groups of MEFs were observed. Vps34-null MEFs displayed a drastic proliferative defect compared with the wild-type control (Fig. S1B). By day 6 after Cre addition, large translucent vacuoles appeared in the Vps34-null MEFs (Fig. 1B). These vacuoles appeared to be decorated at least partially by the late-endosome marker Rab7 and the lysosomal membrane protein LAMP1 but not LC3 (Fig. S2A), and were acidic as indicated by their staining with the lysosomotropic dye Acridine Orange, which emits red fluorescence when protonated (Fig. S2B). Under electron microscopy (EM), these large vacuoles had a single membrane and the content appeared empty, which were different from the autophagosomes induced by serum starvation in wild-type cells (Fig. 1C). These observations were consistent with those described in Vps34 RNAi knockdown cells (12) and indicate that the large-sized vacuoles may be derived from late endosomes and/or lysosomes. Two important functions of PI(3)P-positive early endosomes are sorting of internalized transferrin receptor to recycling endosomes for recycling to the plasma membrane, and sorting of internalized signaling receptors such as the epidermal growth factor receptor (EGFR) to vesicles that bud into the interior of multivesicular bodies for delivery to lysosomes for degradation. We next asked whether Vps34 function was required for either of these functions. Interestingly, although the endocytic recycling of transferrin was not affected (Fig. 1D), similar to that reported in the knockdown study (12), endocytic turnover of EGFR was drastically inhibited in Vps34-null MEFs (Fig. 1E).
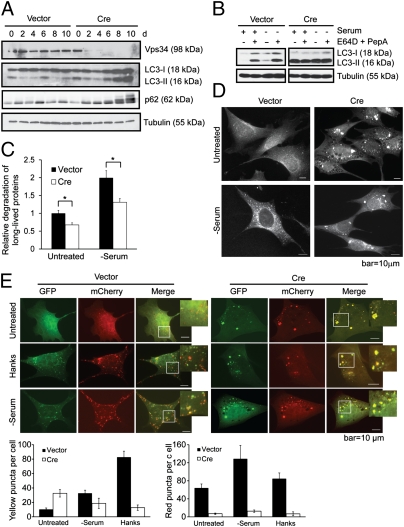
Fig. 2.
Autophagy flux is impaired in Vps34-null MEFs. (A) Vps34f/f MEFs were infected with control or Cre virus. Cell lysates were collected at indicated time points after infection and probed for Vps34, LC3, and p62. The fluctuation of the levels of LC3 and p62 in control cells may be due to the change of confluency during passaging. (B) Vps34f/f MEFs were infected with vector control or Cre. At 7 d after infection, cells were left untreated or serum deprived for 6 h, with or without the addition of lysosomal protease inhibitors E64D (10 μg/mL) and PepA (10 μg/mL). (C) MEFs were labeled with 14C-valine for 24 h and left untreated or cultured in serum-free medium for 6 h. Degradation of long-lived proteins was measured. Average of three independent experiments ± SEM is shown. *P < 0.05. (D) Vps34f/f MEFs infected with vector or Cre virus were cultured in serum-free medium for 6 h. Cells were stained for LC3 for immunofluorescence analysis. Note that although serum deprivation induces LC3 puncta in vector control cells, LC3 forms large-sized structures upon Cre infection, with no further increase upon serum starvation. (E) Vector and Cre-infected Vps34f/f MEFs were transfected with mCherry-GFP-LC3. Forty-eight hours after transfection, cells were starved in Hanks buffer or serum-free medium. Cells were observed under a deconvolution microscope. Representative images are shown. The average numbers of yellow or red puncta were obtained from three countings. Note although nutrient starvation induces both autophagosomes (yellow) and autolysosomes (red) in wild-type cells, both inductions are inhibited in Vps34-null cells.
Autophagy Flux Is Blocked in Vps34-Null MEFs.
