Abstract
Directed cell migration is important for normal animal development and physiology. The process can also be subverted by tumor cells to invade other tissues and to metastasize. Some cells, such as leukocytes, migrate individually; other cells migrate together in groups or sheets, called collective cell migration. Guidance of individually migrating cells depends critically on subcellularly localized perception and transduction of signals. For collective cell migration, guidance could result from cells within a group achieving different signaling levels, with directionality then encoded in the collective rather than in individual cells. Here we subject this collective guidance hypothesis to direct tests, using migration of border cells during Drosophila oogenesis as our model system. These cells normally use two receptor tyrosine kinases (RTKs), PDGF/VEGF-related receptor (PVR) and EGFR, to read guidance cues secreted by the oocyte. Elevated but delocalized RTK signaling in one cell of the cluster was achieved by overexpression of PVR in the absence of ligand or by overexpression of fusion receptors unable to detect Drosophila ligands; alternatively, Rac was photoactivated centrally within a single cell. In each case, one cell within the group was in a high signal state, whereas others were in low signal states. The high signal cell directed cluster movement effectively. We conclude that differences in cell signaling states are sufficient to direct collective migration and are likely a substantial contributor to normal guidance. Cell signaling states could manifest as differences in gene expression or metabolite levels and thus differ substantially from factors normally considered when analyzing eukaryotic cell guidance.
Directed cell migration is important for many aspects of tissue formation during development as well as in physiology, for example in immune responses. For directional migration, cells interpret guidance cues in their environment and mount the appropriate spatially controlled responses. If the cues are gradients of soluble attractants or repellants, the process is also called chemotaxis (1). For eukaryotic cells migrating as singular entities, guidance information is perceived and responded to with subcellular spatial resolution. This helps generate the front and back of a migrating cell or orient the front and back, or both, in a spatially appropriate manner (1–3). Such responses have been extensively studied in tissue culture settings and shown to differ somewhat between cell types but they also display common properties such as localized activation of small GTPases like Rho and Rac (2, 4, 5).
Under physiological conditions (in vivo), many cells migrate not as single entities, but together in groups or sheets as collective cell migration (6, 7). It is of considerable interest to understand whether such collectives behave as a simple collection of single cells in terms of movement and guidance or whether the collective has additional properties. Sheet migration in tissue culture, reflecting wound closure and similar morphogenetic events, is the best-understood type of collective migration (8–10). The two-dimensionality of the process allows good imaging and biophysical measurements to support modeling and new collective properties have been observed in this system (11). In terms of directionality, sheet migration is mainly controlled by the presence of a free edge. A question then is whether guided collective migration, as in groups of cells responding to environmental cues by migrating to a particular place, has unique properties as well. Using border cells of the Drosophila ovary as an in vivo model, we have presented evidence that some guidance responses are mediated at the collective level (12, 13).
Border cells delaminate from an epithelium and migrate invasively and directionally, as cohesive group (14) (Fig. 1A). The border cell cluster consists of about eight cells, the central two of which are nonmigratory polar cells. Live imaging has revealed that the group is dynamic with frequent exchange of cell in the front position (12, 15). Single cell speed appears to be constant during migration (16), but net cluster movement ranges from efficient, sliding forward movement, to more inefficient and disordered forward movement, also described as tumbling.
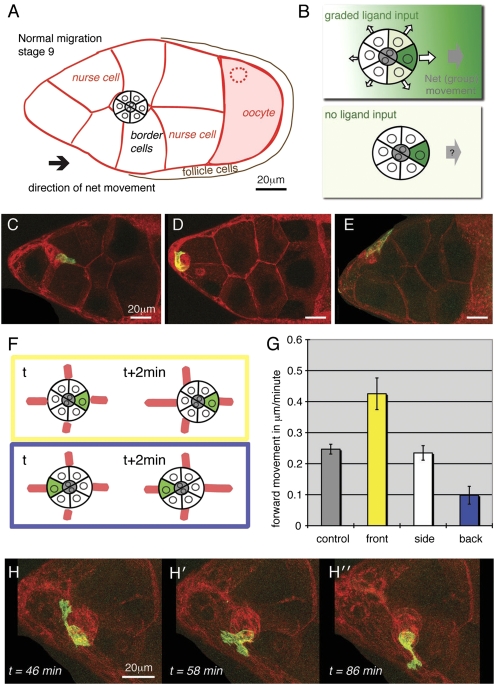
Fig. 1.
