Abstract
Type IV P-type ATPases (P4-ATPases) catalyze translocation of phospholipid across a membrane to establish an asymmetric bilayer structure with phosphatidylserine (PS) and phosphatidylethanolamine (PE) restricted to the cytosolic leaflet. The mechanism for how P4-ATPases recognize and flip phospholipid is unknown, and is described as the “giant substrate problem” because the canonical substrate binding pockets of homologous cation pumps are too small to accommodate a bulky phospholipid. Here, we identify residues that confer differences in substrate specificity between Drs2 and Dnf1, Saccharomyces cerevisiae P4-ATPases that preferentially flip PS and phosphatidylcholine (PC), respectively. Transplanting transmembrane segments 3 and 4 (TM3-4) of Drs2 into Dnf1 alters the substrate preference of Dnf1 from PC to PS. Acquisition of the PS substrate maps to a Tyr618Phe substitution in TM4 of Dnf1, representing the loss of a single hydroxyl group. The reciprocal Phe511Tyr substitution in Drs2 specifically abrogates PS recognition by this flippase causing PS exposure on the outer leaflet of the plasma membrane without disrupting PE asymmetry. TM3 and the adjoining lumenal loop contribute residues important for Dnf1 PC preference, including Phe587. Modeling of residues involved in substrate selection suggests a novel P-type ATPase transport pathway at the protein/lipid interface and a potential solution to the giant substrate problem.
Keywords: membrane asymmetry, protein transport
P-type ATPases are phylogenetically grouped into five subclasses of integral membrane pumps that mediate active transport of substrate across membrane bilayers (1). This family is defined by the formation of a phosphorylated intermediate (P-type) and four characteristic domains: actuator (A), nucleotide-binding (N), phosphorylation (P), and transmembrane (TM). The best characterized subclass, the type II P-type ATPases (P2-ATPases) catalyze the energy-dependent movement of cations across lipid bilayers to establish an electrochemical gradient. The substrate binding sites of P2-ATPases lie within the TM domain, and involve multiple residues that coordinate the cations in a well defined binding pocket formed by charged and polar residues in TM 4, 5, 6, and 8 in the Ca2+ ATPase (2) and TM segments 4, 5, 6, 8, and 9 in the Na+/K+ ATPase (3). Access to the cation binding sites alternates between the cytosolic and exofacial sides of the membrane, and is driven by E1-E2 conformational changes associated with ATP binding, transfer of the γ-phosphate from ATP to a conserved aspartate, and subsequent hydrolysis of the aspartyl-phosphate bond (4). Whether or not this alternating half-channel mechanism of substrate recognition and transport is conserved for all P-type ATPase subclasses is unknown.
In contrast to the cation transporters, members of the type IV P-type ATPase subfamily (P4-ATPases) translocate phospholipid across a membrane bilayer by flipping their substrate from the exofacial leaflet to the cytosolic leaflet (5, 6). The unidirectional flip of specific phospholipid species, such as phosphatidylserine (PS) and phosphatidylethanolamine (PE), creates an asymmetric membrane structure with PS and PE concentrated within the cytosolic leaflet. The P4-ATPase subfamily is conserved among eukaryotes with Saccharomyces cerevisiae expressing five members that are essential for viability (7), whereas mammals have at least 14 members (8, 9). Several of these mammalian flippases are implicated in pathological conditions such as obesity and type II diabetes (10), intrahepatic cholestasis (11, 12), progressive hearing loss (13), male infertility (14), and deficiencies in B cell lymphopoiesis (15, 16). In addition, P4-ATPases play critical roles in vesicle-mediated protein trafficking in the secretory and endocytic pathways (17). Many P4-ATPases associate with Cdc50 family proteins that are potentially analogous to the Na+/K+ ATPase β and γ subunits. For example, the yeast P4-ATPase Drs2 interacts with Cdc50 to allow their mutual export from the endoplasmic reticulum (ER) to the Golgi complex (18).
