Abstract
Transcription of the centromeric regions has been reported to occur in G1 and S phase in different species. Here, we investigate whether transcription also occurs and plays a functional role at the mammalian centromere during mitosis. We show the presence of actively transcribing RNA polymerase II (RNAPII) and its associated transcription factors, coupled with the production of centromere satellite transcripts at the mitotic kinetochore. Specific inhibition of RNAPII activity during mitosis leads to a decrease in centromeric α-satellite transcription and a concomitant increase in anaphase-lagging cells, with the lagging chromosomes showing reduced centromere protein C binding. These findings demonstrate an essential role of RNAPII in the transcription of α-satellite DNA, binding of centromere protein C, and the proper functioning of the mitotic kinetochore.
Keywords: chromatin, noncoding RNA, epigenetics
The centromere is an essential chromosomal structure that mediates microtubule attachment during cell division to ensure correct chromosome segregation. Although centromere function is highly conserved, centromere DNA sequences show no evolutionary conservation. Instead, centromeric chromatin typically is filled with species-specific satellite DNA sequences that lack transcribed genes. The presence of functional ectopic centromeres (neocentromeres) at genomic regions devoid of classical satellite DNA repeats confirmed the epigenetic nature of centromere function (1).
The centromere is organized into two broad domains characterized by distinct sets of epigenetic determinants. The centromere core domain comprises the centromere-specific histone H3 variant centromere protein A (CENP-A) that is essential for kinetochore formation, whereas the pericentric heterochromatin is vital for sister chromatid cohesion (for review, see ref. 2). In yeast, pericentric outermost DNA repeats are transcribed and processed by RNAi machinery into siRNAs, which direct the deposition of heterochromatic markers such as H3K9me3 and HP1 at the pericentric heterochromatin (3). The RNAi pathway also has been shown to be vital for the establishment of pericentric heterochromatin in plant and animal cells (4, 5). However, although the depletion of Dicer in human-chicken hybrid cells causes defective pericentric heterochromatin, it does not affect the binding of CENP-A and centromere protein C (CENP-C) at the centromere core domain (6), suggesting that the RNAi pathway is not required for core centromere function. Our previous studies showed that transcription is permissible within the kinetochore domain of a human neocentromere (7, 8), but others have reported the presence of active genes within the rice kinetochore domain (9). Consistent with these reports, the CENP-A domain in Drosophila and human cells is enriched with the euchromatin-like marker H3K4me2 (10). Such studies suggest that transcription of centromeric chromatin is permissible and compatible with centromere function. Furthermore, we and others have demonstrated the presence of RNA at the mitotic kinetochore (11, 12). These RNA transcripts were essential for the localization of mitotic centromere proteins including CENP-C, but it was unclear if the transcripts were produced during mitosis. Here we demonstrate that RNA polymerase II (RNAPII) and some of its associated transcription factors localize to the mammalian kinetochore during mitosis. We provide evidence of RNAPII-mediated transcriptional activity at the mitotic kinetochore and evidence that inhibition of RNAPII activity during mitosis leads to an increase in lagging chromosomes with reduced CENP-C levels.
Results
RNAPII Localizes to the Mitotic Kinetochore and Is Associated with Kinetochore Activity.
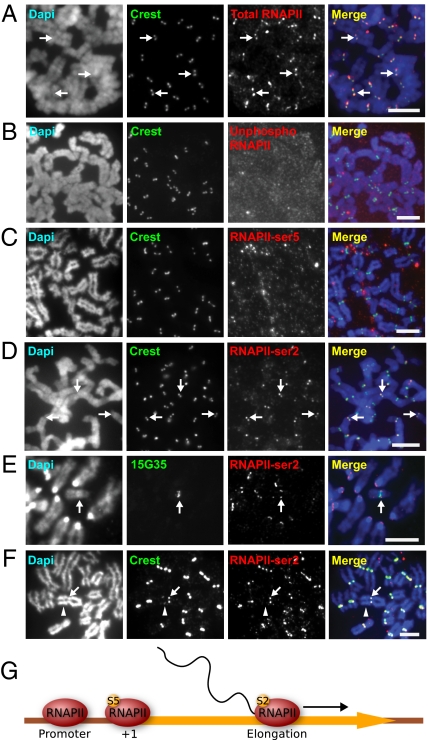
Immunofluorescence analysis of interphase cells showed no significant enrichment of RNAPII at the interphase centromeres (Fig. S1 A–D). To investigate the localization pattern of RNAPII during mitosis, we performed immunofluorescence analysis using an antibody that recognized all forms of RNAPII. Cytospun metaphase HeLa cells showed clear colocalization of RNAPII and the centromere marker Calcinosis, Raynaud phenomenon, Esophageal dysmotility, Sclerodactyly, and Telangiectasia (CREST) antisera signals, indicating the presence of RNAPII at the kinetochores of metaphase HeLa cells (Fig. 1A). Further analysis of other submitotic phases showed no specific enrichment for RNAPII at the kinetochores in prophase cells; however, a small subset of kinetochores in prometaphase cells showed RNAPII localization (Fig. S1 E and F), whereas RNAPII was found at most kinetochores of metaphase and anaphase cells (Fig. 1A and Fig. S1 G–H). Collectively our data suggest that RNAPII is recruited to mitotic kinetochores as early as prometaphase.