As infection of adenoviral Cre led to a progressive abolishment of Vps34 expression, a reciprocal accumulation of LC3-I/II and p62 was observed (Fig. 2A). These changes are indicative of either increased autophagy onset or decreased autophagic protein degradation. The EM analysis indicates that starvation-induced autophagosome formation is blocked in Vps34-null MEFs (Fig. 1C), suggesting the increased LC3-II and p62 is due to decreased autophagic degradation. To verify this decreased autophagic degradation, vector control and Cre-infected MEFs were serum-starved in the presence or absence of lysosomal protease inhibitors E64D and pepstatin A (PepA). E64D and PepA treatment led to increased levels of LC3-II in wild-type MEFs at the basal state and upon serum starvation, indicating an autophagic flux (Fig. 2B). In sharp contrast, the level of LC3-II was high in Vps34-null MEFs at the basal state and was not further increased upon serum starvation and/or E64D plus PepA treatment (Fig. 2B), indicating that although LC3 conjugation to phosphatidylethanolamine (PE) still occurs, autophagy flux is impaired in Vps34-null MEFs. Compromised autophagic flux was also indicated in the long-lived protein degradation assay, at both basal and serum-starved conditions (Fig. 2C). The remaining protein degradation activity in Vps34-null MEFs is likely due to alternative forms of protein degradation such as chaperone-mediated autophagy. It is interesting to note the accumulation of LC3-II in Vps34-null MEFs. Because LC3 puncta formation and LC3 lipidation are well known to be blocked by 3-MA or wortmannin, the accumulation of LC3-II in Vps34-null cells suggests that the molecular action of 3-MA or wortmannin on autophagy is not solely through the inhibition of Vps34.
To visualize autophagy, MEFs were cultured in serum-deprived medium and subjected to immunofluorescence analysis of endogenous LC3. The control cells displayed the conversion of a diffuse cytosolic LC3 pattern to a punctate pattern upon deprivation of serum, indicating the formation of autophagic LC3 puncta (Fig. 2D). In contrast, large-sized LC3 structures were observed in Vps34−/− MEFs even at the basal state (Fig. 2D). These large LC3 structures are morphologically distinct from the autophagic LC3 puncta and are reminiscent of the LC3 aggregates observed when autophago-lysosomal protein degradation is blocked (13, 14). Importantly, the pattern of these large-sized LC3 aggregates did not change upon nutrient starvation (Fig. 2D), consistent with the impaired autophagic flux in Vps34-null cells. This phenomenon was also observed in cells stably expressing GFP-LC3 that were treated with amino acid deprivation, serum deprivation, or cultured in Hanks buffer (Fig. S3A). Immuno-EM analysis demonstrated that the GFP-LC3 gold particles were present both on the autophagosome membrane and in the autophagy cargo in wild-type cells, whereas in Vps34-null cells, the gold particles tend to form aggregates that were not associated with membrane structures, indicating that the large-sized LC3-positive structures are not autophagosomes and are instead part of protein aggregates (Fig. S3B).
To further assess whether functional autophagic flux is inhibited in Vps34-null cells, we used the mCherry-GFP-LC3 construct. Owing to the more stable nature of mCherry compared with GFP in the acidified compartment, functional autophagic flux can be determined by the appearance of more red puncta (15). In wild-type MEFs, a number of red puncta are observed under fed conditions, representing the basal level autophagy (Fig. 2E). Upon nutrient starvation, the numbers of both yellow (as a result of merged green and red signals) and red puncta increased, indicating an increase in autophagosomes and autolysosomes, respectively. In contrast, large-sized yellow aggregates were observed in both fed and nutrient-starved conditions in Vps34-null MEFs, and few red puncta were observed under either fed or starved conditions (Fig. 2E), suggesting that both autophagosome formation and autolysosome function are blocked in Vps34-null cells.
Ablation of Vps34 Leads to Diminished Localized PI(3)P Production and Autophagosome Formation.
It is believed that the localized production of PI(3)P is essential for autophagy. In yeast, Vps34 is the sole PI3K responsible for PI(3)P production (9, 16). However, in metazoans, PI(3)P may be produced via Vps34-independent mechanisms, including dephosphorylation of PtdIns(3,4,5)P3 by PtdIns 5-phosphatases and 4-phosphatase (17) or directly through the Class II PI3K (18). To observe PI(3)P generation, the PI(3)P binding domain FYVE conjugated to GFP (GFP-2xFYVE) was expressed, which forms puncta in wild-type cells (Fig. 3A). In sharp contrast, the formation of FYVE puncta was virtually absent in Vps34-null MEFs (Fig. 3A), indicating the lack of localized production of PI(3)P. Similarly, the PI3K activity associated with Vps34 or Beclin 1 immunoprecipitates was substantially lower in Vps34-null MEFs than in wild-type cells (Fig. S4).