The mechanism of phospholipid transport by P4-ATPases is undefined and has been described as the “giant substrate problem” because fitting a phospholipid into the structurally conserved, but spatially restrictive, substrate binding pockets would be impossible within the confines of the available P-type ATPase crystal structures (19). Moreover, P4-ATPases lack the charged residues that form the cation binding sites in P2-ATPases and no phospholipid binding site has been identified. Translocation of phospholipid is proposed to occur at the E2-P to E1 transition based upon the direction of substrate translocation and the demonstration that phospholipid substrate stimulates dephosphorylation of Atp8a1, a mammalian P4-ATPase (20). Cdc50 preferentially associates with Drs2 in the E2P state, suggesting that Cdc50 might form part of the phospholipid binding pocket (21). However, recent evidence from Arabidopsis thaliana P4-ATPases indicates the Cdc50 subunit does not affect the substrate specificity of the heterodimer, implying that determinants of substrate specificity primarily reside within the catalytic subunit (22). For P4-ATPases, nothing is known about: (i) the residues in the TM domain that recognize the phospholipid substrate, (ii) how different P4-ATPases recognize different phospholipid substrates, and (iii) the pathway phospholipid follows through the TM domain from the exofacial to cytosolic leaflet. The giant substrate problem is one of the paramount questions concerning the function of P4-ATPases.
To gain insight into the mechanism of phospholipid flip by P4-ATPases, we sought to identify residues that define phospholipid substrate specificity. Our strategy was to generate chimeras between two P4-ATPases that differ in substrate preference in order to map residues determining this preference. The yeast P4-ATPase Dnf1 prefers phosphatidylcholine (PC) and PE (23, 24), whereas Drs2 prefers PS (5, 25) with a minor activity towards PE (26). Here we report the identification of sequences in TM segments 3–4 that help determine phospholipid specificity in Dnf1 and Drs2. From a structural model of Dnf1 and the positions of the key residues impacting phospholipid specificity, we suggest an entirely unique mechanism of transport by a member of the P-type ATPase superfamily.
Results
Drs2 TM Segments 3–4 Confer NBD-PS Uptake Activity to Dnf1.
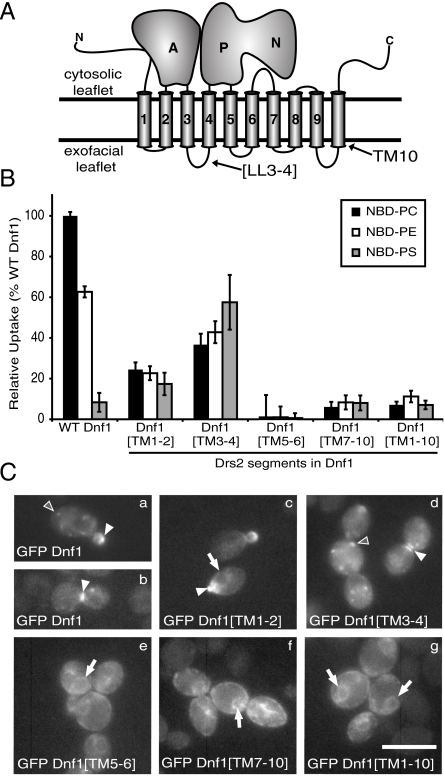
P4-ATPases initially select their substrate from the exofacial leaflet of the bilayer; therefore, residues involved in substrate selection must lie within the exofacial loops or within the transmembrane region (Fig. 1A). We systematically generated chimeras by exchanging pairs of transmembrane segments with intervening lumenal loops from Drs2 into Dnf1 (SI Appendix, Fig. S1) and tested the Dnf1[Drs2] chimeras for a change in substrate specificity. Advantages of this approach include (i) Dnf1 is expressed on the plasma membrane (7), allowing us to easily assay the chimera translocase activity in living cells. (ii) Mutations causing a change in substrate preference, rather than loss, must be compatible with appropriate folding and trafficking of the flippase. (iii) The ratio of PS to PC uptake for each chimera provides a measure of substrate specificity that is independent of translocase number at the plasma membrane.
Fig. 1.