A single border cell expressing PVR–GFP directs cluster movement in the absence of ligand. (A) Schematic of a stage 9 egg chamber (section), germ line cells outlined in red. Border cells move left to right (oocyte). (B) Schematics of border cell clusters. (Upper) Proposed collective guidance mechanisms (12, 13). (Lower) Experimental approach used in this study. (C–E) Still images from three movies with one PVR–GFP positive border cell (green); all cells are outlined by red FM4-64 dye. Clusters are initiating forward movement (C) (Movie S1), rotating at start (D) or taking abnormal “side” route (E) (Movie S2). (F) Outline of how cell position and cluster movement were quantified: yellow, cell in front; blue, cell in back. Examples shown in C–E are not free clusters and could not be used; Movie S3 shows one quantified movie section. (G) Forward speed (X-direction only, toward oocyte) to the next time point for the center of the cluster according to the position of the PVR–GFP positive cell. A total of 296 time points were scored from 11 different clusters; SEM is indicated; the difference between forward speed for front versus side position and for back versus side position are highly significant (P < 0.001). (H–H′′) Three time points from Movie S4 showing one high PVR cell exploring in all directions. Genotype for all panels: Pvf11624, hs-FLP/Pvf11624; slbo-flipout-EGFR-RNAi/+; c522 (Gal4), FRT82, tub-Gal80/FRT82, UAS-PVR-GFP.
Border cells are guided by ligands coming from the oocyte, including the PDGF-related Pvf1 and the TGFα-related Gurken. The two receptor tyrosine kinases (RTKs) PVR (PDGF/VEGF related receptor) and EGFR (17, 18), detect the guidance cues and act redundantly. If both are disrupted, border cells do not find their way to the oocyte. PVR and EGFR control directionality of border cell clusters, but do not affect single cell motility (12, 15, 16). Interestingly, recent experiments have shown that local activation of Rac can be sufficient to direct border cell migration (19), just as it can direct individually migrating cells (20, 21). Direct detection of activated PVR showed that it is locally activated, with highest signal at the front of the leading cell in the presence of the normal Pvf1 ligand gradient (22). PVR activation was also elevated at the back of the rear cell. This enrichment at cell tips required RTK endocytosis and recycling (22, 23), possibly via the exosome (24), but did not strictly require ligand. Thus, with respect to guidance signaling, the cluster reacts neither as one big cell, with the rear cell representing the back, nor as multiple fully independent cells. It is a collective where cluster topology influences signaling, cells affect each other physically, and signaling in one cell affects the behavior of another (19).
In terms of guidance mechanism for the border cell group, we previously suggested that the guidance cues provide spatial information in two ways: by localized signaling as in single cells and by inducing different levels of RTK signaling in different cells (12, 13). In the latter “collective” mode, different signal levels for cells of the group would encode the directional information. As cells are inward/outward polarized by the group interactions, this information can instruct net-directed movement (Fig. 1B, Upper). We initially provided evidence that the collective mode contributed to border cell guidance (12). Indirect support has also come from other systems, in that some cells need to be in a group to perceive and react appropriately to guidance information (25). In this study, we stringently test the principle of collective guidance and find that it is sufficient for guidance of a cell group.
Results
Guidance of Border Cell Clusters by a Single High PVR Cell Without Ligand.
To determine whether a difference in signaling levels between cells would be sufficient to guide movement of a cell group, we decided to create border cell clusters deficient in normal guidance information and supply one cell with higher, but nonlocalized RTK signal. The collective guidance hypothesis predicts that this configuration should give directionality and the cluster should move in the direction given by this cell's position in the cluster (13) (Fig. 1B, Lower). Within the framework of conventional guidance signaling, based on subcellularly localized signal perception and response, this nonlocalized signal should not give directionality.
