Abstract
Surveying genome-wide coding variation within and among species gives unprecedented power to study the genetics of adaptation, in particular the proportion of amino acid substitutions fixed by positive selection. Additionally, contrasting the autosomes and the X chromosome holds information on the dominance of beneficial (adaptive) and deleterious mutations. Here we capture and sequence the complete exomes of 12 chimpanzees and present the largest set of protein-coding polymorphism to date. We report extensive adaptive evolution specifically targeting the X chromosome of chimpanzees with as much as 30% of all amino acid replacements being adaptive. Adaptive evolution is barely detectable on the autosomes except for a few striking cases of recent selective sweeps associated with immunity gene clusters. We also find much stronger purifying selection than observed in humans, and in contrast to humans, we find that purifying selection is stronger on the X chromosome than on the autosomes in chimpanzees. We therefore conclude that most adaptive mutations are recessive. We also document dramatically reduced synonymous diversity in the chimpanzee X chromosome relative to autosomes and stronger purifying selection than for the human X chromosome. If similar processes were operating in the human–chimpanzee ancestor as in central chimpanzees today, our results therefore provide an explanation for the much-discussed reduction in the human–chimpanzee divergence at the X chromosome.
Keywords: Pan troglodytes troglodytes, SNP, site frequency spectrum, distribution of fitness effects, faster X
Quantifying the relative importance of purifying, neutral, and positive selection in shaping divergence between species remains a challenge in evolutionary biology. Beneficial mutations are central for understanding evolution by natural selection. However, the rarity of beneficial mutations has frustrated attempts to characterize their most basic genetic properties in higher organisms (1). One way forward is to combine genome-wide surveys of polymorphism within species and between species divergence to estimate the fraction, α, of mutations that have been fixed by positive selection. Empirical studies, particularly in the Drosophila genus show that mutations fixed by positive selection can make up a sizable fraction (>50%) of the divergence between species in gene coding regions (2) but whether these results are general for mammals, for example, remains unclear. Furthermore we still know very little about the distribution of fitness effects (DFE) of these mutations and even less about their dominance in diploid organisms. Theory predicts and recent empirical studies emphasize that the demographic history as well as variation in mutation and recombination rates can blur footprints of molecular adaptation by Darwinian selection (3, 4). This in turn can complicate the inference of DFE and α (5, 6).
In that context, contrasting the DFE and rates of adaptation in autosomes versus sex chromosomes is an elegant strategy to infer the genetic properties of the mutations underlying adaptation (7). In a panmictic population, autosomes and the X chromosome experience the same demographic history but selection and genetic drift can be different. First, hemizygosity of the X chromosome in males makes natural selection on recessive adaptive mutations more efficient, and these mutations can therefore drive higher rates of adaptive evolution on X-linked relative to autosomal regions (1, 8). Second, depending on the reproductive variance in the two sexes, we expect X chromosome regions to have between 50 and 100% of the effective size of autosomes, undergo more genetic drift, and thus be more prone to accumulate more slightly deleterious mutations. Empirical evidence for increased levels of adaptation in X-linked regions remains elusive (7), except for a unique instance of a newly formed neo-X chromosome in Drosophila miranda (9) and a recent analysis of polymorphism and divergence between Drosophila melanogaster and Drosophila simulans for about 100 genes in X-linked regions (2). In human studies, X-linked variation is lower than autosomal variation close to genes suggesting that selection affects X chromosomes more than autosomes, but Europeans have a genome-wide relative decrease in X-linked variation when comparing with Africans, suggesting that genetic drift has specifically affected X chromosome in Europeans (3, 10–12). Likewise, it has been the subject of much discussion why there is a reduced variation of the X chromosome relative to autosomes in the human–chimpanzee ancestral species (13–16).
Exome capturing efficiently targets the protein coding part of the genome at high coverage and thus allows accurate individual genotyping of synonymous and nonsynonymous diversity. Exome sequencing studies in humans have revealed an abundance of low-frequency nonsynonymous variants and very limited evidence for adaptive evolution (17).