Fig. 1.
RNAPII-ser2 localizes to the mitotic kinetochore and is associated with active kinetochore activity. (A) Antibody 4H8total RNAPII immunostained the kinetochores of HeLa cells, as shown by the colocalization with CREST anti-centromere signals (arrows). (B) Antibody 8WG16unphosphorylated, specific for unphosphorylated RNAPII did not immunostain HeLa mitotic kinetochores. (C) Antibody H14phosphoSer5, which recognizes RNAPII phosphorylated at serine 5 (RNAPII-ser5) associated with transcription initiation, showed no staining of the kinetochores. (D) H5phosphoSer2, specific for elongating RNAPII, stained the kinetochores of mitotic HeLa cells (arrows). (E) Combined immunofluorescence/DNA-FISH (antibody H5phosphoSer2 and 10q25 band-specific 153G5 BAC probe) of mouse ES mardel (10) cells showed the presence of RNAPII-ser2 at the mardel (10) neocentromere (arrow) as well as at endogenous mouse kinetochores. (F) RNAPII-ser2 was present only at the active neocentromere of the PD-NC4 chromosomes (arrow) but not at the inactive native centromere (arrowhead, as indicated by the weaker CREST signals attributed to residual CENP-B). (G) Schematic depicting the change in RNAPII phosphorylation status across the transcription cycle. At the promoter, RNAPII is unphosphorylated. At transcription initiation, RNAPII is phosphorylated at serine 5 (S5), but before the RNAPII complex is competent for transcription elongation it must be phosphorylated at serine 2 (S2). (Scale bars: 5 μm.)
To characterize further the transcriptional status of the kinetochore-bound RNAPII, we used phosphospecific RNAPII antibodies in our immunofluorescence analysis (Table S1), because the transcriptional activity of RNAPII is correlated strongly with its differential phosphorylation state (for review, see ref. 13). The results revealed that the kinetochore-bound RNAPII is phosphorylated at serine 2 but not at serine 5 (Fig. 1 B–D). The serine 5 phosphorylation mark is associated with promoter-bound transcriptional complexes, whereas the serine 2 phosphorylation state (RNAPII-ser2) is an indicator of active transcription (14), suggesting that the RNAPII is transcriptionally active at the mitotic kinetochore. This pattern of RNAPII-ser2 localization at the kinetochore was consistent: 98% (average of n = 19 mitotic spreads) of chromosomes were positive for RNAPII staining. Similar results were obtained using other human and mouse cell lines (Fig. S2 and also Fig. 1E; see below), indicating that the localization of RNAPII-ser2 to the mitotic kinetochore is conserved in mammalian cells.
To establish whether RNAPII is associated with kinetochore function, we assayed for the presence of RNAPII-ser2 at the neocentromeres of two marker chromosomes: the human chromosome 10-derived mardel (10) marker chromosome containing a 10q25 neocentromere (15), and the pseudodicentric-neocentric chromosome 4 (PD-NC4), which contains an inactivated native α-satellite centromere and a functional neocentromere at 4q21 (16). We observed enrichment of RNAPII-ser2 at the active mitotic neocentromeres of both the mardel (10) and the PD-NC4 chromosomes (Fig. 1 E and F) but not at the inactive alphoid centromere [marked by a faint CREST signal because of residual centromere protein B (CENP-B)] of PD-NC4 (16). These results suggest that RNAPII is localized to active kinetochores rather than satellite repeats and demonstrate an association between RNAPII localization at the kinetochore and centromere activity.
RNAPII-Specific Transcription Factors Carboxy-Terminal Domain, RNA Polymerase II, Polypeptide A Phosphatase, Subunit 1 and Structure-Specific Recognition Protein 1 Localize to the Mitotic Kinetochore.