To remove endogenous guidance information, we genetically removed Pvf1, the ligand for PVR in this context (18). This has the same effect as complete removal of PVR from border cells (18, 23). EGFR signaling was reduced by RNAi directed against Egfr in all border cells to bypass the earlier role of the key ligand Gurken (17) and render cells insensitive to other ligands as well (26). These clusters show strongly attenuated but still measurable forward movement, possibly due to residual EGFR activity. In this background we sought to restore RTK signaling in a controlled and traceable manner. Direct measurement of PVR signaling activity has shown that overexpressed PVR autoactivates in the absence of ligand, but with abnormal spatial distribution (22). Importantly, simultaneously increasing expression of PVR in all migratory border cells does not improve directed migration in the Pvf1 mutant background (16). The instructive experiment was therefore to determine whether overexpression of PVR in one cell only, generating one high signal cell, would direct movement. To achieve this situation, PVR was tagged with GFP and mildly overexpressed in random single border cells (Methods). Because the position of the high signal cell would be random, the resulting cluster behavior was analyzed by live imaging (Movies S1, S2, S3, and S4). We found that cluster behavior was affected by the presence of one high signal cell and that the directional response depended on where in the cluster the high signal cell was located (Fig. 1 C–E).
Initial qualitative observations showed that clusters in which the front cell had high signal initiated migration effectively (PVR–GFP positive in Fig. 1C and Movie S1). When the expressing cell was in the back, clusters might attempt to move backward but stayed within the egg chamber (Fig. 1D). Some clusters took an abnormal route along the boundary of the egg chamber (Fig. 1E and Movie S2). These behaviors were difficult to quantify properly due to the physical constraints at the boundary of the egg chamber and of the large nurse cells. For quantification, we used movies in which the cluster was inside the egg chamber (as drawn in Fig. 1A) and measured movement in the x axis only, thus analyzing the least constrained movement (Fig. 1F and example in Movie S3). For each time point, we determined the center of the cluster and the center of the PVR–GFP-expressing cell. The position of the positive cells was noted as front, back, or side quadrant and the movement of the cluster center to the next time point (2 min later) was measured (Fig. 1 F and G). The cluster was followed for as long as quality control allowed for a maximum of 2 h, with the cell position reevaluated at each time point. Control values from clusters without PVR–GFP expression showed some forward movement. However, the presence of a single PVR high cell had significant effects on net cluster movement: When the PVR high cell was in the front, the cluster on average moved more forward. When the PVR high cell was in the back, the cluster moved relatively more backward and when on the side, cluster migration was unaltered (Fig. 1G). These results indicate that, even in the absence of ligand, one cell with a higher level of PVR signaling could direct the movement of the whole cell group.
Individual PVR high cells were quite active and some produced very prominent extensions that dynamically explored alternate directions (Fig. 1H and Movie S4). Overall, clusters containing a PVR high were also dynamic and displayed disordered movement including cluster tumbling (Movie S3). Previous analyses showed that PVR, but not EGFR, could direct the efficient, sliding forward movement typical of the initial phase of border cell migration (16). Also, overexpression of PVR in one border cell allowed this cell to stay in the front (12) and overexpression in all cells allowed whichever cell was in the front to stay there (16). These effects all depended on endogenous (graded) Pvf1. More tumbling and less efficient forward cluster movement is normally seen as border cells approach the oocyte (16). This phase is dominated by EGFR and appears to be more dependent on collective effects (12). Net forward cluster movement is about 0.5 μm/min and “persistence” (net speed cluster speed/single cell speed) only 0.3 (16). Interestingly, a similar net cluster speed was observed in the present experiments when the single high PVR cell was in the front (Fig. 1G). This may reflect the movement bias obtainable by collective guidance. In conclusion, PVR can direct persistent movement in a ligand gradient but PVR is also capable of steering, using the collective mode. The behavior of the migration group reflects the mode of guidance used.
Guidance Using “Ligand-Blind” RTK Fusions Expressed in One Border Cell.
The stringency of the experiments with PVR depends on the Pvf1 mutant background giving no spatial information in border cells. Genetic tests support that the Pvf1 mutant allele retains no function in this context (18), but it is not a gene deficiency and Pvf2 or Pvf3 could potentially contribute. To circumvent potential ligand contributions altogether, we decided to generate RTK derivatives that would signal like PVR or EGFR but not be able to detect endogenous ligands. These fusion receptors had the extracellular domain of the human EGF receptor and intracellular domains of Drosophila PVR or EGFR (hE-PVR and hE-EGFR in Fig. 2A). The mammalian ligands for EGFR, EGF, and TGFα, do not cross-react with the Drosophila receptor (27). To confirm the specificity in vivo, we exploited the fact that co-overexpression of guidance receptor and cognate ligand in border cells severely perturbs directed migration (18). When expressed in border cells, hE-PVR and hE-EGFR responded strongly to coexpression of TGFα, but not to Pvf1 or Vein, ligands for Drosophila PVR and EGFR, respectively (Fig. 2B). The fusions also did not rescue the defect in directional migration induced by expression of both dominant negative PVR and EGFR (DN-PVR + DN-EGFR in Fig. 2C). The fusion receptors localized partly to the cell surface but mostly to intracellular vesicles as observed for EGFR (Fig. 2D). Importantly, as for PVR (22) (Fig. 2E), receptor autoactivation and signaling could be observed upon overexpression of hE-PVR (Fig. 2F). These fusion receptors therefore had the characteristics needed to generate single border cells with high signal that mimicked signal from PVR or EGFR, but where this signal could not be modified by ligands present in the fly.