Here we use exon capture and sequencing to extensively characterize patterns of polymorphism segregating in chimpanzees from Central Africa. (Pan troglodytes troglodytes). Central chimpanzees are genetically two to three times more variable than human populations, allowing more coding variation to be included in analysis, and previous genetic analysis suggests that chimpanzee demographic history is less complex than that of humans (18). We use these data of ∼62,000 coding SNPs to estimate the DFE and amount of adaptive evolution in chimpanzees since their divergence with humans.
Results
Patterns of Polymorphism in Central Chimpanzees.
We captured and sequenced the 12 central chimpanzee exomes to an average depth of 35× (Tables S1–S7). For analysis, we included only exons from nonduplicated areas with a unique mapping coverage against the human genome of at least 20× for each individual, leaving 49% of all exons (Table S8) and a total of 15.7 million exonic base pairs where genotypes for all SNPs could be called in all individuals (19). Fixed differences with the human reference genome were called in the same regions (Table 1). We also mapped reads against the chimpanzee reference genome and found that 96% of SNPs (97% of X-linked SNPs) were also called this way. The discrepancies (Table S4) can be attributed to the recent duplication history and the fragmented assembly of the chimpanzee reference genome (20, 21). The results reported below are based on mapping using the human reference, which contains less error and thus is better suited for interspecific comparison, but our conclusions remain unaltered when excluding ambiguous SNPs (Table S4).
Table 1.
Diversity and divergence for 12 central chimpanzees on autosomes and the X chromosome
| Autosome | X chromosome | X/A | |
| Number of synonymous SNPs | 32,942 | 808 | |
| Synonymous divergence with humans | 32,548 | 1,223 | |
| Number of synonymous sites called | 3,287,414 | 172,476 | |
| Number of nonsynonymous SNPs | 26,462 | 617 | |
| Nonsynonymous divergence with humans | 20,632 | 1,054 | |
| Number of nonsynonymous sites called | 11,380,785 | 600,624 | |
| Watterson´s θ (nonsynonymous) | 0.00062 (4e-06) | 0.00030 (0.00001) | 0.480 (0.019) |
| Watterson´s θ (synonymous) | 0.00268 (0.00001) | 0.00136 (0.00005) | 0.508 (0.017) |
| πN | 0.00046 (4e-06) | 0.00019 (0.00001) | 0.421 (0.024) |
| πS | 0.00204 (0.00002) | 0.00093 (0.00004) | 0.453 (0.022) |
| πN/πS | 0.22417 (0.00250) | 0.20847 (0.0152) | 0.930 (0.067) |
| dN | 0.00072 (0.00001) | 0.00085 (0.00004) | 1.177 (0.054) |
| dS | 0.00415 (0.00004) | 0.00293 (0.00013) | 0.706 (0.032) |
| dN/dS | 0.17317 (0.00242) | 0.28887 (0.0181) | 1.668 (0.107) |
SEM for diversity and divergence estimates were computed assuming a binomial distribution (independence of sites) and are shown in parentheses. SEM for ratios were approximated using the delta method (32). πN, nonsynonymous diversity; πS, synonymous diversity; dN, nonsynonymous divergence on the chimpanzee branch since split with humans; dS, synonymous divergence on the chimpanzee branch since split with humans.
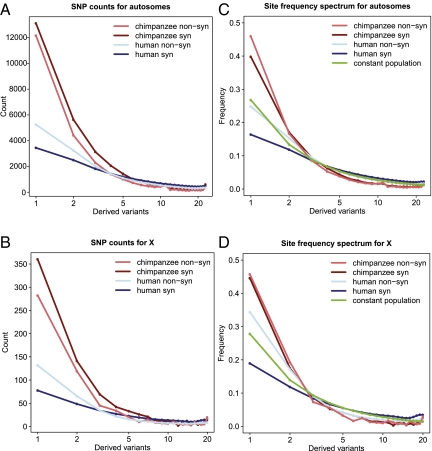
The synonymous nucleotide diversity in central chimpanzees is high (Table 1 and Tables S6–S8) but markedly reduced on the X chromosome where we observe less than half the level of diversity found in autosomes (Table 1 and Fig. S1). In humans, X to autosome diversity ratios range from 0.57 (Europeans) to 0.7 (Africans) near genes and between 0.75 (Europeans) and 0.87 (Africans) far away from genes (3, 10–12). We used the human reference genome to orient SNPs (Methods) and obtain the unfolded site frequency spectrum (SFS) at both synonymous and nonsynonymous sites (Fig. 1). The synonymous SFS is shifted toward lower frequencies relative to what is expected under a constant population model but closely fits the neutral expectation from a population recently experiencing a four- to fivefold expansion (Methods and Fig. S2). This is also reflected in Watterson's estimator of the scaled mutation rate being higher than nucleotide diversity (Table 1 and Fig. S1). The nonsynonymous SFS is slightly shifted toward rare variants relative to the synonymous SFS on the autosomes (Fig. 1C) but not on the X chromosome (Fig. 1D).