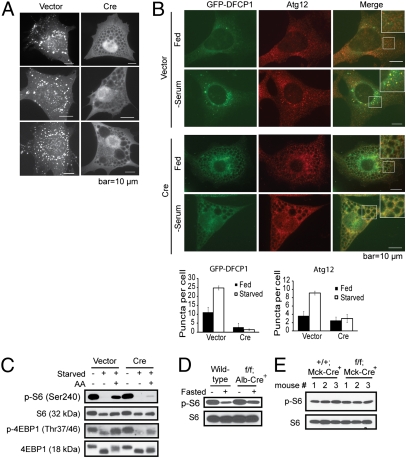
Fig. 3.
Localized production of PI(3)P, autophagosome formation, and mTOR signaling are impaired in the absence of Vps34. (A) Ablation of Vps34 leads to diminished PI(3)P production. Vps34f/f MEFs stably expressing GFP-FYVE were infected with vector control or Cre. Three representative cells from each group are shown. Note the lack of FYVE puncta in Cre-infected MEFs, indicating the absence of PI(3)P. (B) Serum starvation fails to induce DFCP1 and Atg12 puncta in Vps34-null cells. Vps34f/f MEFs were infected with vector control or Cre virus for 5 d. Cells were transiently transfected with GFP-DFCP1. Forty-eight hours after transfection, cells were fixed and immunofluorescence for Atg12 was performed. Representative images are shown. The average numbers of yellow or red puncta were determined from eight countings and are shown with SEM. Note that upon serum withdrawal, vector control cells exhibit colocalization of DFCP1 and Atg12 puncta. In contrast, no puncta formation was observed for either DFCP1 or Atg12 in the Vps34-null cells. (C) Amino acid-stimulated mTOR activation is compromised in the absence of Vps34. Vps34f/f MEFs infected with vector or Cre were starved and stimulated for 30 min with 2× MEM amino acids. mTOR signaling was measured by the levels of phospho-S6 and phospho-4EBP1. (D and E) Steady-state mTOR signaling is not affected by the loss of Vps34. Whole liver lysates from wild-type and Vps34f/f;Alb-Cre+ mice (D) and lysates from cardiomyocytes isolated from wild-type and Vps34f/f;Mck-Cre+ mice (E) were probed for phospho-S6 and total S6.
Because intracellular PI(3)P has been described to play critical roles for both endocytosis and autophagy, we examined the autophagic pool of PI(3)P by determining the localization of GFP-DFCP1 (double FYVE domain-containing protein 1), which localizes to omegasomes in a PI(3)P-dependent manner upon autophagy initiation (19). Although serum starvation induced the punctate pattern of GFP-DFCP1 in wild-type cells, it failed to do so in Vps34-null cells (Fig. 3B). Moreover, although serum starvation induced Atg12 punctate formation and partial colocalization with GFP-DFCP1 in wild-type cells, Atg12 remained diffused in Vps34-null cells (Fig. 3B). These results, together with the observations that cargo-containing autophagosomes were absent in Vps34-null MEFs and livers (Figs. 1C and 4F and Fig. S3B), indicate that early autophagosome formation is blocked in Vps34-null cells. Interestingly, Vps34 has been implicated in regulating Atg5-Atg12 conjugation (20), which complexes with Atg16L and serves as an E3-like enzyme for LC3 conjugation to PE. Although an increase in free Atg5 was observed, the level of conjugated Atg5-Atg12 was not significantly affected in Vps34-null cells (Fig. S5). This finding is consistent with the observation that LC3 lipidation can occur in the absence of Vps34 (Figs. 2 A and B and 4A), indicating that Atg5-Atg12 conjugation is independent of Vps34.
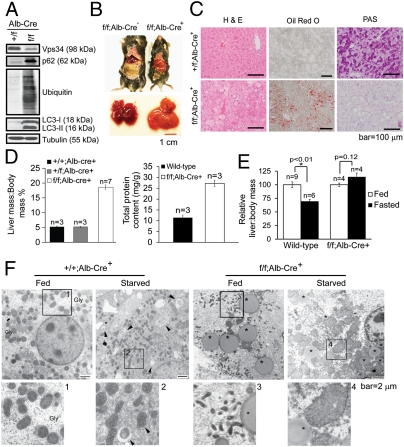
Fig. 4.