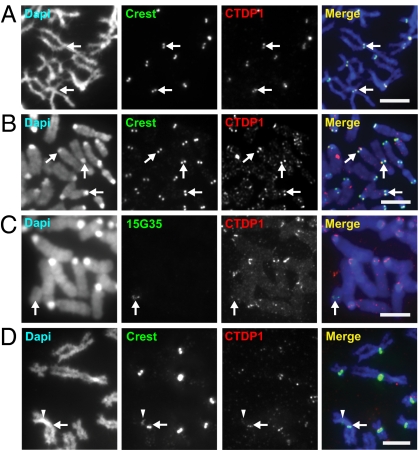
The activity of RNAPII is dependent on the coordinated binding of transcription factors that mediate its activity in vivo. Consistent with active transcription, we found the presence of carboxy-terminal domain, RNA polymerase II, polypeptide A phosphatase, subunit 1 (CTDP1), an RNAPII-specific transcription factor required for transcription elongation (17), at the mitotic kinetochores of human and mouse cells (Fig. 2 A and B). CTDP1 also localized to the mitotic kinetochores of the mardel (10) and PD-NC4 neocentromeres but not to the inactive centromere of PD-NC4 (Fig. 2 C and D). We also detected another key component of the RNAPII machinery, structure-specific recognition protein 1 (SSRP1), a subunit of the histone chaperone facilitates chromatin transcription (FACT) complex, at the mitotic kinetochores of human and mouse chromosomes (Fig. S3). It is interesting that a recent study has implicated FACT in facilitating the deposition of CENP-A at the centromere (18), although the mechanism by which FACT facilitates this deposition remains uncertain.
Fig. 2.
Transcription factor CTDP1 is found at the kinetochores of human and mouse cells as well as at neocentromeres. (A and B) CTDP1 localized to the kinetochores of mitotic human HeLa and mouse NIH 3T3 cells respectively, as evidenced by colocalization with CREST signals (arrows). (C) CTDP1 also localized to the kinetochore of the mardel (10) neocentromere, as shown by the colocalization of CTDP1 and 153G5 BAC DNA-FISH probe specific for the mardel (10) 10q25 neocentromere (arrow). (D) CTDP1 is enriched at the kinetochore of the active PD-NC4 neocentromere, as shown by its colocalization with the brighter CREST signal (arrow), but not at the inactive native alphoid centromere with reduced CREST signal (arrowhead). (Scale bars: 5 μm.)
Mitotic Kinetochore-Bound RNAPII Is Involved in Active Transcription.
The above data suggested the presence of an actively transcribing RNAPII complex at the kinetochore during mitosis. To investigate this possibility further, we used an in situ transcription assay to test the transcriptional competency of the kinetochore-bound RNAPII. NIH 3T3 metaphase cells were cytospun and incubated immediately in a transcription buffer supplemented with FITC-rUTP to visualize sites of active transcription. Analysis of interphase cells showed nucleolar and nuclear incorporation of FITC-rUTP, representative of the transcriptional activity of interphase nuclei (Fig. 3A). In contrast, when mitotic chromosomes were analyzed, signals of active transcription, presenting as discrete FITC-rUTP foci, were enriched at the kinetochore region (Fig. 3B). Subsequent immunostaining with CREST antisera showed direct colocalization of the FITC-rUTP signals with CREST signals (Fig. 3 D and E). No FITC-rUTP labeling of the kinetochore was observed when cells were treated with the RNAPII-specific inhibitor α-amanitin (Fig. 3C), demonstrating that the FITC-rUTP foci are a direct consequence of RNAPII transcriptional activity at the kinetochore.
Fig. 3.
The kinetochore is transcriptionally active during mitosis. (A) In situ transcription assay revealed transcriptional activity in cytospun NIH 3T3 interphase cells, as shown by nuclear and nucleolar FITC-rUTP incorporation (arrow). (B) The in situ transcription assay showed FITC-rUTP incorporation at the kinetochore regions (arrows) of the mitotic chromosomes. (C) FITC-rUTP incorporation at the kinetochore is sensitive to the RNAPII inhibitor α-amanitin. (D) CREST immunofluorescence performed after the in situ transcription assay showed that the FITC-rUTP signals colocalized with CREST signals (arrows). (E) Fluorescence intensity line scans of a single chromosome (boxed in D). Lateral (solid line) and axial (dashed line) fluorescence intensity line-scans through the kinetochores confirmed that the FITC-rUTP signals (green lines) colocalized with CREST signals (red lines); blue lines indicate DAPI staining. (Scale bars: 5 μm in A–D, 2 μm in E.)