Transmembrane segments 3–4 from Drs2 are sufficient to confer a PS activity to Dnf1. (A) Topology diagram of a P4-ATPase; A, actuator domain; N, nucleotide-binding domain; P, phosphorylation domain; LL3-4, lumenal loop between TM3 and TM4; black lines denote the membrane boundaries. (B) NBD-PL uptake across the plasma membrane mediated by Dnf1[Drs2] chimeras. TM3-4 contains important sequence for phospholipid recognition. Results represent averages of at least three independently isolated transformants from at least three independent experiments (mean ± SEM). (C) Localization of N-terminal GFP fused Dnf1 and Dnf1[Drs2] chimeras in S. cerevisiae. GFP-Dnf1 is localized to intracellular punctae in nonbudded cells (a, d), to the bud tip in small-budded cells (a, c) and to the bud neck in large-budded cells (b, d). A subset of GFP Dnf1[Drs2] chimeras mislocalize to the ER (e–g). Arrowheads designate polarized expression patterns (bud tip or bud neck); open arrowheads show punctate localization; arrows indicate perinuclear ER fluorescence. Fluorescence intensities for the images are scaled independently to emphasize localization. (Scale bar: 10 μm.)
To assay flippase activity, chimeras were expressed in a dnf1∆dnf2∆ strain lacking the major plasma membrane P4-ATPases in yeast (23) and we measured uptake of 7-nitro-2–1,3-benzoxadiazol-4-yl phospholipid (NBD-PL) across the plasma membrane by flow cytometry. In each experiment, dnf1,2∆ cells bearing an empty vector control were assayed to subtract chimera-independent uptake of each NBD-PL and the uptake of NBD-PC by wild-type (WT) Dnf1 was used to normalize the data. WT Dnf1 is reported to translocate PC and PE, and debatably PS (23, 27). In these experiments, WT Dnf1 mediated robust uptake of PC and PE and had a minor influence on the uptake of PS (n = 150+) (Fig. 1B). The Dnf1[TM1-2] chimera showed reduced activity for both PC and PE, but no significant change in activity for PS (Fig. 1B). In contrast, Dnf1[TM3-4] had a reduced activity to PC and PE, but gained an activity towards PS, the preferred substrate of Drs2. Dnf1[TM5-6], Dnf1[TM7-10] and Dnf1[TM1-10] each had little to no activity at the plasma membrane for any of the NBD-PLs tested (Fig. 1B).
The lack of activity for some of the chimeras could be caused by a failure to traffic through the secretory pathway to the plasma membrane. We tested this possibility by tagging each chimera with GFP for localization studies. WT Dnf1 exhibits a polarized localization that depends on the cell cycle (7, 18). GFP-Dnf1 localized to intracellular punctae in nonbudded cells, to the bud tip in small-budded cells, and to the bud neck in large-budded cells [Fig. 1 C, a and b arrowheads]. GFP-Dnf1[TM1-2] and GFP-Dnf1[TM3-4] retained similar localization patterns to GFP-Dnf1 regardless of cell cycle stage (Fig. 1 C, c and d), indicating the decrease in activity by Dnf1[TM1-2] is not caused by an inability to localize to the plasma membrane. GFP-Dnf1[TM5-6], GFP-Dnf1[TM7-10], and GFP-Dnf1[TM1-10] exhibited localization patterns characteristic of the ER, as indicated by the ring of fluorescence surrounding the nucleus (Fig. 1 C, e–g, arrows). ER retention is a common problem for improperly folded integral membrane proteins (28), providing a likely explanation for the lack of PM uptake activity for these chimeras.
TM4 Is Required for PS Activity, but Not Specificity.
TM3-4 from Drs2 was sufficient to confer a PS activity to Dnf1, and this Dnf1[TM3-4] chimera also showed a reduced activity for PC and PE (Fig. 1B and 2A). By plotting the ratio of PS to PC uptake, we quantified a nearly eightfold increase in PS preference for the Dnf1[TM3-4] chimera relative to Dnf1 (Fig. 2B). Because Dnf1[TM1-2] displayed reduced PC and PE activities, it was possible that TM1-2 from Drs2 contributed to selection against PC and PE. However, exchanging TM1-4 as a unit, to generate Dnf1[TM1-4], yielded a chimera with a similar PC and PE activity to Dnf1[TM3-4], but with a slightly lower PS activity (SI Appendix, Fig. S2A). This result suggests TM1-2 does not contribute additional substrate specificity to TM3-4.