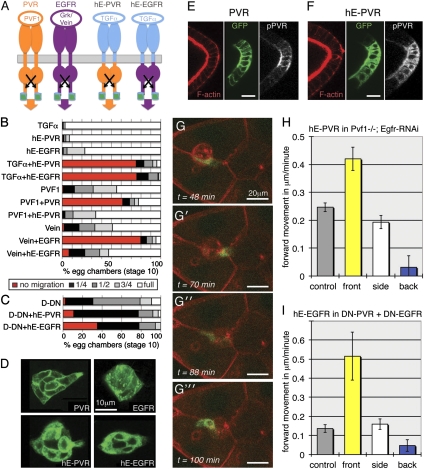
Fig. 2.
RTK fusions with human EGFR extracellular domain cannot detect Drosophila ligands but can direct cluster movement. (A) Schematic of fusion receptors generated; green barrel is GFP. Cognate ligands are indicated for information. (B and C) Quantification of border cell position at stage 10; normal is full migration; the strongest effect is no migration; >100 egg chambers were scored per genotype. Genotypes: slbo-Gal4 plus the indicated UAS or EP transgenes. DN: DN-PVR, DN-EGFR. (D) Distribution of receptors (as in A) expressed in all border cells, detected by the GFP tag. (E) Antiphospho-PVR staining (22) (white) of follicle cells expressing PVR–GFP (green); phalloidin (red) outlines the cells. (F) Antiphospho-PVR staining (22) (white) of follicle cells expressing hE-PVR-GFP (green) showing receptor autoactivation; phalloidin (red). (G) Time-series images from Movie S5; genotype Pvf11624, hs-FLP/ Pvf11624; slbo-flipout-EGFR-RNAi/+; c522 (Gal4), FRT82, tub-Gal80/FRT82, UAS-hE-PVR-GFP. (H) Quantification of cluster movement according to position of the hE–PVR-expressing cell, genotype as in G; 253 time points from seven clusters; difference between front or back and side or control is highly significant (P < 0.001). (I) Quantification of cluster movement when all cells express DN-PVR and DN-EGFR, according to position of single hE–EGFR-expressing cell. Genotype: hs-FLP/+; UAS-DN-PVR, UAS-DN-EGFR/slbo-Gal4; FRT82/ FRT82, UAS-hE-EGFR-GFP; 299 time points from seven cluster; difference between front or back and side or control is highly significant (P < 0.005).
We first tested effects of single-cell expression of the hE-PVR fusion in the same genetic background used for PVR experiments. As for single-cell PVR expression, overall disordered, or tumbling border cell movements were observed in these clusters (Movie S5 and Fig. 2G). More importantly, detailed quantification of cluster movement for each time point showed significant directive effects of the high signal cell, depending on the position of the cell (Fig. 2H). Thus, in this mutant background, a ligand-blind fusion protein could control cluster movement in the same way as PVR.
The heterologous fusions also allowed us to investigate a situation in which all border cells expressed both dominant negative PVR and EGFR (DN-PVR + DN-EGFR). In this background, normal guidance was effectively blocked (Fig. 2I, control). Clusters with only one cell expressing the fusion receptor were rare, but we were able to analyze a sufficient number of movies for hE-EGFR. We observed significant directional effects of hE-EGFR expression, depending on location of the expressing cell (Fig. 2I). So even in the presence of both dominant negative PVR and EGFR, expression of an active but blind RTK fusion could steer group movement. This experiment also confirmed that the intracellular domain of EGFR could mediate the directional effects required in collective guidance. Finally, in both experimental setups using fusion receptors (Fig. 2 H and I), the average net forward speed observed when a high signaling state cell was in the front was similar to that normally observed in the late phase of migration. Together, these experiments confirm that a cell with high RTK signal can direct cluster movement and that it can do so in the absence of graded guidance input.