Ablation of Vps34 in the liver leads to hepatomegaly and steatosis. (A) Lysates from primary hepatocytes with indicated genotypes were probed with indicated antibodies. Note the accumulation of p62, LC3-I/II, and polyubiquitinated proteins in Vps34-null hepatocytes, indicative of blocked autophagic protein degradation. (B) Gross anatomical views of representative mice and livers with indicated genotypes. (C) Livers isolated from mice with indicated genotypes were stained with hematoxylin and eosin (H&E), Oil Red O, or Periodic Acid Schiff (PAS). Representative images are shown. (D) The mass of livers with indicated genotypes was measured and expressed relative to body mass (Left). Total protein content of wild-type and Vps34-null livers was quantified and expressed relative to the body weight (Right). (E) Wild-type and Vps34f/f;Alb-Cre+ mice were fasted for 24 h. Liver/body mass ratio was determined and expressed as normalized to that of respective fed animals. Note the difference between fed and fasted Vps34-null livers is statistically insignificant. (F) Electron micrographic images of livers from fed and 24 h fasted Vps34f/f;Alb-Cre− and Vps34f/f;Alb-Cre+ mice. Higher magnification views of boxed areas are numbered and shown in Lower. Gly, glycogen area; arrowheads point to autophagosomes; asterisks denote lipid droplets. Note the lack of glycogen deposition, accumulation of lipid droplets, and smaller mitochondria in fed Vps34-null livers. Starved Vps34-null livers are devoid of autophagosomes but are noted to have swollen mitochondria.
Vps34 Is Essential for Acute mTOR Activation by Amino Acids but Not for Steady-State mTOR Signaling.
In addition to regulation of membrane trafficking, an undefined function of Vps34 is the activation of TOR signaling. We examined mTOR activation in MEFs upon amino acid stimulation. Activation of mTOR as assessed by phospho-S6 and phospho-4EBP1 was compromised in Vps34-null MEFs (Fig. 3C), indicating that Vps34 plays an essential role in mediating amino acid activation of mTOR. Although this finding is in agreement with studies using Vps34 RNAi silencing (21–23), we also noticed that at the steady state, mTOR activation in MEFs cultured in complete medium was not affected by Vps34 deficiency (Fig. 3C). No apparent defect in mTOR signaling was observed in the Vps34-null liver under either fed or fasted conditions (Fig. 3D). Similarly, mTOR signaling was not affected by the lack of Vps34 in isolated cardiomyocytes cultured in complete medium (Fig. 3E). Therefore, although Vps34 plays an important role in mTOR activation upon acute amino acid stimulation, it is dispensable for mTOR signaling at the steady state.
Vps34 Plays an Essential Role in the Liver.
To study the effect of Vps34 deletion in vivo, we bred Vps34f/f animals with Alb-Cre mice, in which expression of Cre is under the control of the albumin promoter, resulting in deletion of floxed genes specifically in the liver by 6 wk of age (24). Successful deletion of Vps34 was confirmed in hepatocytes isolated from 8-wk-old mice (Fig. 4A). Littermates with either Vps34f/f;Alb-Cre− or Vps34+/+;Alb-Cre+ genotypes were used as controls for the possible effect of either floxed Vps34 or Cre, and did not show apparent differences, hence are both referred to as wild-type mice. In addition, heterozygous Vps34+/f;Alb-Cre+ mice were indistinguishable from wild-type mice. Noticeably, liver-specific Vps34−/− mice were smaller than wild-type mice and exhibited enlarged and pale livers (Fig. 4B). Histological analysis revealed the presence of intracellular vacuolation, and Oil Red O staining indicated an accumulation of lipids in Vps34−/− livers (Fig. 4C). Additionally, Periodic Acid Schiff (PAS) staining revealed a lack of glycogen deposits (Fig. 4C). Both liver mass and liver protein content of Alb-Cre+;Vps34f/f mice were significantly higher than that of wild-type mice (Fig. 4D). After 24 h of fasting, wild-type mice showed a decrease of liver mass of ≈30%, whereas no significant liver weight change was detected in Vps34f/f;Alb-Cre+ mice (Fig. 4E). Increased levels of p62, LC3-I/II, and polyubiquitinated proteins were observed in isolated hepatocytes (Fig. 4A) and in Vps34-null total liver lysates (Fig. S6A). By EM, Vps34-null livers revealed deficient hyaloplasmic glycogen deposits and the presence of large lipid droplets (Fig. 4F), consistent with the PAS and Oil Red O staining results (Fig. 4C). In certain hepatocytes in the Vps34-null liver, large-sized vacuoles representing swollen endosomes/lysosomes were observed, similar to what was seen in the Vps34-null MEFs (compare Fig. S6B and Fig. 1C). Interestingly, a large number of small-sized mitochondria was observed in the Vps34−/− livers under fed conditions (Fig. 4F, box 3), suggesting a possible defect in mitochondrial fusion. Moreover, although a 24-h fast induced autophagosomes in wild-type liver, Vps34-null liver displayed virtually no autophagosomes and many swollen mitochondria (Fig. 4F).
