Abstract
Single strand nicks and gaps in DNA have been reported to increase the efficiency of nucleosome loading mediated by chromatin assembly factor 1 (CAF-1). However, on mismatch-containing substrates, these strand discontinuities are utilized by the mismatch repair (MMR) system as loading sites for exonuclease 1, at which degradation of the error-containing strand commences. Because packaging of DNA into chromatin might inhibit MMR, we were interested to learn whether chromatin assembly is differentially regulated on heteroduplex and homoduplex substrates. We now show that the presence of a mismatch in a nicked plasmid substrate delays nucleosome loading in human cell extracts. Our data also suggest that, once the mismatch is removed, repair of the single-stranded gap is accompanied by efficient nucleosome loading. We postulated that the balance between MMR and chromatin assembly might be governed by proliferating cell nuclear antigen (PCNA), the processivity factor of replicative DNA polymerases, which is loaded at DNA termini and which interacts with the MSH6 subunit of the mismatch recognition factor MutSα, as well as with CAF-1. We now show that this regulation might be more complex; MutSα and CAF-1 interact not only with PCNA, but also with each other. In vivo this interaction increases during S-phase and may be controlled by the phosphorylation status of the p150 subunit of CAF-1.
MMR has evolved to process mismatches arising during replication or recombination (1, 2). That its malfunction leads to cancer (3) bears witness to the importance of its role in the maintenance of genomic stability. In eukaryotes, mismatch repair (MMR) is initiated by the binding of MutSα, a heterodimer of MSH2 and MSH6, to non-Watson–Crick base pairs in DNA. Exchange of ADP for ATP then converts MutSα into a sliding clamp, which diffuses along the DNA contour, possibly together with a heterodimer of MLH1/PMS2 named MutLα, in search of free DNA termini that serve as initiation sites for exonuclease 1-catalyzed degradation of the error-containing DNA strand (1, 2).
In mammalian cells, free termini are generally marked by PCNA, which helps orchestrate replication and repair DNA synthesis through interactions with key players of DNA metabolism, including MSH6 and chromatin assembly factor 1 (CAF-1) (4). The latter complex, composed of p150, p60, and p48 subunits (5), promotes rapid assembly of nucleosomes on newly replicated DNA (6).
Nucleosome loading causes supercoiling, which compacts the genome and increases its stability, but which makes DNA less accessible to metabolic processes. Thus, during nucleotide excision repair, efficient processing of UV-induced DNA damage requires chromatin remodeling (7, 8) and CAF-1-mediated chromatin reassembly upon repair completion (9). Whether similar transactions take place during MMR is unknown. As the sliding function of MutSα was reported to be blocked by nucleosomes (10, 11), albeit not in all sequence contexts (12), we argued that mismatches arising during replication would have to be repaired before nucleosome assembly takes place. However, given that nucleosomes are loaded on newly replicated DNA approximately 250 base pairs behind the replication fork (13), the time window for repair is extremely small. We therefore postulated that mismatch correction and chromatin assembly in the immediate vicinity of the replication fork must be coordinated to either prevent the assembly of nucleosomes on mismatch-containing DNA, or, alternatively, to enable the MMR system to displace them. Using combined in vitro MMR/chromatin assembly assays, we show that ongoing MMR interferes with nucleosome deposition. In vivo, the interplay between MMR and chromatin assembly may be regulated also by a direct interaction between MutSα and the p150 subunit of CAF-1, which we identified in the course of this study and which increases during S-phase of the cell cycle, or upon certain types of DNA damage.
Results
A Combined MMR/Chromatin Assembly Assay.
To study the interplay of MMR and chromatin assembly under identical experimental conditions, we modified existing (14, 15) in vitro systems (Fig. S1A).
We used nuclear extracts of MMR-deficient LoVo (MutSα-deficient), or 293T-Lα- [MutLα-deficient (16)] human epithelial cells. The extract amounts were adjusted to contain comparable amounts of CAF-1 (Fig. S1B). We also supplemented the extracts with [α-32P] dCTP, to visualize DNA repair synthesis.
Our MMR substrate is a plasmid heteroduplex containing a single G/T mismatch within an AclI restriction site and a nick 304 nucleotides 3′ from the mispaired G (15). The mismatch makes the plasmid refractory to AclI digestion at that site, but correction of the G/T mispair to A/T regenerates it. Thus, AclI digestion of the plasmid DNA recovered from the MMR reaction generates two or three restriction fragments of different lengths from the unrepaired or the repaired plasmid, respectively (Fig. S1C).