Guidance by Photoactivatable (PA)-Rac Activated Centrally in One Border Cell.
As a final approach to testing the ability of cells with different signal levels to direct collective migration, we used the recently generated PA-Rac. This elegant tool is a genetically encoded, activated but caged form of the small GTPase Rac, which can be transiently uncaged to expose active Rac in a spatially controlled manner by a focused light beam (20). Because the uncaging is transient, it also provides temporal control. Activation of PA-Rac was first shown to guide movement of single cells (20, 21), as predicted from analyses of Rac function in the front of migrating cells (2). Subsequent experiments showed that local activation of Rac was also sufficient to guide a whole border cell cluster in both wild-type and guidance-signaling defective backgrounds (19). In the border cell cluster experiments, photoactivation appeared to be centered on the outer part of a cell (front of front cell or back of back cell). It is therefore not clear whether the observed effects are due to local activation of Rac within a cell or different levels of Rac activation in different cells, or both. To distinguish between these possibilities, we decided to redo the experiments but deliberately produce high Rac-activity cells without subcellular bias. This was achieved by activating PA-Rac centrally in a target cell (Fig. 3 A and C). If such activation could steer a cluster, this would indicate that cell-based difference in level of Rac activity is sufficient to give directional movement of a cluster.
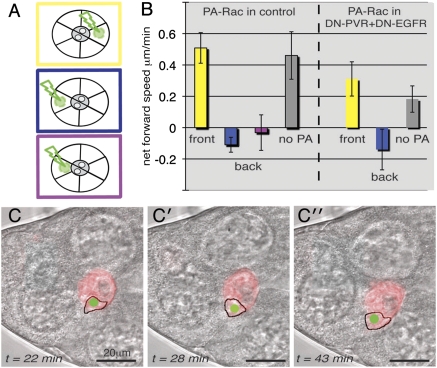
Fig. 3.
Activation of Rac, on a whole-cell basis, directs cluster movement. (A) Schematic of experimental setup with Rac-PA where the photoactivation area is indicated in green. (Top, yellow) Midcell activation of the front cell. (Middle, blue) Midcell activation of the back cell. (Bottom, purple) Activation in the inner part of the back cell. (B) Average net cluster movement (per movie) in different scenarios, with SD indicated. For each photoactivation (PA) scenario, five to seven movies were analyzed. Differences between mock and rear cell activation and, for DN-PVR+DN-EGFR, between mock and front cell activation were statistically significant (P < 0.01). Genotypes: UAS-mCherry-PA-RacQ61L/+; slbo-Gal4/+ (Rac-PA, Movies S6, S7, and S8) and UAS-mCherry-PA-RacQ61L/UAS-DN-PVR, UAS-DN-EGFR; slbo-Gal4/+ (Rac-PA in DN-PVR+DN-EGFR, Movies S9 and S10). (C) Example where Rac photoactivation in the middle of a back cell causes the cluster to move backward between two nurse cells (stills from Movie S7). The transmission image is combined with red fluorescent channel (Rac-PA-Cherry expression); the selected cell is outlined and the actual photoactivation area is indicated in green.
For these experiments, PA-Rac was expressed in all cells and visualized by its red fluorescent tag. The photoactivation region was limited to 4 μm diameter in the single confocal section used and was repositioned, as cluster movement required, to maintain activation in the middle of the target cell (Fig. 3C and Movies S6, S7, S8, S9, and S10). Clusters were followed for 60 min and the final net movement in the x axis was measured for each experiment (Fig. 3B). Photoactivation of the front cell had negligible effect in normal guided clusters (compare “front” to “no PA”; Movie S6), but activation in the middle of the back cell could drive net cluster movement backward (Movie S7). Thus, the endogenous guidance signals informing the front cell apparently could not be significantly improved by a nonlocalized increase in Rac activity, but could be balanced or even overridden by high Rac activity in another cell. In a separate set of experiments, we placed the center of the photoactivation spot in the part of the back cell opposite to its prospective leading edge (purple in Fig. 3 A and B and Movie S8). Again, back cell activation counteracted the endogenous forward dominance confirming that what matters is which cell is activated, not where in the cell activation occurs. In the guidance defective background (dominant negative PVR and EGFR), midcell activation of the front cell or back cell promoted forward movement or backward movement of the cluster, respectively (Fig. 3B and Movies S9 and S10). These experiments confirm that differential cell-based signaling states—in this case providing one cell with a higher level of active Rac overall—can effectively guide migration of a cell group.