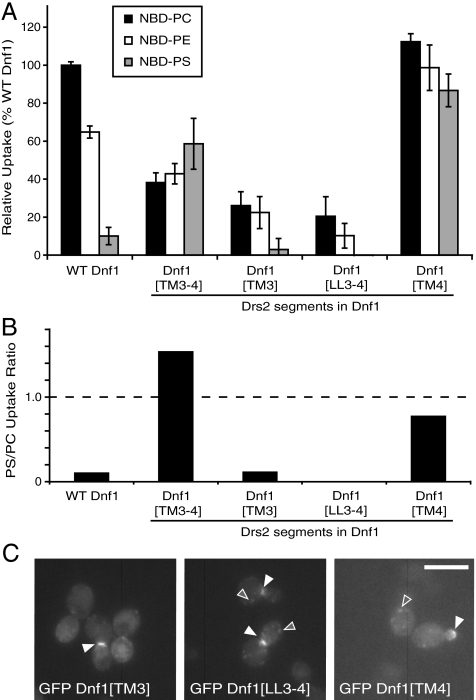
Fig. 2.
Drs2 TM3-4 confers PS preference to Dnf1, while TM4 confers a PS activity without affecting the recognition of PC or PE. (A) NBD-PL uptake shows TM4 from Drs2 is sufficient to confer a PS activity to Dnf1, while TM3 and LL3-4 are important for NBD-PC and NBD-PE selection (mean ± SEM). (B) The ratio of PS to PC uptake from (A) was plotted to provide a measure of substrate preference that is independent of expression level. (C) Localization of GFP tagged Dnf1[TM3], Dnf1[LL3-4], and Dnf1[TM4] chimeras in S. cerevisiae indicate these chimeras localize and traffic similarly to WT Dnf1. (Scale bar: 10 μm.)
We analyzed each section of TM3-4 individually to determine their relative contribution to this change in substrate specificity. Dnf1[TM3] and Dnf1[LL3-4] showed reduced PC, PE, and PS activity and so these segments failed to confer PS activity to Dnf1 (Fig. 2A). Dnf1[TM4] retained WT activity for PC, showed a modest increase in activity for PE, but had a substantial increase in PS activity (Fig. 2A). Thus, TM4 from Drs2 was sufficient to confer a PS activity to Dnf1, but still allowed for translocation of PC and PE. Relative to WT Dnf1, Dnf1[TM3-4] showed the greatest preference for PS (Fig. 2B) even though Dnf1[TM4] showed a greater increase in PS activity (Fig. 2A). The localization pattern of each of these chimeras was similar to WT Dnf1 (Fig. 2C, fluorescence at the bud tip or bud neck) and was therefore unlikely to be responsible for any difference in activity or substrate preference. These results suggest that TM3 and LL3-4 in Dnf1 contribute to PC and PE selection, and TM4 from Drs2 contributed residues important for PS selection.
TM4 contains three sequence blocks that differ between Dnf1 and Drs2 (Fig. 3A), so we exchanged each Drs2 TM4 block independently into Dnf1 to determine their contribution to the PS activity of Dnf1[TM4]. Of these three chimeras, only Dnf1[YIS → FVT] displayed an increased activity for PS (Fig. 3B) and a substrate preference equivalent to Dnf1[TM4] (Fig. 3C). Therefore, we exchanged each residue individually (Y618F, I619V, and S620T) and in pairwise combinations. The Dnf1 Y618F point mutation had the greatest influence on net PS activity although Dnf1 I619V and Dnf1 S620T also showed slightly enhanced PS transport relative to WT Dnf1 (Fig. 3B). Dnf1[YIS → FVT] had a greater net PS activity compared to any of the individual point mutations but this chimera had no increased preference for PS (Fig. 3C). Dnf1 Y618F displayed specificity for PS comparable to Dnf1[TM4] and Dnf1[YIS → FVT] (Fig. 3C) without the general increase in activity of Dnf1[YIS → FVT]. Y618, I619, and S620 were changed to Drs2 residues in pairs, with no additional specificity for PS being conferred, indicating Y618F is the key substitution enabling PS translocation (SI Appendix, Fig. S3 A and B). The closest mammalian homolog of Drs2, Atp8a1, has a leucine in the TM4 position analogous to Y618 in Dnf1 and F511 in Drs2 (Fig. 3A), so we generated a Y618L substitution in Dnf1 to determine if this could also allow PS uptake (Fig. 3B). However, the Y618L substitution did not confer a PS activity and instead reduced Dnf1 activity against all three substrates.
Fig. 3.