Fig. 1.
Comparison of site frequency spectra in central chimpanzees versus humans. (A) Brown and red: Counts of the number of synonymous and nonsynonymous SNPs on the autosomes as a function of the frequency of the derived variant established using humans as outgroup. Blue and light blue: Corresponding expected counts for a sample of 24 exomes of the 200 human exomes (17) (Methods). (B) Corresponding counts on the X chromosome. (C) Autosomal site frequency spectrum of synonymous and nonsynonymous SNPs derived from Fig. 1A and compared with the expectations from a constant size population without selection (green). (D) X chromosome site frequency spectrum for humans and chimpanzees.
Comparison with Human Coding Diversity.
We then compared our data to a dataset of 200 human exomes of European origin (17) by calculating the expected SFS for human autosomes and X chromosome for a sample corresponding to the size of our dataset (Methods). A larger fraction of polymorphisms in humans is nonsynonymous (50%) relative to that of central chimpanzees (45%) (Fig. 1 A and B and Fig. S3). The human synonymous SFS has fewer rare variants, suggesting different demographics, but the nonsynonymous SFS has a stronger shift toward rare variants in humans (Fig. 1C). This is even more striking for the X chromosomes (Fig. 1D). Assuming selective neutrality of synonymous polymorphisms, an explanation for these differences is more efficient selection against nonsynonymous mutations in chimpanzees, particularly on the X chromosome. To understand further the processes underlying differences in relative frequencies of rare nonsynonymous polymorphisms between human and chimpanzee, we classified the functional consequences of nonsynonymous mutations into benign, possibly damaging, and probably damaging and contrasted the proportion of singletons among these categories (Fig. S4). In both chimpanzee and human autosomes the proportion of singletons increases in categories predicted to be more damaging (Fisher's exact test, P < 10−8 for both species). For the X chromosome, there is no enrichment for chimpanzees (Fisher's exact test, P = 0.73) in contrast to humans (Fisher's exact test, P = 0.002) (Fig. S4).
Positive Selection Targets the Chimpanzee X Chromosome.
We then used the Sumatran orangutan (Pongo abelii) genome (22) to infer which of the human–chimpanzee fixed differences occurred on the chimpanzee branch (Table S8) and combined these with our chimpanzee polymorphism data. This allows measurement of the amount of adaptive evolution on the branch leading to chimpanzees, using the approach of ref. 6. All autosomes have an excess of nonsynonymous polymorphisms segregating compared with nonsynonymous fixed differences, leading to a neutrality index (NI) >1 and no evidence for adaptive evolution (Table S8). In contrast, the X chromosome shows evidence for adaptive evolution (NI = 0.76) and a proportion of amino acid differences fixed by adaptive evolution of α = 29% [95% confidence interval (CI) 0.1–0.36]. Last, we examined the 37 most abundant gene ontology categories in our data (58% and 38% of the exome data on the autosome and the X, respectively). We found a significant heterogeneity in α among categories on autosomes (Λ = 184, P < 1e-6) and on the X chromosome (Λ = 54, P < 0.003), suggesting that the differences above are genuine (6).