Discussion
The experiments presented in this study show that one cell within a group with elevated signaling level, i.e., in a signaling state different from the other cells, can direct group migration. This means that the collective guidance mode proposed previously (12, 13) is sufficient to guide group movement in vivo. The average directional bias observed when a high signal cell expressing PVR or hE-PVR was in the front was similar to that seen during normal late phase of migration (16). Also, the effect achieved by activation of Rac-PA in the middle of a cell (Fig. 3) was both qualitatively and quantitatively similar to those obtained previously by activation of Rac at the edge of the cluster (19). Thus, cell-based signaling differences can give functional outputs that are quantitatively similar to that achieved by normal guidance. Although this does not preclude additional contributions from local reactions to guidance cues, it indicates that, at least in this case, collective information is likely a major contributor to guidance.
The key significance of cell-based or collective guidance is that it changes our view of what guidance information is. Allowing differences at the whole cell level to transmit spatial information means signal transduction mechanisms in addition to those normally associated with guided cell migration have to be considered. For example, difference in gene expression, in level or state of a small diffusible protein or even in levels of metabolites may provide directional information. In conventional eukaryotic guidance characterized in single cells, based on localized signaling, such mechanisms are not possible as the spatial information is lost.
How might whole-cell–based features of migratory cells be modulated in such a way that directional group movement results? The high signal cell could be more prone to making cellular extensions that reach outward from the cluster and grab the substrate. Its extensions might also be more productive by adhering better to the substrate and generating more force. Both extension formation and productivity of extensions have been shown to be guidance regulated in this system (16). For technical reasons, it is not feasible to do the detailed quantitative analysis required to reveal how each of these features are affected by the manipulations performed in this study. However, when considering cellular mechanisms, the key point is that guidance information serves as a bias acting on a group of cells, each of which intrinsically polarized due to the geometry of the cell–cell interactions within the group (illustrated in Fig. 1B). So, whereas the guidance information is cell based, the outputs can be local.
In addition to the guidance input, differences in behavior between cells may also be accentuated by inhibitory interactions. In experiments with PA-Rac, activation in one part of the cluster was shown to negatively affect extensions from another (19). Inhibitory interactions may be signaling effects, for example through Delta/Notch signaling (28, 29), Eph/Ephrin signaling (30, 31), or other pathways. The cell–cell interactions could also be based on mechanically transmitted information. The border cell cluster displays what appears to be considerable cell cohesiveness within the migrating group, which would be expected to reflect some mechanical coupling. Cluster cohesiveness is likely one determinant of how much collective behavior dominates in a group. However, in less cohesive groups such as Xenopus neural crest cells, cell–cell interactions can also control directionality of movement (31). Thus, collective guidance effects appear not to be restricted to the most cohesive cell groups.
Cell migration is a dynamic process. Border cells continuously exchange places, including the lead position (12, 15). Interestingly, exchange of cells in the lead position was also recently observed when sprouting angiogenesis was analyzed by live imaging (29). Dynamics in cell positions within a migratory group can be viewed as a reflection of the probing nature of the guidance process at the group level, not unlike regional dynamics in individually migrating cells. Dynamic border cell groups are seen under normal conditions and also when guidance signals are reduced (12, 15, 16). The group can be “locked” into a sliding mode with high level of graded and polarized PVR signaling (16, 22). This suggests that, mechanistically, group dynamics could require decay or inactivation of guidance signaling or outputs. The group is not locked by simply biasing one cell with higher PVR signaling level (this study). Thus, highly polarized dominant guidance signals may be uniquely refractory to this inactivation, highlighting additional differences in the modes of guidance.