When the LoVo extracts were supplemented with purified recombinant MutSα, approximately 80% of the heteroduplex G/Tnicked substrate were repaired after 30 min, as shown by UV shadowing of an ethidium bromide-stained (EtBr) agarose gel (Fig. S1D, Upper). An autoradiograph of the same gel (Lower) indicated that the [α-32P]dCMP was incorporated preferentially into the 813 and 735 bp fragments resulting from AcII cleavage at the corrected mismatch site. No repair and only background levels of radioactivity could be detected upon incubation of the substrate with extracts that were not supplemented with MutSα.
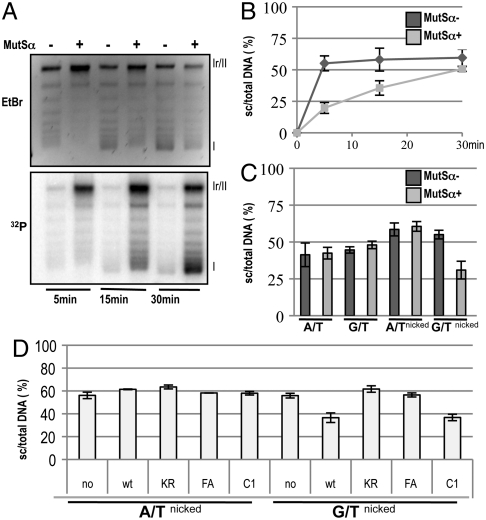
In this assay, we could also examine the extent of supercoiling generated by nucleosome loading onto the plasmid DNA (14). Winding of DNA around a nucleosome introduces torsional strain. In nicked or gapped molecules, this strain is released. However, repair of the gap and/or sealing of the nick blocks the relaxation, such that covalently closed circular molecules carrying nucleosomes become supercoiled once the proteins are removed. The 3.2 kb phagemid used in this assay could accommodate up to a maximum of 16 nucleosomes. Protein extraction followed by agarose gel electrophoresis and poststaining with EtBr should thus detect 17 bands: the 16 different topoisomers and the open circular (nicked or gapped) molecules, which migrate the slowest. In the absence of MMR, more than 50% of the G/Tnicked substrate were converted to covalently closed circular topoisomers already at the earliest time point (Fig. 1A, Upper; for quantitation see Fig. 1B, dark gray line). This supercoiling was less efficient in MutSα-supplemented LoVo extracts (Fig. 1A, Upper, Fig. 1B, light gray line). Interestingly, the autoradiograph of the same gel revealed that the molecules which incorporated the highest levels of [α-32P]dCMP at the 30 min. time point were almost fully supercoiled (Fig. 1A, Lower). Taken together, the above findings showed that chromatin assembly on the G/Tnicked substrate was delayed in MMR-proficient extracts, but that successful repair was accompanied by efficient nucleosome loading.
Fig. 1.
Ongoing MMR delays chromatin assembly. (A) Kinetics of supercoiling of nicked mismatch-containing (G/Tnicked) DNA substrates incubated with nuclear extracts of human MutSα-deficient LoVo cells, supplemented where indicated with recombinant MutSα. The figure shows a UV shadowing of an agarose gel poststained with EtBr (Upper) and its autoradiograph (32P, Lower). Migration positions of the open/nicked circular (Ir/II) and supercoiled DNA (I) isoforms are indicated. (B) Quantitation of three independent G/Tnicked supercoiling experiments (A shows a representative example), as a ratio of the sum of all covalently closed topoisomers (cc) versus total DNA. (C) Supercoiling of homoduplex (A/T, A/Tnicked) and heteroduplex (G/T, G/Tnicked) substrates incubated with nuclear extracts of human MutSα-deficient LoVo cells, supplemented where indicated with recombinant MutSα. Data from three independent experiments are shown. The error bars represent the standard deviation from the mean. (D) Quantitation of supercoiling assays of A/Tnicked and G/Tnicked substrates after incubation with nuclear extracts of human MutSα-deficient LoVo cells, supplemented where indicated with recombinant MutSα—either wild type, or its variants KR (ATPase mutant), FA (DNA binding mutant), or C1 (PCNA interaction mutant). Data from three independent experiments were analyzed. The error bars represent the standard deviation from the mean.
