Abstract
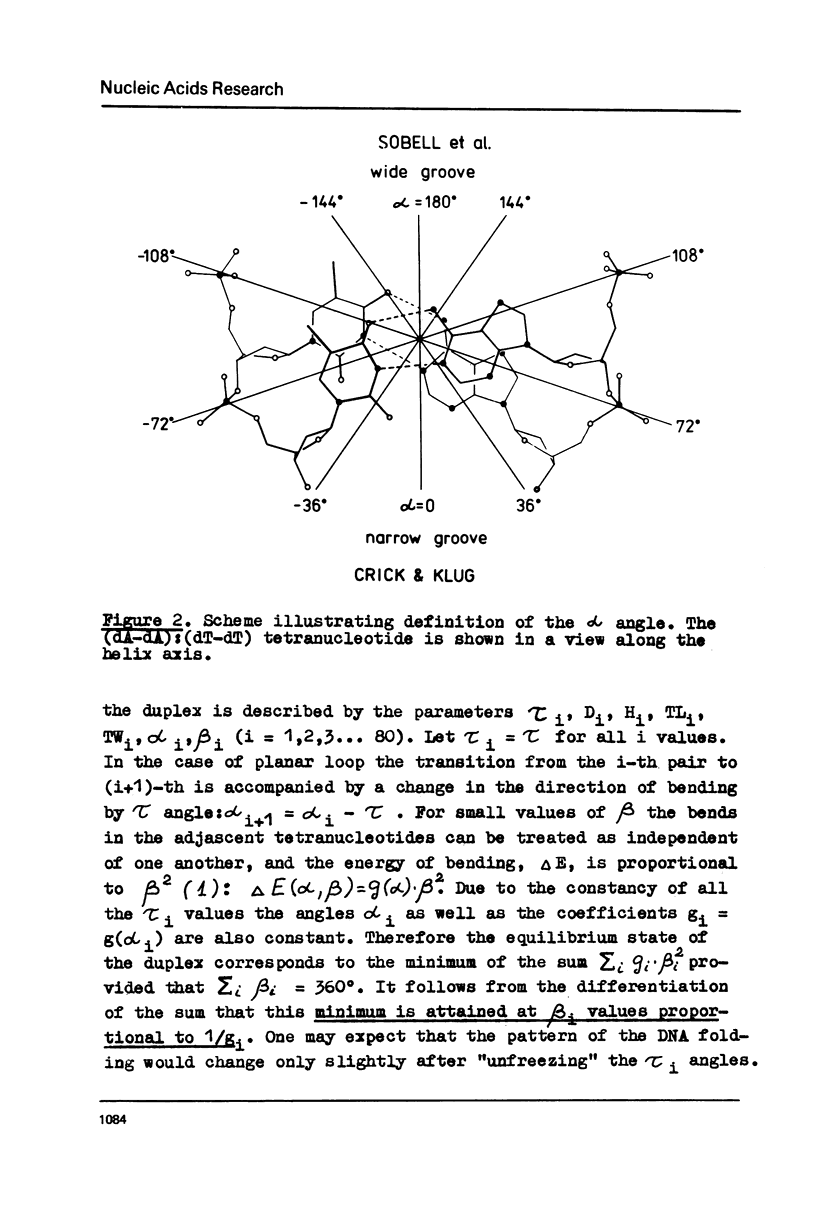
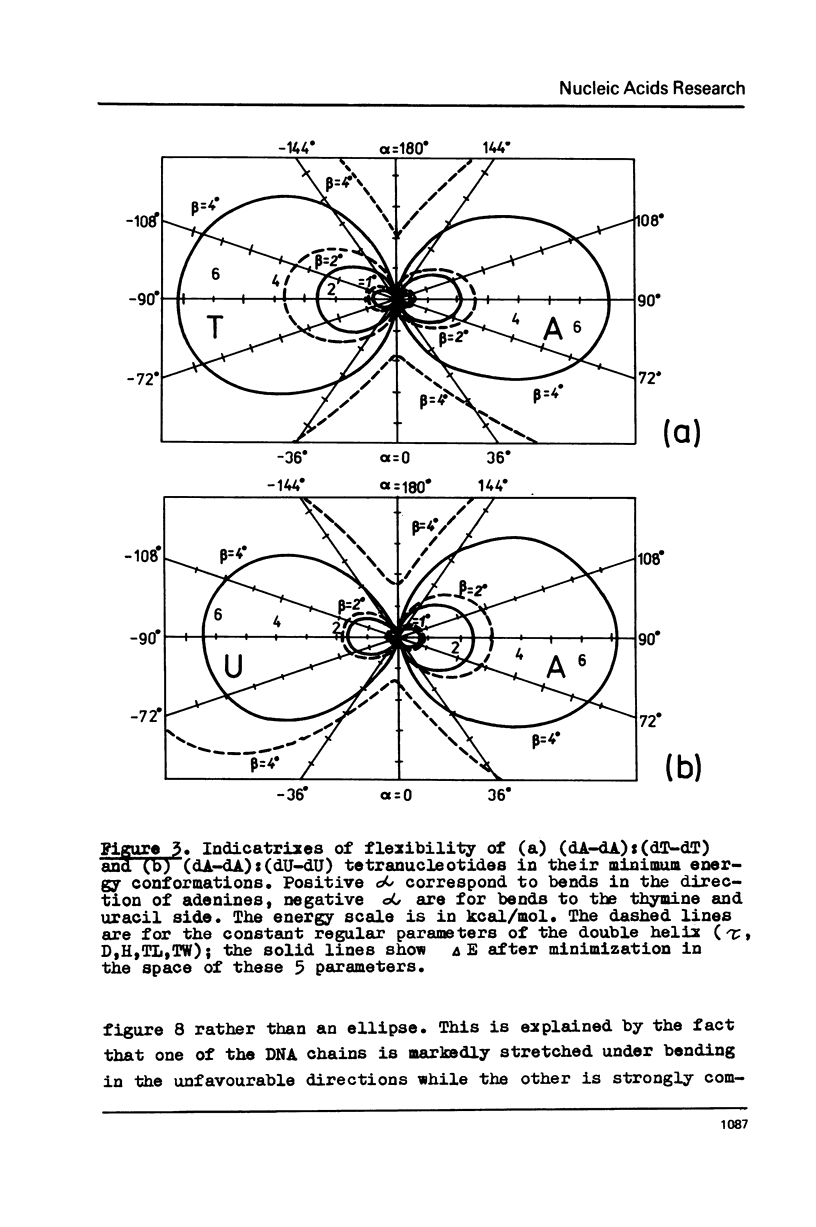
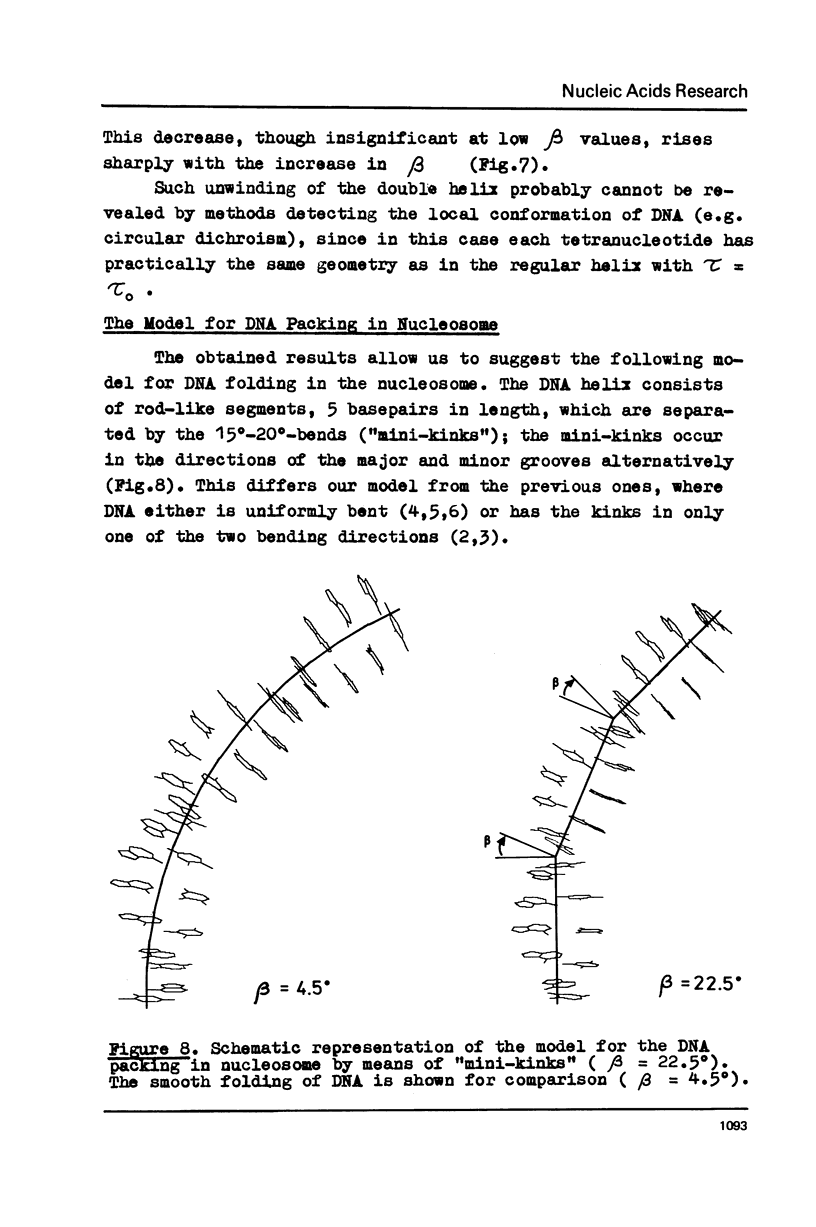
Potential energy calculations of the DNA duplex dimeric subunit show that the double helix may be bent in the direction of minor and major grooves much more easily than in other directions. It is found that the total winding angle of DNA decreases upon such bending. A new model for DNA folding in the nucleosome is proposed on the basis of these findings according to which the DNA molecule is kinked each fifth base pair to the side of the minor and major grooves alternatively. The model explains the known contradiction between a C-like circular dichroism for the nucleosomal DNA and the nuclease digestion data, which testify to the B-form of DNA.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S. The geometry of nucleic acids. Prog Biophys Mol Biol. 1970;21:265–319. doi: 10.1016/0079-6107(70)90027-1. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M. Folding of 140-base pair length DNA by a core of arginine-rich histones. J Biol Chem. 1978 Jul 25;253(14):5213–5219. [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. A simple model of DNA superhelices in solution. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1708–1712. doi: 10.1073/pnas.75.4.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Godfrey J. E., Eisenberg H. The flexibility of low molecular weight double-stranded DNA as a function of length. I. Light scattering measurements and the estimation of persistence lengths from light scattering, sedimentation and viscosity. Biophys Chem. 1976 Sep;5(3):301–318. doi: 10.1016/0301-4622(76)80042-7. [DOI] [PubMed] [Google Scholar]
- Govil G. Conformational structure of polynucleotides around the O-P bonds: refined parameters for CPF calculations. Biopolymers. 1976 Nov;15(11):2303–2307. doi: 10.1002/bip.1976.360151119. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Schyolkina A. K., Poletayev A. I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12(1):89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- Johnson R. S., Chan A., Hanlon S. Mixed conformations of deoxyribonucleic acid in intact chromatin isolated by various preparative methods. Biochemistry. 1972 Nov 7;11(23):4347–4358. doi: 10.1021/bi00773a023. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namoradze N. Z., Goryunov A. N., Birshtein T. M. On conformations of the superhelix structure. Biophys Chem. 1977 Jun;7(1):59–70. doi: 10.1016/0301-4622(77)87015-4. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Renugopalakrishnan V., Lakshminarayanan A. V., Sasisekharan V. Stereochemistry of nucleic acids and polynucleotides. 3. Electronic charge distribution. Biopolymers. 1971;10(7):1159–1167. doi: 10.1002/bip.360100707. [DOI] [PubMed] [Google Scholar]
- Röder O., Lüdemann H., Von Goldammer E. Determination of the activation energy for pseudorotation of the furanose ring in nucleosides by 13-C nuclear-magnetic-resonance relaxation. Eur J Biochem. 1975 May 6;53(2):517–524. doi: 10.1111/j.1432-1033.1975.tb04094.x. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. Flexibility of DNA. Biopolymers. 1974 Jan;13(1):217–226. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Trifonov E. N. Possibility of nonkinked packing of DNA in chromatin. Proc Natl Acad Sci U S A. 1978 Jan;75(1):103–107. doi: 10.1073/pnas.75.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tselikova S. V., Lobachev V. M., Mandrugin A. A., Ivanov V. I. Osobennosti zakruchivaniia DNK bifunktsional'nymi kationamn. Mol Biol (Mosk) 1976 Sep-Oct;10(5):1153–1158. [PubMed] [Google Scholar]