Mitotic Inhibition of RNAPII Causes a Decrease in α-Satellite Transcripts and a Concomitant Increase in Lagging Chromosomes with Decreased CENP-C Levels.
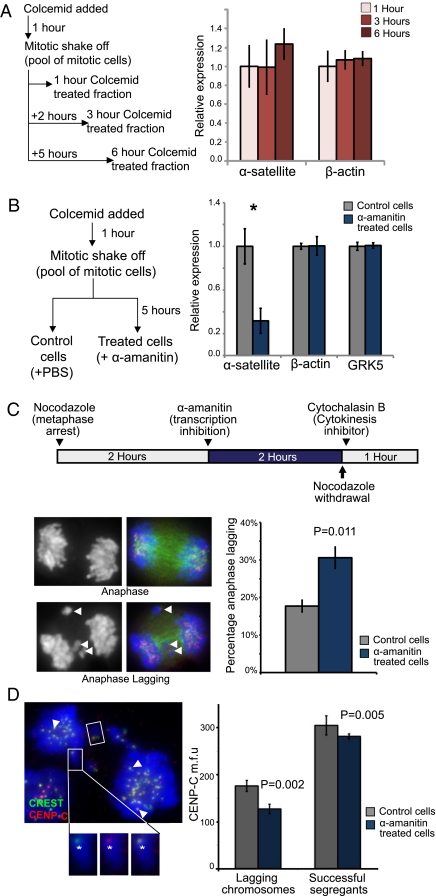
A predicted outcome of active RNAPII transcription at the metaphase kinetochore is the production of centromeric transcripts in mitotic cells. Quantitative real-time reverse transcriptase-PCR (real-time qRT-PCR) was used to compare the levels of centromeric α-satellite transcripts in human 14ZBHT cells (19) that had been arrested in mitosis for 1–6 h. 14ZBHT cells were treated with colcemid for 1 h and subjected to mitotic shake-off to isolate a pool of metaphase-enriched cells (average mitotic index of 70%; n = 5), and were held in mitosis for a further 2 or 5 h (to give 1-, 3-, and 6-h mitotic-arrested fractions; Fig. 4A). Although α-satellite transcripts were detected successfully in mitotic 14ZBHT cells, they were of low abundance, being ∼200-fold less abundant than in GAPDH transcripts (in which levels remained high throughout mitotic arrest). However, no significant increase in α-satellite transcripts (or of the other housekeeping gene, β-actin) was detected in the 1-, 3-, or 6-h mitotic-arrested cells (Fig. 4A), indicating that α-satellite transcripts do not accumulate during mitotic arrest and may have a short half-life. This result resembles the report of transcripts derived from the CENP-A domain in fission yeast, which were subjected to constant turnover (20).
Fig. 4.
Transcription of α-satellite DNA during mitosis is mediated by RNAPII and is required for correct kinetochore function. (A) A mitotic shake-off protocol and real-time qRT-PCR were used to measure the levels of α-satellite RNA in metaphase-arrested cells. Briefly, 14ZBHT cells were treated with colcemid for 1 h, and mitotic shake-off was performed to isolate a population of mitotic-enriched cells (average mitotic index = 70%; n = 5). Cells were maintained in mitotic arrest for an additional 0, 2, or 5 h to give 1-, 3-, and 6-h mitotic-arrested populations. Total RNA was reverse-transcribed into cDNAs for real-time qRT-PCR using the ΔΔCT method (normalized against the mean of three housekeeping genes, β-actin, GAPDH, and HPRT). No significant change in the levels of α-satellite or actin transcripts was detected. (B) The effect of RNAPII inhibition on centromere transcription was assayed. A pool of mitotic 14ZBHT cells was isolated via mitotic shake-off and split into two fractions, an α-amanitin–treated fraction (incubated with 50 μg/mL α-amanitin) and a control fraction (PBS added), for a further 5 h. RT-PCR quantification was performed as described previously. α-Amanitin–mediated RNAPII inhibition caused a significant decrease in α-satellite levels (by 68%; n = 4; *P = 0.016; Student's t test). Transcripts of the control genes, G protein-coupled receptor kinase 5 and β-actin, were unaffected. (C) A protocol was designed to target RNAPII inhibition to mitotic cells, and an anaphase-lagging assay was used to detect kinetochore dysfunction. 14ZBHT cells were treated with nocodazole for 2 h to enrich for mitotic cells; then cells were treated with α-amanitin (50 μg/mL) or PBS for a further 2 h. Cells then were released from mitotic arrest and placed in cytochalasin B-containing medium to inhibit cytokinesis. One hundred anaphase events were scored as normal or anaphase lagging. Arrowheads indicate examples of lagging chromosomes. α-Amanitin treatment caused a significant increase in lagging anaphases (n = 3; P = 0.011; Student's t test). (D) An anaphase-lagging cell. Lagging chromosomes are indicated by a box, and successfully segregated sister kinetochores are indicated by arrowheads. Insets show close-up images (CREST in green, CENP-C in red) (Left) and merged images (Right) of a lagging chromosome. The fluorescence intensities of CENP-C at the kinetochores of lagging chromosomes (an example is indicated by asterisk) and successfully segregated chromosomes were measured. After α-amanitin inhibition, the mean fluorescence intensity of CENP-C at the kinetochores of the successfully segregated chromosomes was reduced slightly (by 7%; P = 0.005); an even greater reduction of CENP-C was seen at the kinetochores’ lagging chromosomes (28%; P = 0.002; based on 77 control and 107 α-amanitin–treated chromosomes from three biological replicates). Error bars in A–D represent SEM.