In control reactions, we wanted to differentiate between the effect of the nick and the mismatch on chromatin assembly. We therefore examined DNA repair synthesis and supercoiling on the homoduplex substrates A/T and A/Tnicked, as well as on an unnicked G/T. As shown in Fig. 1C, Fig. S1E, after 15 min, the covalently closed circular A/T and G/T substrates were supercoiled to similar extents in both MMR-proficient and -deficient extracts. The A/Tnicked and G/Tnicked substrates incubated with MMR-deficient extracts were supercoiled slightly more efficiently, which agrees with the finding that nicks accelerate nucleosome loading on circular DNA molecules (17). In contrast, supercoiling of the G/Tnicked substrate was inhibited in the MMR-proficient extracts. Coupled with the fact that this substrate incorporated substantial amounts of [α-32P]dCMP (Fig. S1E, Lower) already at this 15 min time point, and that it was generally efficiently repaired (Fig. S1D), the present evidence indicates that active MMR inhibits nucleosome loading.
Supercoiling Inhibition Requires Functional MMR Factors.
To confirm that the inhibition of supercoiling on the G/Tnicked substrate required active MMR, we first made use of MutSα variants C1, KR, or FA (Fig. S1F) (18). The C1 variant lacks 80 N-terminal amino acids of MSH6 that contain the PCNA interaction peptide (PIP) motif, but is able to complement the MMR defect in the LoVo extracts on 3′-nicked substrates (19) (see also Fig. S1G). As shown in Fig. 1D (see also Fig. S1H), it inhibited supercoiling of the G/Tnicked substrate similarly to the wild type protein. The KR mutant carries mutations in the Walker A motifs of both MSH2 and MSH6. It is unable to undergo the ATP-driven transition to a sliding clamp and hence remains bound at the mismatch (20). The FA mutant carries a mutation at the mismatch binding site of MSH6 and fails to form a stable complex with DNA (18). Both these latter proteins were inactive in our MMR assay. As anticipated, they failed to promote incorporation of [α-32P] dCMP into the substrates tested (Fig. S1H, Lower) and did not interfere with supercoiling in the complemented extracts (Fig. 1D; see also Fig. S1H, Upper).
We also tested the requirement for MutLα. The MMR-proficient 293T-Lα+ cells can be induced to shut off MLH1 expression by doxycycline treatment (16). As shown in Fig. S1 I and J, chromatin assembly on the G/Tnicked substrate upon incubation with the 293T-Lα+ extracts was inhibited similarly to that seen in the LoVo extracts supplemented with wild-type MutSα; no inhibition was seen in the MMR-deficient 293T-Lα- extracts.
Taken together, the above experiments further confirmed that nucleosome deposition on the heteroduplex substrate is delayed solely during mismatch processing and not by mismatch binding alone, as in the case of the KR-complemented LoVo extracts, or by sequestration of a factor controlling chromatin assembly by an MMR protein, as in the case of the FA variant of MutSα.
CAF-1 does not Overcome MMR-Dependent Inhibition of Nucleosome Assembly.
The active inhibition of nucleosome assembly on the G/Tnicked substrate could mean either that MMR in the extracts was more rapid than nucleosome loading, or that MMR was able to displace nucleosomes from the repair tract. In an attempt to answer which scenario was more likely, we added purified recombinant CAF-1 to our assays, to accelerate nucleosome assembly. As anticipated, CAF-1 addition increased the number of fully supercoiled molecules on both A/Tnicked and G/Tnicked substrates (Fig. 2A, Fig. S2A). However, nucleosome loading on the actively processed substrate was somewhat less efficient than on the same substrate incubated with the MMR-deficient extract. This difference was small, but that is what would be anticipated, given that active MMR on this substrate would involve a stretch of approximately 400 nucleotides that could bind only two nucleosomes. Moreover, the efficiency of MMR was not affected by the addition of CAF-1 (Fig. 2B, Fig. S2B), which suggested that active MMR indeed inhibited nucleosome deposition on the heteroduplex substrate.
Fig. 2.
Effect of CAF-1 on the efficiency of chromatin assembly and MMR. (A) Representative supercoiling/repair assay of A/Tnicked and G/Tnicked substrates in nuclear extracts of LoVo cells supplemented with MutSα and/or with recombinant CAF-1 as indicated. (B) MMR efficiency in the extracts was estimated by recovering the G/Tnicked substrates following incubation with the extracts and digestion with AclI. As shown, addition of CAF-1 to the assay did not alter MMR efficiency. (C) After 10 min preincubation (+) of the A/Tnicked and G/Tnicked substrates with LoVo extracts supplemented (+) or not (−) with MutSα. In the control experiments (−), the extract was incubated under identical conditions, and the substrates were added after 10 min together with the MutSα. The extent of supercoiling was examined 15 min later. Data from three independent experiments were analyzed. The error bars represent the standard deviation from the mean. (D) MMR assay of the G/Tnicked substrate preincubated with (+) or without (−) MutSα as indicated.