It is clear that individual cells can be guided, in vitro and in vivo, by localized signaling effects. This is exquisitely displayed during the efficient directional movement by cells of the immune system (32, 33). For border cells (22, 23), and for other cells that migrate collectively (34, 35), localized signaling also occurs and contributes to directional movement. So why is collective guidance used, when a well-functioning single cell guidance system exists? One broad rationale could be that multiple information-gathering systems are better than one for ensuring biological robustness. Also, the cell-state–based measurements used in collective guidance may allow averaging of signal input over larger areas or longer timescales. This may be relevant during morphogenesis of complex systems. More specific explanations could include the possibility that some receptor/ligand pairs are well placed for providing directional information in the tissue but not well suited for subcellularly polarized signals. Drosophila EGFR seems to be one such example (16, 17). Overall, the cell biology of guidance responses acquires an addition dimension in collective migration as these cells interact with each other as well as with their substrate and environment. Collective guidance is an emergent property of this added dimension.
Methods
Fusions Constructs.
PVR and EGFR were tagged by introducing an XbaI site in place of the stop codon and inserting 6xGly linker and GFP. For hE-PVR-GFP and hE-EGFR-GFP, an NcoI site was placed after the transmembrane coding sequence of human EGFR and of PVR (for EGFR the endogenous NcoI site was used) and the domains exchanged. Constructs were cloned into pUAST-attB, with attP landing site at 86Fb used, and pUAST (a random integration for one set of UAST-PVR-GFP experiments). To reduce Egfr expression in all border cells, slbo-flipout-EGFR-RNAi was constructed: a 550-bp fragment was amplified using primers TAGCTCTAGAGCGACTGGAGGTGTTCTC and TAGCTCTAGACTCCTGGCAGTGATCTG (extracellular) and cloned into pWIZ (36) tail to tail. The 2.6-kb slbo border cell enhancer (37) was inserted in HindIII and EcoRI of pUAS-attB, the EGFR inverted repeats cloned into AvrII site, and flip-out lacZ cassette (38) inserted into Asp718 site in forward direction. For transgenics, the attP landing site at 51D was used. After imaging, standard Xgal staining was performed and only egg chambers with complete removal of the lacZ cassette were used.
Genetics and Analysis of Fixed Samples.
For border-cell–specific expression, slbo-Gal4 (37) or c522 (Bloomington) were used. The Pvf11624 mutant background, EPg-Pvf1 and EPg-Vein and dominant negative EGFR and PVR were described previously (18). After hs-FLP mediated excision, slbo-flipout-EGFR-RNAi gave migration defects like DN-EGFR (17), but weaker. Human TGFα was cloned with a C-terminal HA-tagged into pUAST. UAS-mCherry-PA-RacQ61L (UAS-PA-Rac) is described in ref. 19 and expression was driven with slbo-Gal4, alone or with UAS-DN-PVR and UAS-DN-EGFR. Fixed sample staining and analysis was done as described previously (18, 22). Detailed genotypes are given in legend to Figs. 1–3 and Movies S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10.
Live Imaging for Single-Cell RTK Expression and Image Analysis.
Females within 24 h after eclosion were heat shocked for 30 min at 37 °C and dissected for imaging after 2 d. Live imaging was done as previously described (12, 16). Images were acquired by confocal microscopy (SP5; Leica) with a 63× objective, sections 2.5 μm apart, covering the cluster every 2 min for up to 2 h. Only fully detached clusters, at stage 9 and with only one GFP positive cell were analyzed.
Images were processed with ImageJ and customized macros. Egg chambers were realigned by the stackreg plug-in (39) using transmission images, which also corrected for the slight backward substrate movement (16). The center of the cluster and the GFP-expressing cell were manually tracked. For position of the expressing cell, a line was drawn from its center to the center of the cluster and the angle defined relative to the x axis. Front was defined as 0° ± 45°, back as 135°–225°, and the rest as side. Each analysis was done per time frame and cluster speed (x axis movement of the center of the cluster) measured to the next time frame. For statistical significances in speed differences, t tests were used. The variances of data were tested by f test. If they were unequal, Welch's t test was used. If not, Student t test was used. P values were from the two-tailed test.
Rac-PA Experiments.
Rac-PA experiments were done by slight modifications to published setup (19) and methods above except for no FM4-64. Argon and DP561 laser intensity were set for 50% power. To photoactivate, the 458-nm laser line was set at 10% intensity and a 4-μm circular spot in a single Z plane defined as the region of interest (ROI) and was scanned 25 times at 200 Hz. Then red (573–667 nm) and transmission image (DIC) were collected followed by a pause. Photoactivation cycles 80 s apart were continued for 1 h, with the ROI manually moved to keep it within the cell and close to the cell center, with a slight inward bias. For deliberate inward bias, only movies the PA spot was centered in the inner half of the cell at all time points were used. Speed was calculated as the distance of the center of the cluster (toward oocyte) between first and last frame divided by the time elapsed; statistical analysis was as above.