A single amino acid change, Y618F, specifically confers PS uptake activity to Dnf1. (A) Primary sequence alignment of TM4 from Dnf1, Dnf2, Drs2, and Atp8a1. Underlined sequences are the predicted TM4; boxes are regions differing between Dnf1 and Drs2. (B) NBD-PL uptake by Dnf1[Drs2] chimeras. Dnf1[YIS → FVT] and Dnf1 Y618F both transport PS (mean ± SEM). (C) PS/PC uptake ratio representing the relative preference of each chimera for either PC or PS independent of expression. (D) NBD-lipid uptake suggests Dnf1[TM3-4], Dnf1[TM4], and Dnf1 Y618F retain specificity for the phospholipid headgroup and glycerol backbone. Dnf1[YIS → FVT] is less selective than other PS transporting chimeras based on its activity for NBD-SM. Each reported value (B–D) is the average of at least three independent samples from at least three independent experiments (mean ± SEM).
The increased activity for PS by several of the chimeras could result from the specific acquisition of a new substrate, or a complete loss of specificity such that any NBD-PL would be translocated. Therefore, we tested if the chimeras could catalyze the uptake of NBD-phosphatidic acid (NBD-PA) or NBD-sphingomyelin (NBD-SM). WT Dnf1 cannot translocate NBD-SM or NBD-PA, and Dnf1[TM3-4], Dnf1[TM4], and Dnf1 Y618F retained comparable specificity (Fig. 3C). Surprisingly, not only did Dnf1[YIS → FVT] display an increased activity for PC, PE, and PS (Fig. 3B), it also conferred a modest increase in NBD-SM uptake (Fig. 3D). However, Dnf1[YIS → FVT] was not capable of NBD-PA transport (Fig. 3D), indicating it had not completely lost phospholipid selectivity nor did this chimera selectively recognize the NBD moiety. The expression level of WT Dnf1, Dnf1[YIS → FVT], and Dnf1 Y618F were similar and unlikely to account for the increased uptake observed in Dnf1[YIS → FVT] (SI Appendix, Fig. S4). To rule out the possibility that the mutations in Dnf1 somehow altered the localization or plasma membrane activity of Drs2, we localized GFP-Drs2 in strains harboring various Dnf1 chimeras (SI Appendix, Fig. S5A) and measured Dnf1 chimera NBD-PL uptake in a dnf1,2Δdrs2Δ strain (SI Appendix, Fig. S5B). In no case was the enhanced NBD-PS uptake observed attributable to Drs2.
The Reciprocal Drs2 F511Y Substitution Disrupts PS Asymmetry.
Because the Y618F substitution allowed PS recognition by Dnf1 without disrupting PE or PC flippase activity, we predicted that the reciprocal F511Y exchange in Drs2 would perturb its PS flippase activity without affecting recognition of PE. To assess Drs2 recognition of endogenous, unmodified phospholipid, we tested the influence of the Drs2 F511Y substitution on PS and PE asymmetry of the plasma membrane. Disruption of Drs2 causes a loss of membrane asymmetry and exposure of PS and PE on the extracellular leaflet of the plasma membrane (29). Loss of membrane asymmetry can be measured by hypersensitivity to pore-forming toxins that bind selectively to PS (papuamide B, PapB) (30) or PE (duramycin) (31) exposed on the outer leaflet of the plasma membrane. As predicted, Drs2 F511Y caused a partial loss of PS asymmetry, as indicated by the intermediate sensitivity to PapB relative to the WT and drs2∆ strains (Fig. 4A). In contrast, Drs2 F511Y conferred WT resistance to duramycin and therefore a normal asymmetric distribution of PE (Fig. 4B). Thus, Drs2 F511Y is specifically defective in establishing PS asymmetry. We also generated a strain expressing the Drs2 F511L variant and found these cells displayed a partial loss of asymmetry for both PE and PS (Fig. 4 A and B). Even though Dnf1 Y618F can translocate NBD-PS across the plasma membrane, it was unable to suppress the PapB sensitivity of a drs2∆ strain (SI Appendix, Fig. S6). We assume that the highly polarized localization may not allow Dnf1 to correct a membrane asymmetry defect across the entire plasma membrane of drs2∆ cells. Each form of Drs2 (WT, F511Y, and F511L) was purified and assayed for ATPase activity as previously described (5). While Drs2 F511Y retained WT activity, Drs2 F511L had a specific activity that was 35% that of WT Drs2 (Fig. 4C). These data imply that the F511Y substitution causes a change in Drs2 substrate specificity without altering the activity, while F511L reduces the overall activity.