To test the effect of RNAPII inhibition on α-satellite transcription specifically in mitotic cells, colcemid-treated cells were subjected to mitotic shake-off to isolate a pool of mitotic cells that were split into control (untreated) and α-amanitin–treated groups and were held in mitosis for a further 5 h. Treatment with α-amanitin resulted in a significant reduction (by 68%) of α-satellite transcripts compared with control mitotic cells (P = 0.016; n = 4) (Fig. 4B). This result indicates that RNAPII is engaged in active transcription of centromeric α-satellite DNA during mitosis. Despite the presence of a small percentage of interphase cells in the mitotic-enriched fraction, no detectable decrease in the transcript levels of control housekeeping genes (i.e., β-actin) was observed in the α-amanitin–treated mitotic cells. This result could be attributed to both the abundance of the housekeeping gene transcripts (even in mitotic cells; Fig. 4A) and their relative stability [GAPDH, hypoxanthine phosphoribosyltransferase (HPRT), and β-actin transcripts have half-lives longer than 20 h (21–23)].
Because RNAPII is essential for cellular viability, RNAi knockdown of the RNAPII complex was not a feasible strategy for studying the role of RNAPII-mediated α-satellite transcription in kinetochore function. Instead, a nocodazole/cytochalasin B-cell synchronization protocol was designed to determine the effect of specific inhibition of RNAPII activity in mitotic cells. The integrity of kinetochore function was measured using a lagging-anaphase assay (24). 14ZBHT cells were treated with nocodazole to enrich for mitotic cells before the addition of α-amanitin (or PBS for control cells) for another 2 h. Cells were released from mitotic arrest into fresh cytochalasin B-containing medium to inhibit cytokinesis. The synchronization protocol and α-amanitin treatment ensured that a large proportion of the accumulated anaphase cells assayed had been targeted for RNAPII inhibition. One hundred anaphase events were scored as either normal or anaphase lagging. The results indicated that inhibition of RNAPII transcriptional activity during mitosis caused a significant increase in anaphase cells with lagging chromosomes, from 18% in control cells to 31% in α-amanitin–treated cells (P = 0.011) (Fig. 4C). A similar increase in the proportion of lagging-anaphase cells (from 6.4% in control cells to 14.6% in α-amanitin–treated cells) was observed when ATCC-CCL 171 cells (a minimally transformed human lung fibroblast cell line with a normal, stable karyotype) were used in the lagging-anaphase assay. The increase in anaphase-lagging cells was observed with or without the presence of cytochalasin B, confirming that the anaphase-lagging phenotype was not an artifact of cytochalasin B (Fig. S4A). Importantly, immunofluorescence experiments and deconvolution microscopy showed that the kinetochores of the lagging chromosomes remain attached to the mitotic spindle (Fig. S4D). This result suggests that the increase in chromosome missegregation is not caused by a failure of the kinetochore to attach to the mitotic spindle.