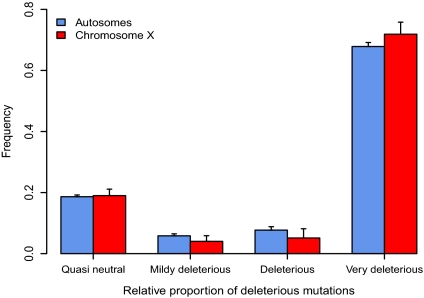
To further qualify the differences in the intensity of purifying and positive selection in chimpanzees, and to examine whether our inference above could be biased by changes in the recent demographic history, we used the SFS and divergence data to estimate jointly the DFE and α, using the approach of ref. 5. This model assumes an expansion model that also yielded the best fit to our data (Fig S2). Note that when estimating DFE from SFS data, the strength of selection against mutations is measured by its effective selection coefficient (|Nes|), which incorporates the actual fitness effect of the mutation (s) and effective size (Ne). A slightly higher fraction of sites with strongly deleterious mutations (|Nes| > 100) is found on the X chromosome relative to autosomes (Fig. 2). This finding suggests marginally more efficient purifying selection on the X chromosome, considering also that its effective size is expected to be smaller than for the autosomes. Using this approach, the fraction of nonsynonymous mutations fixed by positive selection on the X chromosome is estimated at α = 38% (95% CI 0.22, 0.51), whereas it is estimated at α ∼ 0 (95% CI −0.09, 0.07) for the autosomes. Interestingly, in human populations, the fraction of nonsynonymous mutations that are strongly deleterious (estimated |Nes| > 100) is estimated to be in the range of 30–50% (5, 23), whereas we find estimates above 70% (Fig. 2).
Fig. 2.
Efficacy of purifying selection against deleterious mutations in central chimpanzees. The strength of purifying selection is measured by the product Nes, where Ne is the effective population size and s the selection coefficient against a heterozygous deleterious mutation. Mutations are divided into four categories: quasineutral mutations (deleterious mutation with 0 < |Nes| ≤ 1), mildly deleterious mutations (1 < |Nes| ≤ 10), deleterious mutations (10 < |Nes| ≤ 100), and strongly deleterious mutations (|Nes| > 100). The proportions are estimated separately in autosomes and chromosome X. Error bars denote one SE around the estimates of each proportion.
Extreme Selective Sweeps Associated with Immunity Genes.
Given the striking difference in adaptive evolution detected using divergence, we used our diversity data to query whether selective sweeps have occurred preferentially on the X chromosome. We searched for the occurrence of extreme selective sweeps causing reduction of polymorphism over megabase-wide regions by scanning the genome using windows of 10 kb of exon data, in which we contrasted the number of polymorphic sites with the number of synonymous differences on the chimpanzee branch. The most striking example is found on chromosome 3 where a 6-Mb region spans 12 consecutive windows of low polymorphism (Fig. 3A). This region contains a cluster of immunity-related genes under positive selection in humans (24) (Fig. 3B) as well as the CCR5 gene involved in HIV resistance (25). The second and third most prominent sweeps are found on chromosome 11 and chromosome 16 (Fig. S5). These sweeps are also associated with immunity gene clusters reported to be under positive selection in human diversity genome scans (24). Chromosome X does not exhibit any clear instances of recent sweeps but its diversity is generally reduced throughout the chromosome (Fig. 3C). Given that our dataset covers 65% of human X-linked exons, and assuming an α of 1/3, we can estimate that around 1054/(3*0.65) = 540 fixations have occurred by positive selection on the X chromosome on the chimpanzee branch during the last 4 million years. That amounts to roughly one adaptive fixation every 500 generations. Of these, we expect only adaptive fixations during the last ∼0.5 million years to have affected present day levels of diversity. This amounts to only 540/8 = 67 out of 1,054/0.65 = 1,600 expected nonsynonymous substitutions and this may explain why we do not observe reduced synonymous diversity in 10kb windows with more nonsynonymous substitutions (Fig. S6), a pattern previously reported in Drosophila as evidence for recurrent selective sweeps (26). It is possible that many of the X-linked adaptive substitutions in the chimpanzee happened on standing variation as recently reported for humans (27). However, in that case the fixation rate should not depend on the dominance of new mutations and we would not expect the striking difference between X and autosomes that we report here (28). Fixation of new recessive mutations therefore remains the likely explanation for our observations.
Fig. 3.