Fig. 4.
Drs2 F511Y specifically perturbs plasma membrane PS asymmetry. (A) Growth of a drs2Δ strain expressing an empty vector, WT Drs2, Drs2 F511Y, or Drs2 F511L in the presence of papuamide B demonstrates a partial loss of PS asymmetry with Drs2 F511Y and Drs2 F511L (mean ± SEM). (B) Growth of a drs2Δ strain expressing the same constructs in the presence of duramycin indicates plasma membrane PE asymmetry is maintained by WT Drs2 and Drs2 F511Y but is perturbed with Drs2 F511L (mean ± SEM). (C) Drs2 F511Y retains an ATPase specific activity comparable to WT Drs2 while Drs2 F511L has a reduced ATPase activity (mean ± SD). (D) Drs2 F511Y fully complements cold-sensitive growth defect of a drs2Δ strain but Drs2 F511L only partially complements.
Under certain conditions, such as low temperature, DRS2 is essential for growth because of its function in supporting protein trafficking from the trans-Golgi network. The drs2-F511Y allele fully complemented the cold-sensitive growth defect of drs2∆ (Fig. 4D), indicating that Drs2 F511Y must localize properly and that the loss of PS asymmetry is not a secondary consequence of a protein trafficking defect or an inactive enzyme. The drs2-F511L allele was only able to partially complement the cold-sensitive growth defect of drs2∆ (Fig. 4D), consistent with it being a hypomorphic allele. Strains carrying deletions of DRS2 and all three DNF genes (dnf1,2,3∆drs2∆) are inviable and while drs2-F511Y was able to complement this growth defect, drs2-F511L was unable to do so (SI Appendix, Fig. S7B).
The Rate of NBD-PL Uptake by Dnf1 Y618F Is Comparable to WT Dnf1.
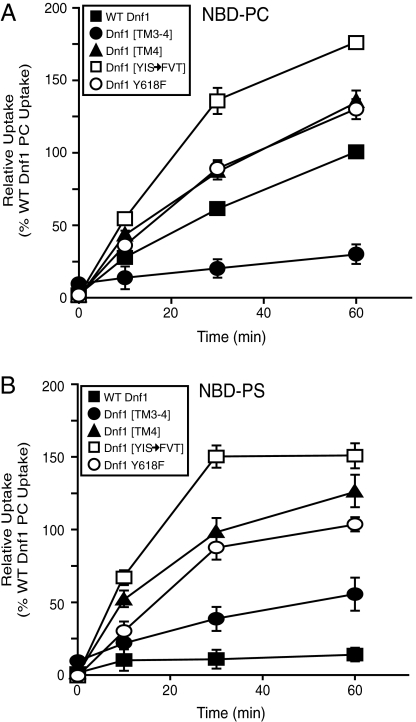
To better define the substrate preferences and transport properties of the Dnf1 [Drs2] chimeras, we examined the kinetics of NBD-PC and NBD-PS uptake. As expected, Dnf1[TM3-4] displayed a reduced rate of NBD-PC uptake compared to WT Dnf1, while Dnf1[YIS → FVT] had an increased rate of NBD-PC uptake. Dnf1[TM4] and Dnf1 Y618F showed a similar rate of NBD-PC uptake relative to WT Dnf1 (Fig. 5A). Thus, acquisition of the ability to translocate PS did not alter the activity toward PC.
Fig. 5.
The rate of NBD-PC and NBD-PS uptake mediated by Dnf1 Y618F is comparable, suggesting a similar transport mechanism. (A) NBD-PC uptake at 0, 10, 30, and 60 min where 100% corresponds to WT Dnf1 NBD-PC uptake at 1 h (mean ± SEM). (B) NBD-PS uptake where 100% corresponds to WT Dnf1 NBD-PC uptake at 1 h (mean ± SEM).