Interestingly, we also observed reduced CENP-C binding at the kinetochores of both lagging and successfully segregated chromosomes in α-amanitin–treated 14ZBHT cells. α-Amanitin inhibition of RNAPII activity caused a small but significant reduction (7%; P = 0.005) of CENP-C levels at the kinetochores of the successfully segregated chromosomes. More importantly, α-amanitin treatment resulted in further reduction (28%; P = 0.002) of CENP-C levels at the kinetochores of lagging chromosomes compared with levels in control lagging chromosomes (Fig. 4D). The reduced CENP-C association may compromise kinetochore function and could explain the increased anaphase lagging observed in the α-amanitin–treated cells. Interestingly, no significant change in the levels of NDC80 kinetochore complex component homolog (S. cerevisiae) (HEC1), an outer kinetochore plate protein, was detected at the kinetochores of either control or α-amanitin–treated cells. This result suggests that inhibition of RNAPII activity does not appear to interfere with the recruitment of HEC1 at the outer kinetochore plate; however we cannot rule out the possibility that the association of other members of the outer kinetochore KMN complex could be disrupted.
Discussion
In this study, we show that actively transcribing RNAPII and its associated transcription factors, CTDP1 and SSRP1, localize at the mammalian mitotic kinetochore. The specific enrichment of these components at the kinetochores of active neocentromeres, but not at inactive alphoid DNA-based centromeres, establishes a link between RNAPII binding and kinetochore activity. It also indicates that RNAPII binding at the kinetochore is DNA sequence independent and does not require large tracts of repeat DNA. With few exceptions, mitosis has long been assumed to be a period of transcriptional silence, characterized by the eviction of both RNAPII and transcription factors from the bulk mitotic chromatin (25–27). In view of the dogma of mitotic gene repression, our demonstration of specific retention of RNAPII, CTDP1, and SSRP1 at the mitotic kinetochore and evidence of centromeric mitotic transcription is striking.
More importantly, we provide evidence that RNAPII is engaged in active transcription at the mitotic kinetochore and that inhibition of RNAPII activity in mitotic cells is sufficient to cause chromosome missegregation. It is intriguing that these lagging chromosomes in the α-amanitin–treated mitotic cells are still attached to the mitotic spindle, indicating that the outer kinetochore is minimally affected (as shown by unaffected levels of HEC1 recruitment). A recent study showed that Aurora kinase B (AUKB) activity is potentiated by the presence of noncoding RNA (28). The loss of transcripts resulting from the inhibition of RNAPII transcription at the mitotic kinetochore could impact the role of AUKB in correcting erroneous kinetochore–microtubule attachments (29) and thus account for the increase in lagging-anaphase cells in the α-amanitin–treated cells. Future analysis of other outer kinetochore proteins, as well as other regulatory and structural kinetochore proteins, will reveal more about the disruption to the overall kinetochore structure.
We demonstrate that the kinetochores of lagging chromosomes in α-amanitin–treated mitotic cells show reduced CENP-C levels. In particular, the lagging chromosomes of α-amanitin–treated cells showed an even greater reduction in CENP-C association, compared with control cells. In an earlier study, we reported that α-satellite RNA is a key component for the assembly of CENP-C at the metaphase centromere (11). Interestingly, a recent study in maize also showed that the in vitro binding of CENP-C to DNA is enhanced and stabilized by the presence of long, single-stranded RNA (12). Fluorescence recovery after photobleaching analysis of CENP-C cell-cycle dynamics revealed that although CENP-C undergoes dynamic exchange throughout most of the cell cycle, CENP-C is stably bound at the mitotic kinetochore (30). The mitotic transcripts generated by the kinetochore-associated RNAPII reported here could function to stabilize the association of CENP-C at the kinetochore during mitosis. The results of our study thus bring together the above findings by showing that RNAPII-mediated active transcription of α-satellite DNA at the mitotic mammalian centromere is important for the generation of α-satellite transcripts, CENP-C binding, and the function of the mitotic kinetochore.
Although earlier studies have shown that centromere transcription is important for pericentric heterochromatin dynamics (3, 6), more recent studies have provided further evidence of the importance of centromere transcription at the centromere core region (20, 31). It has been shown that the chromatin of a synthetic human kinetochore contains histone modification profiles consistent with active chromatin. Long-term loss of H3K4me2 (a marker of open chromatin) at kinetochore chromatin resulted in decreased centromeric transcripts and defects in CENP-A assembly because of the loss of recruitment of a CENP-A chaperone, Holliday junction recognition protein (HJURP), and subsequent chromosome missegregation. It was suggested that transcription at the kinetochore region could be directly coupled to CENP-A deposition or be indirectly responsible for maintaining a chromatin environment favorable for CENP-A deposition (31). The role of RNAPII in promoting CENP-A deposition also is implied in studies that show the association of CENP-A with centromeric transcripts (8, 20) and the association of the chromatin remodeler FACT with CENP-A chromatin (18). Interestingly, CENP-C also has been implicated recently in the recruitment of Mis18-binding protein 1 and HJURP to the centromere (32). Our demonstration that inhibition of mitotic transcriptional activity results in the destabilization of CENP-C suggests that RNAPII transcription at kinetochore also may have an upstream role in CENP-A loading, via the stabilization of CENP-C binding. Our observations, together with work from others, implicate RNAPII transcription in determining kinetochore structure and remodeling of the centromere chromatin during mitosis.