An alternative explanation could be that nucleosomes present no barrier to MMR (12). To test this possibility, we preincubated the substrates with the MMR-deficient LoVo extracts for 10 min prior to adding MutSα. Under these conditions, nucleosome loading in the absence of added MutSα was similar on the A/T and G/T substrates, and addition of MutSα did not alter the situation. In contrast, in the control experiments, where the extract was preincubated for the same period, but the substrates were added together with MutSα, the presence of MutSα inhibited nucleosome deposition to a significant extent (Fig. 2C, Fig. S2C). Moreover, MMR efficiency was inhibited by the preincubations (Fig. 2D, Fig. S2D), which confirmed that nucleosomes hinder the repair process (10, 11).
Repair Synthesis is Accompanied by Efficient Chromatin Assembly.
We noticed that molecules which had been repaired and which therefore contained high levels of [α-32P]dCMP generally contained more superhelical turns than the unlabeled topoisomers (see, e.g., Fig. 1A, Right). This result implied that active MMR increased the efficiency of nucleosome loading. Because repair synthesis involves polymerase-δ, which requires PCNA for processivity, and because both pol-δ and CAF-1 interact with PCNA, we argued that PCNA might recruit CAF-1 to the repaired molecules and thus increase the efficiency of nucleosome deposition. However, MutSα and MutLα also interact with PCNA (19, 21, 22), as does DNA ligase I (23). These findings raised the possibility that the choice of the metabolic pathway (MMR, repair synthesis, or nick ligation), might be controlled through access to PCNA. We therefore compared the efficiency of supercoiling (Fig. S2E) and MMR (Fig. S2F) of the G/Tnicked substrate in LoVo extracts supplemented with either wild-type or C1 MutSα. Unexpectedly, there were no appreciable differences. We therefore set out to examine whether MutSα and CAF-1 can bind to PCNA concurrently, or whether they form distinct complexes.
CAF-1 Interacts with MutSα via its p150 Subunit.
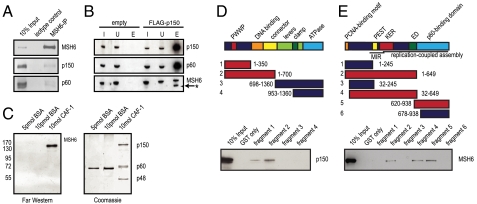
When MutSα was immunoprecipitated from the cell extracts, we were able to identify the CAF-1 subunits p150 and p60 (Fig. 3A). Independently, MSH6 was found among polypeptides pulled-down with FLAG-tagged CAF-1 p150 subunit (Fig. 3B).
Fig. 3.
MutSα interacts directly with the p150 subunit of CAF-1. (A) Anti-MSH6 coimmunoprecipitations were analyzed by Western blotting for CAF-1. Ten percent HeLa nuclear extracts served as the input control. (B) Extracts of 293 cells stably expressing FLAG-CAF-1 p150 were incubated with anti-FLAG beads. Elution was done using FLAG peptides. The control was 293 cells expressing FLAG only (empty). The input control was 0.5% of eluted material. Input fraction (I), Unbound material (U), Eluate (E). Asterisk marks leftover signal of previous p150 blot. (C) Far Western blot showing a direct interaction between MutSα and CAF-1. The CAF-1 trimer was separated by SDS-PAGE, transferred onto a membrane, incubated with MutSα and hybridized with an anti-MSH6 antibody. BSA was used as negative control. (D) Schematic representation of human MSH6. The PWWP (red), DNA-binding (orange), and ATPase (light blue) domains are indicated. The clamp region (dark green) is located within the lever domain (light green) that follows the connector domain (yellow). The GST-MSH6 fusion fragments that interacted with purified p150 in a GST pull-down experiment are shown in red, fragments that did not interact are in blue. (E) Schematic representation of the p150 subunit of human CAF-1. MIR (MOD1-interacting region), PEST (yellow), as well as KER (red), and ED (green) histone interacting domains are shown. The PCNA-binding motif (orange) as well as the p60-interacting region (light blue) are also indicated. The C-terminal half of p150 is needed for replication-coupled assembly. The GST-p150 fragments that interacted with purified MutSα in a GST pull-down experiment are indicated in red, those that did not interact are shown in blue.