Scanning for selective sweeps reveals a 6-Mb wide sweep on chromosome 3 and generally reduced polymorphism on chromosome X. (A) Synonymous diversity (measured as Watterson's θ) and divergence (on the chimpanzee branch) in windows of 10 kb of accumulated exon base pairs where SNPs were called. A 6-Mb region with suppressed diversity is marked by vertical lines. (B) Zooming in on this region reveals a cluster of genes involved in immunity and associated with positive selection in humans plus a gene (CCR5) involved in HIV resistance in humans. (C) Diversity and divergence in 10-kb windows on the X chromosome.
Selection Strongly Reduces Coding X Polymorphisms in Chimpanzees.
If the reproductive variance in males and females is equal, we expect a relative ratio of synonymous diversity on the X chromosome and autosomes [πs(X)/πs(A)] of 0.75. The chimpanzee mating system makes it likely that the reproductive variance is higher among males than females, increasing this ratio (in humans it is estimated at 0.81) (7). However, we observe a ratio of only 0.46–0.51 in chimpanzees (Table 1). A lower mutation rate on the X chromosome caused by male biased mutation (29) can explain only part of this reduction, e.g., even with a male-to-female mutation rate of 4, the diversity ratio is reduced only from 0.75 to 0.6 (8). Finally, we note that the ratio we observed is close to the one reported in a recent study (30), surveying a sample of six western chimpanzees for nucleotide variation on the X chromosome and chromosome 21 (π(A) = 0.081%, π(X) = 0.034%, π(X)/π(A) = 0.42). Demographic effects have the potential to temporarily alter the ratio of synonymous diversity between X and autosomes (31). We investigated whether any realistic demographic scenario has the potential to produce the observed ratio of diversity and found that only a recent dramatic reduction in population size, or corresponding bottleneck ending recently, has the potential to produce a ratio of synonymous diversity between X and autosomes near the observed. Such scenarios, however, are incompatible with the enrichment of rare synonymous diversity that we observe. Here demographics alone (Fig. S2) are unlikely to explain this pattern, and rather we argue that the reduced variation on the X chromosome is driven by a combination of more efficient selection against detrimental variants and selection for advantageous recessive mutations. Recent studies in humans have also reported a reduced X/A ratio near genes and interpreted this as Hill–Robertson effects (10, 12, 27). Purifying selection is, if anything, marginally stronger on the X chromosome than on autosomes in chimpanzees (Fig. 2), and a sizable fraction (10–50%) of X-linked nonsynonymous changes in the chimpanzee lineage has been fixed by positive selection after divergence from humans. Thus, the previously noted (7, 32) large X-linked dN/dS ratio for the chimpanzee lineage, confirmed in this larger study, appears largely driven by positive selection rather than relaxed purifying selection. This represents a clear example of faster X evolution (1, 7) driven by adaptive evolution and is our main finding.
Discussion
Using a much larger dataset, we confirm previous reports that central chimpanzees harbor two to three times more synonymous polymorphism than human populations and that the population has undergone expansion (18, 33, 34). We report here that purifying selection, as measured by the DFE (Fig. 2), is comparatively stronger than in humans (5, 23). Consistent with this finding, PolyPhen predicts that a smaller fraction of nonsynonymous mutations that segregate at low frequencies will have harmful effects (Fig. S4).
Our study suggests that both deleterious and beneficial mutations are at least partly recessive. Assuming that new mutations have the same underlying effects on fitness on the X chromosome and the autosomes, a larger number of slightly deleterious mutations are expected to segregate on the X due to increased genetic drift. If we use synonymous diversity as a proxy for the amount of genetic drift on the X chromosome and autosomes we would expect slightly deleterious mutations (|Nes| < 10) to be far more abundant on the X chromosome. This is clearly not the case (Fig. 2) and suggests that a sizable fraction of these mutations are partially recessive and thus removed more efficiently on the X chromosome (35).
Theory predicts that recessive beneficial mutations should result in faster X evolution. Increased fixation of slightly deleterious mutations on the X chromosome can also contribute to nonsynonymous divergence and drive a faster X evolution even with partial dominance (7). However, our data clearly rule out this latter possibility and leave the partial recessive beneficial mutations as a parsimonious explanation. The only alternative could be an extreme bias in the gene repertoire of the X relative to autosomes with the X chromosome harboring genes with biological functions that are most prone to adaptive evolution. However, when comparing rates of evolution within biological function (as defined by the 37 most abundant Gene Ontology (GO) categories) we find that X-linked genes have higher rates of evolution than their autosomal counterparts (paired Wilcoxon rank test, P value < 0.007, Fig. S7). Interestingly, the GO categories that exhibit the highest rates of evolution include “regulation of transcription, DNA-dependent,” “negative regulation of transcription from RNA polymerase II promoter,” and “multicellular organismal development.” This squares nicely with previous reports arguing that many of the extant differences between modern humans and chimpanzees involve changes in gene regulations and neoteny (36).