To further study the involvement of Vps34 in liver autophagy, Vps34f/f mice were bred with GFP-LC3 transgenic mice (25), then with the Alb-Cre mice. As expected, a diffused pattern of GFP-LC3 was observed in the wild-type liver. A 24-h fast caused the formation of GFP-LC3 puncta in the wild-type liver, representative of starvation-induced autophagy (Fig. S6C). However, in the Vps34−/− livers, large GFP-LC3 aggregates were observed (Fig. S6C), similar to those in Vps34−/− MEFs (Fig. 2 D and E and Fig. S3). The amount and pattern of GFP-LC3 aggregates appeared unaffected by fasting (Fig. S6C). The majority of the Alb-Cre;Vps34f/f mice died within 1 yr (Fig. S6D). Taken together, these results indicate that functional autophagy flux is blocked in Vps34−/− livers. The appearance of hepatomegaly, hepatic steatosis, increased liver protein content, lack of glycogen deposition, and increased fatality are highly consistent with the phenotype observed in the autophagy-deficient Atg7−/− and Atg5−/− livers (26–28).
Vps34 Plays an Essential Role in the Heart.
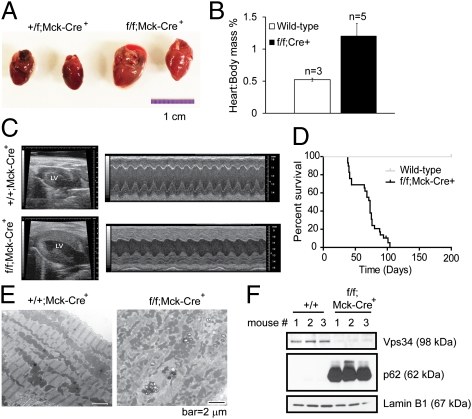
Autophagy also plays an important role in the heart. As such, Vps34f/f mice were bred to Mck-Cre mice (29) to delete Vps34 in cardiomyocytes (Fig. 5F and Fig. S7A). Gross anatomical analysis of Mck-Cre;Vps34f/f hearts revealed marked cardiomegaly (Fig. 5A) and increased heart to body weight ratio (Fig. 5B). Echocardiography of the animals in vivo demonstrated an increase in left ventricular wall thickness and mass, and decreased cardiac contractility with lower ejection fraction and fractional shortening (Fig. 5C and Table 1). Although they appeared normal at birth, Mck-Cre;Vps34f/f mice died between 5 and 13 wk of age (Fig. 5D). Electron microscopy revealed disorganized mitochondria and Z-lines in Vps34−/− hearts, and an increased number of small-sized mitochondria and enlarged vacuoles (Fig. 5E). Consistent with the Vps34-null MEFs and liver, Vps34−/− hearts and cardiomyocytes displayed elevated levels of p62, LC3-I/II, and polyubiquitinated proteins (Fig. 5F and Fig. S7A), and the presence of large GFP-LC3 aggregates (Fig. S7B). These data indicate that Vps34 is necessary for autophagic flux in the heart, and that Vps34 ablation leads to heart failure and death.
Fig. 5.