That the above interaction was direct rather than PCNA-mediated was shown by Far Western analyses using purified, baculovirus-expressed proteins. We first showed that CAF-1 spotted directly onto the membrane interacted with MutSα in the absence of PCNA (Fig. S3A). When the CAF-1 heterotrimer was first resolved into its individual subunits by SDS-PAGE and then transferred onto the membrane, MutSα was seen to bind exclusively to the CAF-1 p150 subunit (Fig. 3C). In GST pull-down experiments, we narrowed down the interaction domains to amino acid residues 1–350 in MSH6 (Fig. 3D) and 620–649 in CAF-1 p150 (Fig. 3E). Given that the C1 MutSα variant also interacts with CAF-1 on Far Western blots (Fig. S3B), the interaction region on MSH6 can be assigned to amino acid residues 81–350. However, we were unable to pull down MutSα with the GST/CAF-1620–649 fusion peptide and we therefore conclude that this domain is necessary, but not sufficient, to facilitate a stable interaction.
MutSα and p150 CAF-1 Interact During S-Phase.
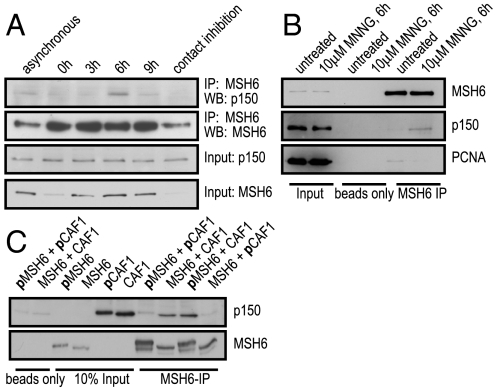
Both CAF-1-mediated chromatin assembly and MMR are coupled to replication. We therefore wanted to test whether the interaction between CAF-1 p150 and MutSα is S-phase-specific. To this end, we arrested U2OS cells with hydroxyurea (HU) at the G1/S boundary and prepared cell extracts at different time points after release. Progression of the cells through the cell cycle was followed by FACS analysis (Fig. S4A) and protein levels were analyzed by Western blotting (WB) (Fig. 4A). Although CAF-1 p150 levels remained constant during the cell cycle (24), MSH6 levels increased during S-phase (25). As anticipated, the interaction between CAF-1 p150 and MutSα was substantially greater 6 h post release (Fig. 4A), when most of the cells were in S-phase (Fig. S4A). The S-phase specificity of the interaction was further supported by a substantial colocalization of MSH6 and CAF-1 p150 in replication foci of early and late S-phase U2OS cells stably expressing GFP-p150 (Fig. S4B).
Fig. 4.
Interactions between MutSα, CAF-1, and PCNA are differentially affected by treatment with DNA damaging agents. (A) Cell extracts from different cell cycle stages (Fig. S4A) were immunoprecipitated with an anti-MSH6 antibody and analyzed by Western blotting. (B) U2OS cells were pretreated with O6-benzylguanine to inhibit methylguanine methyl transferase before treatment with 10 μM MNNG. Anti-MSH6 immunoprecipitates were analyzed for MSH6, CAF-1 p150, or PCNA. (C) Lambda phosphatase treated or untreated purified, recombinant MutSα and CAF-1 p150 were used in immunoprecipitations with an anti-MSH6 antibody. The prefix p indicates polypeptides endogenously phosphorylated in Sf9 cells.
MutSα Interaction with CAF-1 is Enhanced by DNA Damage.
We asked whether CAF-1 interacts with MutSα within the context of mismatch repair, the primary function of which is to remove mismatches introduced during replication. As it is not possible to experimentally induce mismatches in vivo, we treated the cells with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), an SN1-type methylating agent that generates primarily N3-methyladenine, N7-methylguanine, and O6-methylguanine (meG) in DNA. The latter methylated nucleotide pairs with C or T during replication and thus gives rise to meG/C and meG/T mispairs, which are recognized by MutSα and activate the mismatch repair machinery (1).
In untreated or MNNG-treated MMR-proficient U2OS cells, MSH6 and CAF-1 p150 levels remained constant (Fig. 4B, input). However, when MSH6 was immunoprecipitated, we detected significantly higher levels of CAF-1 p150 in the coimmunoprecipitate of the MNNG-treated cells (Fig. 4B, MSH6 immunoprecipitate). This finding suggested that the formation of the MutSα-CAF-1 complex was MNNG-inducible. The interaction was DNA-independent, as it was unaffected by the addition of benzonase.