Recently, there has been much debate on the causes of the reduced divergence of the X chromosome between human and chimpanzee relative to autosomes (13–16). The reduced divergence can be accounted for by an effective size of the X chromosome of about 50% of that of the autosomes in the common ancestor of human and chimpanzees (13). Our study demonstrates that a substantial amount of adaptive evolution is targeting the X chromosome and that selection against deleterious mutations is more efficient on the X than on the autosomes in central chimpanzees. In the light of these results, a similar process of X-linked adaptation and stronger efficiency of purifying selection in the common ancestor of human and chimpanzee appears to be an attractive hypothesis that may account for reduced divergence on the X chromosome.
Methods
Sample Acquisition.
Blood samples were collected from 12 wild-born unrelated chimpanzees from Gabon, Equatorial Guinea, and zoos in Europe (Table S1). All necessary permits from the Convention on International Trade in Endangered Species were obtained. Blood-derived DNA was used to minimize somatic and cell-line–derived false positives.
Capture and Sequencing.
Each of the 12 qualified genomic DNA samples were randomly fragmented by Covaris and DNA fragments with a peak at 150–200 bp were selected for ligation, with adapters ligated to both ends. The adapter-ligated templates were purified using Agencourt AMPure SPRI beads and fragments with insert size about 250 bp were excised. Extracted DNA was amplified by ligation-mediated PCR (LM-PCR), purified, and hybridized to the SureSelect Biotinylated RNA Library (BAITS) for enrichment. Hybridized fragments were bound to the streptavidin-coated beads, whereas nonhybridized fragments were washed out after 24 h. Captured LM-PCR products were subjected to Agilent 2100 Bioanalyzer to estimate the magnitude of enrichment. Each captured library was then loaded on the Hiseq2000 platform, and high-throughput sequencing was performed for each captured library independently to ensure that each sample met the desired average fold coverage. Raw image files were processed using Illumina base-calling software 1.7 for base calling with default parameters and the sequences of each individual were generated as 90-bp paired end reads.
Mapping and Quality Filtering.
SOAPaligner (Soap 2.20) was used to align the clean reads to the human reference genome (National Center for Biotechnology Information build 36.3) as well as the chimpanzee reference genome (PanTro2) allowing a maximum of two mismatches per 90-bp fragment. Full SOAP options were: -a -b -D -o -2 -r 1 -t -n 4 –v 2.
On the basis of SOAP alignment results, the software SOAPsnp was used to assemble the consensus sequence and call genotypes in target regions. The following options were set: -i -d -o -r 0.00005 -e 0.0001 -M -t -u -L -s -2 –T (consult http://soap.genomics.org.cn/ for details).
For SNP calling we chose to include only exons with an average coverage of >20 for all 12 individuals. For chromosome X we required females to have mean coverage of >20. Using this strict criterion we maintain 49% of all exons where individual genotypes can be called in all individuals. In these regions we included SNPs if they had a quality score of >20. We called genotypes if coverage of the alternative allele (i.e., not in the reference chimpanzee genome or in the human genome) was >4.
We then excluded 1,886 SNPs from duplicated regions, yielding our final Dataset S1.
SNP Orientation.
SNPs in chimpanzee were oriented using the reference human genome sequence. Adding the Sumatran orangutan genome sequence as an extra outgroup gave conflicting results for 5,712 SNPs, equivalent to 9.4% of all SNPs, but the SFS based on SNP's concordant was very similar to the one reported. The orangutan genome sequence was also used to place fixed differences between human and chimpanzee on the chimpanzee and human branch, respectively.
Comparison with 200 Human Exomes.