Cardiac dysfunction in Mck-Cre;Vps34f/f mice. (A) Hearts isolated from 6-wk-old Mck-Cre;Vps34+/f and Mck-Cre;Vps34f/f mice were photographed. (B) Hearts were weighed, and the heart mass is normalized to body mass. (C) Representative B-mode (left ventricle labeled) and M-mode echocardiographic images of control and Mck-Cre;Vps34f/f mice. (D) Kaplan–Meier survival curve of Mck-Cre;Vps34f/f mice. (E) Representative EM images of control and Mck-Cre;Vps34f/f hearts. Note that wild-type heart section shows characteristic alignment of mitochondria and organization of Z-lines, whereas the Vps34-null tissue shows disorganized mitochondria and Z-lines. (F) Cardiomyocytes were isolated from control and Mck-Cre;Vps34f/f hearts. Cell lysates were probed with indicated antibodies.
Table 1.
Echocardiographic measurements
| Measurement | Wild-type | Vps34f/f;Cre+ |
| LVEDD, mm | 3.17 ± 0.06 | 2.93 ± 0.18 |
| LVESD, mm | 1.58 ± 0.03 | 2.03 ± 0.14 |
| SWT, mm | 0.13 ± 0.01 | 0.43 ± 0.13 |
| PWT, mm | 0.96 ± 0.03 | 0.94 ± 0.16 |
| FS, % | 50.0 ± 3.4 | 30.1 ± 3.4* |
| EF, % | 82.3 ± 3.2 | 57.7 ± 5.2* |
| LV mass, mg | 34.8 ± 4.5 | 49.8 ± 6.4* |
EF, ejection fraction; FS, fractional shortening; LV, Left ventricular; LVEDD, LV end-diastolic diameter; LVESD, LV end-systolic diameter; PWT, posterior wall thickness; SWT, septal wall thickness. Values are mean ± SEM.
*Significantly different from the control group, P < 0.05, Student's t test. n = 4 for control; n = 6 for Vps34f/f;Cre+.
Discussion
Class III PI3K Vps34 is the only PI3K that is evolutionarily conserved from yeast to mammals (30). It has been implicated as a critical regulator for a number of molecular and cellular events including endocytic trafficking, autophagy, and mTOR signaling. Its versatile molecular function has been attributed to its various binding partners and distinct subcellular localizations (31–33). Its importance is exemplified by the fact that germ-line loss of Vps34 leads to embryonic lethality (10). Although Vps34 has been shown to play an essential role in autophagy in yeast and Drosophila (34, 35), its precise molecular function in mammalian autophagy remained elusive. A recent loss-of-function study in mouse sensory neurons suggested that functional autophagy flux still exists upon Vps34 deletion (10). Using a conditional loss-of-function mouse model of Vps34, we have examined the role of Vps34 in MEFs, liver, and heart. In agreement with the literature, endocytic protein turnover was markedly deficient in Vps34-null cells (Fig. 1E). Importantly, marked accumulation of p62, LC3, and polyubiquitinated proteins was observed in both Vps34-null liver and heart. Acute deletion of Vps34 in MEFs resulted in blocked autophagic degradation as assessed by various biochemical assays. Furthermore, large GFP-LC3 aggregates were observed in Vps34-null cells and tissues, similar to what was reported (10). Characterization of autophagy flux clearly indicated the accumulation of large-sized LC3 structures results from abrogated flux rather than induction of the onset of autophagy. Therefore, the abnormalities that we observe in Vps34-null liver and heart such as hepatomegaly, hepatic steatosis, cardiac hypertrophy, and defective contractility can be at least partially explained by defective autophagy because these phenotypes were also observed in Atg7−/− and Atg5−/− animals (26, 27, 36). In comparison with the Atg5- and Atg7-deficient settings, the more severe phenotype caused by loss of Vps34 may be due to the loss of autophagy-independent functions such as endocytosis.
Although the precise role of Vps34 throughout the full autophagic process including formation of omegasomes, maturation of autophagosomes, and autophago-lysosome fusion still needs to be determined, our study indicates that the formation of functional autophagosomes is largely impaired in Vps34-null cells. This conclusion is supported by the lack of electron-dense material in intracellular vacuoles observed by EM (Figs. 1C and 4F and Fig. S3B) and by the failure of GFP-DFCP1 and Atg12 puncta formation (Fig. 3B). It is interesting to note that several phenomena that we observed in Vps34-null cells (accumulation of LC3 aggregates and LC3 lipidation) are distinct from what is observed by using PI3K inhibitors such as 3-MA or wortmannin, suggesting Vps34-independent effects of these inhibitors in suppressing autophagy.