MutSα Interaction with CAF-1 is Modulated by Phosphorylation.
Both MSH6 and CAF-1 p150 are phosphoproteins. Phosphorylation of MSH6 was reported to affect its localization (26), whereas the phosphorylation status of CAF-1 was shown to change during the cell cycle (27, 28). Because the interaction of CAF-1 with MutSα increased during S-phase (Fig. 4A), we wanted to test whether this change was dependent on the phosphorylation status of the two proteins. We expressed CAF-1 and MutSα in the baculovirus system, where both proteins are constitutively phosphorylated, as witnessed by their slower migration in SDS-PAGE when compared with the same proteins after dephosphorylation with lambda phosphatase (Fig. 4C, input). In coimmunoprecipitation experiments, we found that only dephosphorylated CAF-1 p150 was able to interact with MutSα. In contrast, the ability of dephosphorylated CAF-1 p150 to interact with MutSα was independent of the phosphorylation status of MSH6 (Fig. 4C). Phosphorylation of CAF-1 p150 by Cdc7/Dbf4 was previously shown to increase its affinity for PCNA at the onset of replication (28). Based on this evidence, our findings imply that MutSα associates predominantly with the unphosphorylated form of CAF-1.
Discussion
All processes of DNA metabolism that need to unwind the two strands of the helix have to remodel chromatin, which reduces their efficiency (6). The effect of chromatin on MMR has not attracted much interest because nucleosomes are removed ahead of the advancing replication fork and mismatches therefore arise in naked DNA. However, CAF-1-catalyzed chromatin reassembly on newly synthesized DNA is rapid, such that only approximately 250 base pairs behind the replication fork are free of nucleosomes (13). Because mismatch-activated MutSα must be able to slide along the DNA contour to a free DNA terminus where the degradation process of the error-containing strand initiates, and because this process is inhibited by nucleosomes (10, 11), we argued that mismatch repair would have to take place prior to chromatin assembly, or, alternatively, that it might interfere with this process. If, as postulated earlier (29), MutSα diffuses away from the mismatch once it has undergone the ATP-driven conversion to a sliding clamp, the mismatch position will become free for the loading of additional MutSα molecules. In this scenario, the stretch between the mismatch and the nick where MMR initiates would be constantly traversed by sliding clamps, such that no nucleosomes would be able to assemble. That MutSα can displace nucleosomes from DNA was shown recently (12). Although this phenomenon was questioned (11), we believe that even if the sliding clamp were unable to displace preformed nucleosomes, it would almost certainly displace assembling histone dimers. However, our finding that MutSα and CAF-1 interact implies that MMR and chromatin assembly are coordinated in a more complex manner. Thus, for example, MutSα may alter the affinity of CAF-1 p150 for its substrates, as the interaction region was mapped to the vicinity of the acidic ED domain (Fig. 3E) that is responsible for binding of CAF-1 to newly synthesized histones (30). Alternatively, mismatch-activated MutSα may interfere with CAF-1 recruitment to the replication fork. The latter process involves changes in CAF-1 p150 phosphorylation status (27, 31) and thus alters the equilibrium between the phosphorylated and the underphosphorylated forms of p150, which in turn would impact on CAF-1 and PCNA interactions (28). Our finding that MutSα affinity for CAF-1 is augmented by methylation damage (Fig. 4B) and that MutSα interacts preferentially with the underphosphorylated form of CAF-1 p150 (Fig. 4C) implies that activated MutSα reduces the amount of CAF-1 available for binding to PCNA.
Based on the above evidence, we propose that activation of MMR might affect chromatin assembly on damage- or mismatch-containing DNA in several stages. First, MutSα sliding clamps might prevent deposition of nucleosomes by simple steric interference (12). Second, MutSα might deny CAF-1 access to PCNA (either through direct competition or via the interaction described above). Once repair is completed, MutSα no longer binds to DNA; CAF-1 can then freely interact with PCNA and resume chromatin assembly. Our findings that MMR delays chromatin assembly agree with recently reported data, where nucleosome deposition on heteroduplex substrates was shown to be similarly affected in the minimal reconstituted MMR system (32).