We obtained the human SFS from Li et al. (17) who sequenced 200 human exomes with a mean coverage depth of 14.1. In their cleaned data, there are 11,273 synonymous and 12,586 nonsynonymous autosomal SNPs. We used the reported frequencies to obtain a SFS for 24 human chromosomal synonymous and nonsynonymous autosomal SNPs and for 21 human chromosomal synonymous and nonsynonymous X-linked SNPs. We obtained the SFS for the smaller sample size by calculating, for each SNP, the binomial probability distribution for calling a SNP at a certain frequency. If the reported autosomal frequency is p then the probability for not calling a SNP is p24 + (1 − p)24 and the probability for the SNP being at frequency x is b(24,x) px(1 − p)24-x, where x = 1, …, 23.
Fitting of SFS.
We fitted five alternative demographic models to the synonymous SFS data using DaDi (37). These include a constant population size model, as well as bottleneck, expansion, and growth models. We used Akaike's information criterion (AIC) to perform model selection.
Estimation of Positive and Purifying Selection.
To infer the strength of purifying selection from patterns of polymorphism and divergence, we binned the data in a series of adjacent genomic windows. Each window comprised 10 kb of exon material. Starting on a new window, we added contiguous exons one at a time, counted the number of nucleotides called in each, and added it to the window count. When the window count exceeded 10,000 we switched to a new window. Because we did not split exons into two windows, all windows contained slightly more than 10,000 sites, and because the exon density varied along the genome, the genomic region each window spanned varied as well.
Within each window, we recorded the number of synonymous and nonsynonymous positions. The orangutan sequence was used to obtain the number of sites contributing to divergence from human, and specifically on the chimpanzee branch. For each polymorphic position in our sample of central chimpanzees, we recorded the number of chromosomes that carried each alternative allele (derived versus ancestral; see SNP orientation for further details) and used that information to build the unfolded SFS for each window. Counts were then summed over the window to obtain genome-wide or chromosome-wide SFSs.
We relied on two complementary approaches to infer the strength of purifying selection and quantify the role of positive selection in driving divergence in coding regions of the chimpanzee genome. First, using the approach implemented by Welch (6), we inferred jointly the fraction 1 − f of mutation under strong purifying selection and the proportion α of nonsynonymous nucleotide divergence driven by positive selection from the counts of polymorphism and divergence. To make a more robust estimation, we first used a series of models of increasing complexity ranging from a simplistic pure neutrality (f = 1, α = 0) model to models incorporating a variable intensity of positive or purifying selection (f and α were allowed to vary according to each chromosome or gene category). Estimation of parameters for each gene or category of gene was made using a likelihood framework as implemented in the MKtest software (6). To compare the various models, we used AIC as suggested by Welch (6). When models differed by less than 5 AIC units, we used a model averaging procedure to obtain robust estimates of α and f parameters as the weighted average on the basis of differences in AIC of the individual estimates obtained under each model. Likelihood profiles were used to obtain an approximate 95% confidence interval for α. Heterogeneity among k GO classes for α was tested using a likelihood ratio test (LRT) comparing a model specifying a different α and f value for each class versus a reduced model still fitting k parameters for f but a single α value. The models are nested and, under the hypothesis of no heterogeneity in α, the LRT statistic (Λ) should be χ2 distributed (k − 1 df).
Second, for a more fine-grained picture of the intensity of purifying selection, we used data on divergence together with the SFS of synonymous and nonsynonymous sites to infer jointly the underlying distribution of scaled selection coefficient (Nes) against new deleterious mutations, the proportion α of the nonsynonymous divergence driven by positive selection on nonsynonymous mutations, and the parameters of a simple expansion model (5). We estimated the fraction of mutations expected to fall within four classes of intensity of selection, effectively neutral (|Nes| range 0–1): weakly deleterious (|Nes| range 1–10), strongly deleterious (|Nes| range 10–100), and very strongly deleterious (|Nes| > 100). Only effectively neutral and weakly deleterious are expected to contribute to nonsynonymous divergence as other classes of deleterious mutations have vanishingly small probabilities to go to fixation and thus contribute to nonsynonymous divergence. Confidence intervals and SEs around estimates of both α and the proportion of mutations within each class were obtained by bootstrap. Bootstrap datasets for the X-linked data were obtained by resampling X-linked 10 kb windows with replacement, whereas bootstrap datasets were obtained by stratified resampling windows across autosomes.