Experimental Procedures
Cell Culture and Cell Synchronization.
All cells were cultured under standard conditions. Human 14ZBHT cells (16) were cultured with 100 μg/mL Zeocin (Invitrogen). Mouse ES monochromosomal cell hybrid ESmar10 cells (33) were cultured in DMEM with 15% FBS (vol/vol), 103 U/mL leukemic inhibitory factor, 0.1 mM β-mercaptoethanol, and 100 μg/mL Zeocin. Colcemid (Gibco) was added to the cell medium at 0.1 μg/mL for 1–2 h before harvesting of mitotic cells.
Immunofluorescence.
Primary antibodies used were mouse monoclonal antibodies against RNAPII (H5, H14, 8WG16, and 4H8; Abcam) (Table S1), CTDP1 (Abnova), SSRP1, CENP-C, and HEC (Abcam), FITC-conjugated mouse monoclonal anti-tubulin (Abcam), and human polyclonal CREST antisera (15). Secondary antibodies used were Alexa Fluor 488 goat polyclonal anti-human, Alexa Fluor 594 donkey polyclonal anti-mouse IgM, and Alexa Fluor 594 chicken anti-rabbit antibodies (Molecular Probes).
Cells were fixed in PBS/0.5% Triton X-100/4% (vol/vol) paraformaldehyde, blocked in PBS/1% BSA (vol/vol), and incubated with relevant primary and secondary antibodies. Metaphase spreads were processed by immunofluorescence analysis as previously described (11).
All immunofluorescence analyses and image processing were performed using a Zeiss Axioimager.M1 microscope/AxioCam MRm camera/AxioVs40v4.6.1.0 software. Z-stack images were imaged at 0.25-μm intervals, and deconvolution was applied using the iterative algorithm native to AxioVs40v4.6.1.0 software.
Immunofluorescence and DNA-FISH.
Combined FISH and immunofluorescence analyses were carried out as previously described (8). The 10q25 neocentromere-specific 153G5 BAC DNA probe was nick-translated with biotin-dUTP according to the manufacturer's instructions (Boehringer Mannheim). Detection of the biotin probe was carried out with Alexa Fluor 488-conjugated avidin.
In Situ Transcription Assay.
All reagents used were RNase-free. Cytospun mitotic cells were incubated immediately in transcription buffer [100 mM KCl, 50 mM Tris⋅HCl (pH 7.4), 10 mM MgCl2, 0.5 mM EGTA, 25% glycerol (vol/vol), 2 mM rATP, 0.5 mM rCTP, 0.5 mM rGTP, 0.5 mM FITC-rUTP (Sigma), 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 20 U/mL RNase inhibitor], rinsed with KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl, 0.5 mM sodium-EDTA, 0.1% Triton X-100), and fixed with 4% (vol/vol) paraformaldehyde.
Real-Time RT-PCR Assessment of Mitotic Transcripts.
14ZBHT cells were treated with 100 ng/mL colcemid (Gibco) for 1 h before mechanical detachment of mitotic cells (average mitotic index of harvested cells = 70%; n = 5). The mitotic cells were treated with 50 μg/mL α-amanitin (Sigma-Aldrich) or PBS (control cells) for a further 5 h. RNA was isolated (SV Total RNA Isolation System; Promega) and subjected to two rounds of RQ1 RNase-Free DNase (Promega) treatment for maximal removal of genomic DNA. cDNA was synthesized (High-Capacity cDNA Reverse Transcription Kit; Applied Biosystems), and real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) in an Applied Biosystems 7900HT Real-Time PCR System. cDNA equivalent to 10 ng RNA was amplified with 200-nM primers (Primer sequences are given in Table S2) Primers were designed using the PerlPrimer software package (.34).
The comparative cycle threshold (CT) method was used for data analyses. Background values (no reverse transcriptase) were subtracted, and CT values were normalized against the average CT value of three housekeeping genes (HPRT, GAPDH, and β-actin) to give the ΔCT value. Relative fold difference was expressed as 2−ΔΔCT in transcription assays.