This study provides experimental evidence that implicates MMR in the attenuation of chromatin assembly through modulating CAF-1 function. It opens a time window necessary to efficiently correct errors of replication. Should this interaction be disregulated, MMR efficiency might be reduced, which would result in increased mutagenesis. Ultimately, this rise in genomic instability might lead to increased incidence of disease and cancer, as well as to accelerated aging.
Materials and Methods
Cell lines, extract preparations, protein purifications, and other standard procedures are described in SI Materials and Methods.
DNA Substrates and Supercoiling/Mismatch Repair Assay.
Isolation of the supercoiled homo- and heteroduplex substrates (24) and the MMR assays were carried out as described in ref. 33, using 100 ng (47.5 fmol) heteroduplex DNA substrate and 50–100 μg of nuclear extracts (depending on CAF-1 level) in a total volume of 25 μL, 20 mM Tris·HCl pH 7.6, 110 mM KCl, 5 mM MgCl2, 1 mM glutathione, 1.5 mM ATP, 50 μg/mL BSA, 100 μM of dATP, 100 μM dGTP, 100 μM dCTP, 100 μM dTTP, and 0.12 mCi/mL and 80 nM of [α-32P] dCTP for 15 min, followed by a 60 min incubation with stop solution (final concentrations: 0.5 mmol/L EDTA, 1.5% SDS, 2.5 mg/mL proteinase K). The DNA was purified using Qiagen MinElute Reaction Cleanup Kit followed by a treatment with 200 μg/mL RNase A (Sigma-Aldrich) and DNA was separated on a 1% agarose gels in Tris acetate EDTA (TAE) buffer. Total DNA was visualized by EtBr poststaining, and 32P incorporation was analyzed by exposing the dried gels to PhosphoImager screens or X-ray films. The MMR efficiency was measured based on an AclI digestion and separation on 1% agarose gels stained with GelRed.
For the preincubation experiment, nuclear extract was incubated for 10 min in the reaction buffer in the presence (or absence) of DNA prior to addition of MutSα (and DNA), followed by incubation for 15 min.
Antibodies.
The primary antibodies were MSH6 (610919, BD Transduction Laboratories, 1∶2,000 WB, 1∶100 IF, where IF is immunofluorescence), PCNA (sc-65, Santa Cruz, 1∶4,000 WB), p150 CAF-1 (ab7655, Abcam, 1∶1,000 WB), p60 CAF-1 (9) (1∶1,000 WB), MSH2 (NA-27 Oncogene, 1∶500 WB), MLH1 (554073, BD Transduction Laboratories, 1∶1,000 WB), β-tubulin (sc-5274, Santa Cruz, 1∶4,000 WB), GST (ab9085, Abcam, 1∶500 WB), 66H6 (34), and anti-FLAG M2 Agarose from mouse (Sigma).
The secondary antibodies were sheep anti-mouse IgG horseradish peroxidase-linked (GE healthcare, 1∶5,000 WB), donkey anti-rabbit IgG horseradish peroxidase-linked (GE healthcare 1∶5,000 WB), and sheep polyclonal to mouse IgG Texas Red (ab6806, Abcam, 1∶200 IF).
Supplementary Material
Acknowledgments.
We thank Kalpana Surendranath for MutSα wild type, or its variants KR, FA, and C1; Fritz Thoma and Katja Kratz for critical reading of the manuscript; Mariela Artola and Myriam Marti for expert technical assistance; and Alain Verreault and Bruce Stillman for the generous gift of the CAF-1-expressing baculoviruses. We also gratefully acknowledge the award of a European Molecular Biology Organization Short-Term Fellowship and a grant from the ProMedica Stiftung (B.S.). The research in the J.J. laboratory leading to these results has received funding from the Swiss National Science Foundation Grant 3100/068182.02/1, the European Community's Sixth Framework Programme Grant LSHC-CT-2005-512113, the European Community's Seventh Framework Programme (FP7/2007-2013) Grant HEALTH-F4-2008-223545, and the Strategic Japanese-Swiss Cooperative Program. The G.A. laboratory is supported by la Ligue Nationale Contre le Cancer (Equipe Labellisée la Ligue), Programme Incitatif et Collaboratif (Replication, Instabilite Chromosomique et Cancer), the European Commission Network of Excellence Epigenome Grant LSHG-CT-2004-503433, ACI-2007-Cancéropôle IdF Breast Cancer and Epigenetics, Agence Nationale Recherche FaRC PCV06_142302. The F.K. laboratory is supported by start-up funds from the Southern Illinois University School of Medicine.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106696109/-/DCSupplemental.