Our study also indicates a positive role of Vps34 in regulating the mTOR signaling in response to acute amino acid stimulation (Fig. 3C). This notion is in agreement with previous reports using RNAi and overexpression approaches (21–23), and in Vps34-null embryos (11), because the early stage embryo depends on nutrients rather than growth factors to activate mTOR (37). However, consistent with a study in Drosophila showing that TOR activity is not affected by loss of Vps34 in the whole larvae (35), we observe here that the steady-state mTOR activation is not affected by loss of Vps34 (Fig. 3 D and E). Therefore, the effect of Vps34 on TOR may depend on the duration (acute vs. chronic) and/or type (nutrient vs. hormonal) of stimuli.
Our study suggests that cautions should be taken when using Vps34 inhibitors in research and in the clinic. The role of autophagy in the initiation, development, and maintenance of human cancers is under intensive investigation (2, 38–41). Pharmacological inhibitors of autophagy to be used in conjunction with traditional anticancer therapies are under investigation (42). Our study, together with other reports (10, 11), shows that complete ablation of Vps34 leads to profound cellular and organ damage. Hence, systemic use of Vps34 inhibitors as anticancer drugs may have serious side effects. Molecules that dictate Vps34 for distinct cellular activities might be more viable drug targets.
Materials and Methods
Mice.
Vps34 (Pik3c3) gene conditional knockout ES cells in the C57BL/6 background were purchased from Sanger Institute. See SI Materials and Methods for further details for the creation of Vps34f/f mice. GFP-LC3 transgenic mice were a gift from Noboru Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) (25). Alb-Cre and Mck-Cre mice were purchased from the Jackson Laboratory. For fasting studies, mice were deprived of food while having free access to water. All mouse experiments were done in compliance with the Stony Brook University and University of Alabama at Birmingham Institutional Animal Care and Use Committee guidelines.
Statistics.
Student's t test was used to compare the differences between two groups. Significance was judged when P < 0.05. Kaplan–Meier curves for mouse survival were constructed using the Log-rank (Mantel–Cox) test.
For other materials and methods, please see SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank Drs. Deborah Brown, Noboru Mizushima, Zhenyu Yue, Qing Zhong, and Hong Chen for reagents and insightful suggestions; Namratha Sheshadri, Shengnan Liu, and Debjani Pal for technical assistance; and Susan van Horn and Edward Philips for assistance on EM. Work was supported by National Institute of Health Grants CA129536 (to W.-X.Z.), DK62722, CA136754 (to R.Z.L.), and NS064090 (to J.Z.), Susan Komen for the Cure Grant KG081538 (to W.-X.Z.), Department of Veterans Affairs merit awards (to R.Z.L. and J.Z.), and the Carol Baldwin Breast Cancer Research Foundation (to W.-X.Z.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112848109/-/DCSupplemental.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 7.Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584:1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Backer JM. The regulation and function of Class III PI3Ks: Novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci USA. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Takatoh J, Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS ONE. 2011;6:e16358. doi: 10.1371/journal.pone.0016358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson EE, Overmeyer JH, Gunning WT, Maltese WA. Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J Cell Sci. 2006;119:1219–1232. doi: 10.1242/jcs.02833. [DOI] [PubMed] [Google Scholar]
- 13.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni HM, et al. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanhaesebroeck B, et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 17.Shin HW, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDougall LK, Domin J, Waterfield MD. A family of phosphoinositide 3-kinases in Drosophila identifies a new mediator of signal transduction. Curr Biol. 1995;5:1404–1415. doi: 10.1016/s0960-9822(95)00278-8. [DOI] [PubMed] [Google Scholar]
- 19.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 22.Nobukuni T, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulati P, et al. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 25.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brüning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 30.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 31.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 33.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 35.Juhász G, et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 37.Martin PM, Sutherland AE. Exogenous amino acids regulate trophectoderm differentiation in the mouse blastocyst through an mTOR-dependent pathway. Dev Biol. 2001;240:182–193. doi: 10.1006/dbio.2001.0461. [DOI] [PubMed] [Google Scholar]
- 38.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hippert MM, O'Toole PS, Thorburn A. Autophagy in cancer: Good, bad, or both? Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 40.Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 2010;584:1427–1435. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenfeldt MT, Ryan KM. The multiple roles of autophagy in cancer. Carcinogenesis. February 11, 2011 doi: 10.1093/carcin/bgr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.