Scanning for Selective Sweeps.
To scan for signals of selective sweeps, we used the same 10 kb window approach and calculated the fraction of observed synonymous changes because the human/chimpanzee ancestor compared with the total number of possible synonymous changes and the Watterson's θ restricted to synonymous sites.
For each chromosome, we plotted these two summary statistics for each window and queried regions where the synonymous polymorphism is unusually small, whereas the synonymous divergence is not similarly reduced. The rationale for this strategy was to search for regions exhibiting a reduced level of variation that was not trivially driven by a reduced level of mutation rate.
Supplementary Material
Acknowledgments
We thank Brian Charlesworth, Aida Andrés, Kay Prüfer, Freddy B. Christiansen, Nick Patterson, and two anonymous reviewers for comments on the manuscript; Qing Zhang for technical assistance; and Claire Neesham for language editing. This work was supported by the Copenhagen Zoo and grants from the Danish Natural Science Research Council (to M.H.S. and H.R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106877109/-/DCSupplemental.
References
- 1.Orr HA. The population genetics of beneficial mutations. Philos Trans R Soc Lond B Biol Sci. 2010;365:1195–1201. doi: 10.1098/rstb.2009.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andolfatto P, Wong KM, Bachtrog D. Effective population size and the efficacy of selection on the X chromosomes of two closely related Drosophila species. Genome Biol Evol. 2011;3:114–128. doi: 10.1093/gbe/evq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. doi: 10.1371/journal.pgen.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keinan A, Reich D. Can a sex-biased human demography account for the reduced effective population size of chromosome X in non-Africans? Mol Biol Evol. 2010;27:2312–2321. doi: 10.1093/molbev/msq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- 6.Welch JJ. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the Faster-X effect: Empirical results and their interpretation. Evolution. 2010;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- 8.Vicoso B, Charlesworth B. Evolution on the X chromosome: Unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 9.Bachtrog D, Jensen JD, Zhang Z. Accelerated adaptive evolution on a newly formed X chromosome. PLoS Biol. 2009;7:e82. doi: 10.1371/journal.pbio.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer MF, et al. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat Genet. 2010;42:830–831. doi: 10.1038/ng.651. [DOI] [PubMed] [Google Scholar]
- 11.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2009;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottipati S, Arbiza L, Siepel A, Clark AG, Keinan A. Analyses of X-linked and autosomal genetic variation in population-scale whole genome sequencing. Nat Genet. 2011;43:741–743. doi: 10.1038/ng.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobolth A, Christensen OF, Mailund T, Schierup MH. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 2007;3:e7. doi: 10.1371/journal.pgen.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 15.Presgraves DC, Yi SV. Doubts about complex speciation between humans and chimpanzees. Trends Ecol Evol. 2009;24:533–540. doi: 10.1016/j.tree.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakeley J. Complex speciation of humans and chimpanzees. Nature. 2008;452:E3–E4, discussion E4. doi: 10.1038/nature06805. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 18.Wegmann D, Excoffier L. Bayesian inference of the demographic history of chimpanzees. Mol Biol Evol. 2010;27:1425–1435. doi: 10.1093/molbev/msq028. [DOI] [PubMed] [Google Scholar]
- 19.Li R, et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 20.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Z, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 22.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: How selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 25.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 26.Macpherson JM, Sella G, Davis JC, Petrov DA. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics. 2007;177:2083–2099. doi: 10.1534/genetics.107.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez RD, et al. 1000 Genomes Project Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orr HA, Betancourt AJ. Haldane's sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makova KD, Li WH. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- 30.Perry GH, Marioni JC, Melsted P, Gilad Y. Genomic-scale capture and sequencing of endogenous DNA from feces. Mol Ecol. 2010;19:5332–5344. doi: 10.1111/j.1365-294X.2010.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pool JE, Nielsen R. Population size changes reshape genomic patterns of diversity. Evolution. 2007;61:3001–3006. doi: 10.1111/j.1558-5646.2007.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Wu CI. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc Natl Acad Sci USA. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hey J. The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Mol Biol Evol. 2010;27:921–933. doi: 10.1093/molbev/msp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- 35.Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 36.Somel M, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.