References
- 1.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiricny J, et al. Mismatch repair defects in cancer. Curr Opin Genet Dev. 2000;10:157–161. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 4.Moldovan GL, et al. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Smith S, et al. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 6.Groth A, et al. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Smerdon MJ, et al. Nucleosome rearrangement in human chromatin during UV-induced DNA-repair synthesis. Proc Natl Acad Sci USA. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moggs JG, et al. Chromatin rearrangements during nucleotide excision repair. Biochimie. 1999;81:45–52. doi: 10.1016/s0300-9084(99)80037-6. [DOI] [PubMed] [Google Scholar]
- 9.Green CM, et al. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorman J, et al. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, et al. Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. J Biol Chem. 2009;284:33056–33061. doi: 10.1074/jbc.M109.049874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javaid S, et al. Nucleosome remodeling by hMSH2-hMSH6. Mol Cell. 2009;36:1086–1094. doi: 10.1016/j.molcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sogo JM, et al. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 14.Gaillard PH, et al. Nucleotide excision repair coupled to chromatin assembly. Methods Mol Biol. 1999;119:231–243. doi: 10.1385/1-59259-681-9:231. [DOI] [PubMed] [Google Scholar]
- 15.Baerenfaller K, et al. Characterization of the mismatch repairosome and its role in the processing of modified nucleosides in vitro. Methods Enzymol. 2006;408:285–303. doi: 10.1016/S0076-6879(06)08018-9. [DOI] [PubMed] [Google Scholar]
- 16.Cejka P, et al. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moggs JG, et al. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufner P, et al. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSα is abolished by a single mutation in the hMSH6 subunit. J Biol Chem. 2000;275:36550–36555. doi: 10.1074/jbc.M005987200. [DOI] [PubMed] [Google Scholar]
- 19.Iyer RR, et al. The MutSα-proliferating cell nuclear antigen interaction in human DNA mismatch repair. J Biol Chem. 2008;283:13310–13319. doi: 10.1074/jbc.M800606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iaccarino I, et al. Mutation in the magnesium binding site of hMSH6 disables the hMutSα sliding clamp from translocating along DNA. J Biol Chem. 2000;275:2080–2086. doi: 10.1074/jbc.275.3.2080. [DOI] [PubMed] [Google Scholar]
- 21.Kleczkowska HE, et al. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer RR, et al. MutLα and proliferating cell nuclear antigen share binding sites on MutSβ. J Biol Chem. 2010;285:11730–11739. doi: 10.1074/jbc.M110.104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song W, et al. The DNA binding domain of human DNA ligase I interacts with both nicked DNA and the DNA sliding clamps, PCNA and hRad9-hRad1-hHus1. DNA Repair. 2009;8:912–919. doi: 10.1016/j.dnarep.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marheineke K, et al. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J Biol Chem. 1998;273:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- 25.Szadkowski M, et al. Identification and functional characterization of the promoter region of the human MSH6 gene. Genes Chromosomes Cancer. 2002;33:36–46. doi: 10.1002/gcc.1211. [DOI] [PubMed] [Google Scholar]
- 26.Christmann M, et al. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutSα mismatch-binding activity. Nucleic Acids Res. 2002;30:1959–1966. doi: 10.1093/nar/30.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith S, et al. Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. J Biol Chem. 1991;266:12041–12047. [PubMed] [Google Scholar]
- 28.Gerard A, et al. The replication kinase Cdc7-Dbf4 promotes the interaction of the p150 subunit of chromatin assembly factor 1 with proliferating cell nuclear antigen. EMBO Rep. 2006;7:817–823. doi: 10.1038/sj.embor.7400750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gradia S, et al. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 30.Ridgway P, et al. CAF-1 and the inheritance of chromatin states: At the crossroads of DNA replication and repair. J Cell Sci. 2000;113:2647–2658. doi: 10.1242/jcs.113.15.2647. [DOI] [PubMed] [Google Scholar]
- 31.Keller C, et al. Requirement of Cyclin/Cdk2 and protein phosphatase 1 activity for chromatin assembly factor 1-dependent chromatin assembly during DNA synthesis. J Biol Chem. 2000;275:35512–35521. doi: 10.1074/jbc.M003073200. [DOI] [PubMed] [Google Scholar]
- 32.Kadyrova LY, et al. CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair. Proc Natl Acad Sci USA. 2011;108:2753–2758. doi: 10.1073/pnas.1015914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer F, et al. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007;133:1858–1868. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Marra G, et al. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci USA. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.