Mitotic RNAPII Inhibition Study.
Asynchronous cells were treated with nocodazole for 2 h before the addition of 50 μg/mL α-amanitin (Sigma-Aldrich) for another 2 h. α-Amanitin inhibition was conducted for only 2 h to minimize spurious inhibition of RNAPII transcription in nonmitotic cells. Cells were released from nocodazole arrest and incubated for 1 h in fresh medium containing 10 μM cytochalasin B to inhibit cytokinesis. Cells were fixed in PBS/0.5% Triton X-100/4% (vol/vol) paraformaldehyde and scored for lagging anaphases.
Quantification CENP-C/HEC1 Signals on Lagging Chromosomes.
Z-stacks were taken at 0.5-μm intervals to span the depth of the mitotic cells to generate a maximal image projection image. This image was used to measure the fluorescence intensities of CENP-C and HEC1 at the anaphase kinetochores. A circular area (0.2-μm diameter) was selected around the kinetochore of interest, and the mean intensity (I) within the delineated area was determined using the AxioVs40v4.6.1.0 software. Nonspecific background signal (IBK) for each cell was calculated by the average intensity of five randomly selected regions on chromatid arms. The average signal intensity of 10 successfully segregated sister chromatid kinetochores (ISUCCESS) (randomly selected in the CREST channel) from each cell was used as a control for the hybridization variation. IBK was subtracted from the mean intensity of kinetochores of lagging chromosomes (ILAG) and ISUCCESS. The ratio of CENP--C/HEC1 fluorescence intensities (R) of lagging kinetochores to the 10 successfully segregated sister chromatid kinetochores was calculated as follows: R = (ILAG − IBK)/(ISUCCESS − IBK). CENP-C/HEC1 mean fluorescence intensity (M) of lagging chromosomes was calculated using the following equation: M = R × ISUCCESS.
Statistical Analysis.
Comparisons between qRT-PCR datasets were performed using a two-tailed, two-sample unequal variance Student's t test. Statistical analysis of the anaphase-lagging data was performed using a two-tailed, paired Student's t test. Analyses of CENP-C and HEC1 mean fluorescence intensity were conducted using two-tailed, two-sample equal variance Student's t test. For all tests α was assumed to be 0.05.
Supplementary Material
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia and the Victorian Government's Operational Infrastructure Support Programme. L.H.W. received a Career Development Award from the NHMRC and K.H.A.C. received an NHMRC Senior Principal Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108705109/-/DCSupplemental.
References
- 1.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ekwall K. Epigenetic control of centromere behavior. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- 3.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 4.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1:e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukagawa T, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 7.Saffery R, et al. Transcription within a functional human centromere. Mol Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- 8.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagaki K, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong LH, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 15.du Sart D, et al. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 16.Amor DJ, et al. Human centromere repositioning “in progress”. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal SS, Cho H, Kim S, Cabane K, Reinberg D. FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol Cell Biol. 2002;22:7543–7552. doi: 10.1128/MCB.22.21.7543-7552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-a in cooperation with FACT and CHD1. Mol Biol Cell. 2009;20:3986–95. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saffery R, et al. Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation. Proc Natl Acad Sci USA. 2001;98:5705–5710. doi: 10.1073/pnas.091468498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi ES, et al. Identification of non-coding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem. 2011;286:23600–7. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang E, et al. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lekas P, Tin KL, Lee C, Prokipcak RD. The human cytochrome P450 1A1 mRNA is rapidly degraded in HepG2 cells. Arch Biochem Biophys. 2000;384:311–318. doi: 10.1006/abbi.2000.2115. [DOI] [PubMed] [Google Scholar]
- 23.Sharova LV, et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong NC, et al. Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet. 2006;2:e17. doi: 10.1371/journal.pgen.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christova R, Oelgeschläger T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 26.Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delcuve GP, He S, Davie JR. Mitotic partitioning of transcription factors. J Cell Biochem. 2008;105:1–8. doi: 10.1002/jcb.21806. [DOI] [PubMed] [Google Scholar]
- 28.Ferri F, Bouzinba-Segard H, Velasco G, Hubé F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009;37:5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 30.Hemmerich P, et al. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergmann JH, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong LH, et al. Analysis of mitotic and expression properties of human neocentromere-based transchromosomes in mice. J Biol Chem. 2005;280:3954–3962. doi: 10.1074/jbc.M410047200. [DOI] [PubMed] [Google Scholar]
- 34.Marshall OJ. PerlPrimer: Cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.