Supplementary Material
Acknowledgments
We thank Denise Montell for sharing the Rac-PA stocks, Graham Wright for help with the PA experiments, and Hsin-Ho Sung for anti-pPVR staining. This work was supported by the Agency for Science, Technology and Research in Singapore and in an early phase by Temasek Life Sciences Laboratory, Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115260109/-/DCSupplemental.
References
- 1.Van Haastert PJ, Devreotes PN. Chemotaxis: Signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;8:215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Wong K, Pertz O, Hahn K, Bourne H. Neutrophil polarization: Spatiotemporal dynamics of RhoA activity support a self-organizing mechanism. Proc Natl Acad Sci USA. 2006;103:3639–3644. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 7.Rørth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 8.du Roure O, et al. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poujade M, et al. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci USA. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitorino P, Meyer T. Modular control of endothelial sheet migration. Genes Dev. 2008;22:3268–3281. doi: 10.1101/gad.1725808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trepat X, Fredberg JJ. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 2011;21:638–646. doi: 10.1016/j.tcb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianco A, et al. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- 13.Rørth P. Collective guidance of collective cell migration. Trends Cell Biol. 2007;17:575–579. doi: 10.1016/j.tcb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Montell DJ. Border-cell migration: The race is on. Nat Rev Mol Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- 15.Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Poukkula M, Cliffe A, Changede R, Rørth P. Cell behaviors regulated by guidance cues in collective migration of border cells. J Cell Biol. 2011;192:513–524. doi: 10.1083/jcb.201010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duchek P, Rørth P. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 2001;291:131–133. doi: 10.1126/science.291.5501.131. [DOI] [PubMed] [Google Scholar]
- 18.Duchek P, Somogyi K, Jékely G, Beccari S, Rørth P. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12:591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo SK, et al. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssens K, Sung HH, Rørth P. Direct detection of guidance receptor activity during border cell migration. Proc Natl Acad Sci USA. 2010;107:7323–7328. doi: 10.1073/pnas.0915075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jékely G, Sung HH, Luque CM, Rørth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Assaker G, Ramel D, Wculek SK, González-Gaitán M, Emery G. Spatial restriction of receptor tyrosine kinase activity through a polarized endocytic cycle controls border cell migration. Proc Natl Acad Sci USA. 2010;107:22558–22563. doi: 10.1073/pnas.1010795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theveneau E, et al. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald JA, Pinheiro EM, Kadlec L, Schupbach T, Montell DJ. Multiple EGFR ligands participate in guiding migrating border cells. Dev Biol. 2006;296:94–103. doi: 10.1016/j.ydbio.2006.04.438. [DOI] [PubMed] [Google Scholar]
- 27.Zak NB, Shilo BZ. Biochemical properties of the Drosophila EGF receptor homolog (DER) protein. Oncogene. 1990;5:1589–1593. [PubMed] [Google Scholar]
- 28.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 29.Jakobsson L, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- 30.Poliakov A, Cotrina M, Wilkinson DG. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev Cell. 2004;7:465–480. doi: 10.1016/j.devcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Carmona-Fontaine C, et al. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 33.Mempel TR, Scimone ML, Mora JR, von Andrian UH. In vivo imaging of leukocyte trafficking in blood vessels and tissues. Curr Opin Immunol. 2004;16:406–417. doi: 10.1016/j.coi.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 34.Ruhrberg C, et al. Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes Dev. 2002;16:2684–2698. doi: 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arboleda-Estudillo Y, et al. Movement directionality in collective migration of germ layer progenitors. Curr Biol. 2010;20:161–169. doi: 10.1016/j.cub.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 36.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 37.Rørth P, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 38.Wimmer EA, Cohen SM, Jäckle H, Desplan C. buttonhead does not contribute to a combinatorial code proposed for Drosophila head development. Development. 1997;124:1509–1517. doi: 10.1242/dev.124.8.1509. [DOI] [PubMed] [Google Scholar]
- 39.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.