Each Dnf1[Drs2] chimera tested displayed a significant increase in NBD-PS uptake activity relative to Dnf1 (Fig. 5B). Dnf1[TM3-4] had the lowest rate of NBD-PS uptake (Fig. 5B) and Dnf1[YIS → FVT] showed the highest rate of NBD-PS uptake compared to the other Dnf1[Drs2] chimeras examined. Importantly, the rate of NBD-PS uptake by Dnf1[TM4] and Dnf1 Y618F was comparable to the rate of NBD-PC uptake by these chimeras (and WT Dnf1), suggesting a similar transport mechanism for PC and PS (Fig. 5B).
PC Recognition by Dnf1.
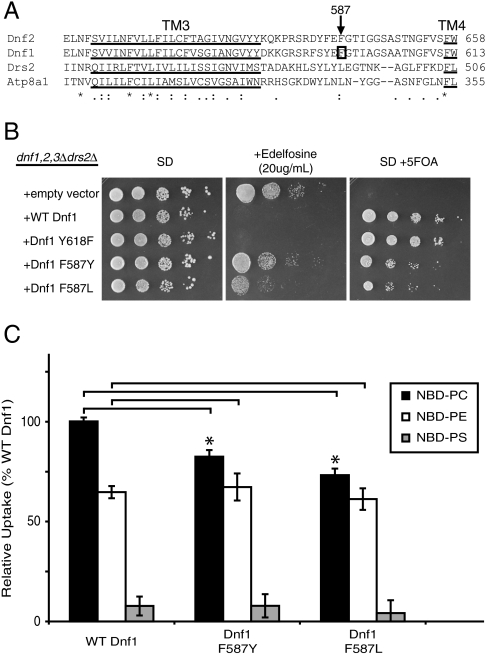
To identify residues involved in phospholipid selection by Dnf1, we searched for residues specifically involved in recognition of PC. Based on the role of TM3 and LL3-4 in PC/PE selection (Fig. 2A) we generated random mutations in sequences encoding TM3-4 of Dnf1 and transformed this collection of DNF1 mutant alleles into a dnf1,2,3∆drs2∆ pRS416-DRS2 strain. These mutants were initially screened for resistance to edelfosine, a toxic PC analog (32), and then counterscreened for the ability to support growth of the dnf1,2,3∆drs2∆ strain by selection on 5-fluoroorotic acid (5FOA, which selects for loss of URA3 encoded on pRS416-DRS2). This screen should select for a functional Dnf1 with a reduced ability to flip PC. In two independent clones carrying point mutations, Phe587 (within LL3-4) was mutated to either a Tyr or Leu residue (Fig. 6A). Dnf1 F587L and F587Y supported growth of dnf1,2,3∆drs2∆ cells, although not as well as WT Dnf1 or Dnf1 Y618F (Fig. 6B, SD + 5FOA). However, strains expressing Dnf1 F587L and F587Y were able to grow at a concentration of edelfosine that killed strains expressing WT Dnf1 or Dnf1 Y618F (Fig. 6B, SD + edelfosine). When assayed for NBD-PL uptake in a dnf1,2∆ strain, both Dnf1 F587Y and F587L showed a statistically significant reduction in PC uptake (p < 0.05) with no change in PE uptake (p > 0.20) relative to WT Dnf1 (Fig. 6C). The normal PE activity of these Dnf1 mutants suggests they are properly expressed and localized to the plasma membrane. We conclude that F587 is specifically involved in PC selection by Dnf1.
Fig. 6.
F587 substitutions specifically perturb PC uptake activity by Dnf1. (A) Primary sequence alignment of TM3-LL3-4 from Dnf1, Dnf2, Drs2, and Atp8a1. Underlined sequences are the predicted TM3 and TM4; box indicate F587 in Dnf1. (B) Growth of dnf1,2,3Δdrs2Δ expressing the Dnf1 constructs indicated on synthetic defined (SD) media, SD plus edelfosine, SD plus 5FOA. (C) NBD-PL uptake indicates a defect in PC uptake by Dnf1 F587Y and Dnf1 F587L (p < 0.05) and no defect in PE uptake (p > 0.20) (mean ± SEM).
Function of Dnf1[Drs2] Chimeras In Vivo.
DNF1,2,3 and DRS2 form an essential gene family and strains harboring mutations in all four of these genes are inviable (7). We examined the ability of Dnf1[Drs2] chimeras to support growth of a dnf1,2,3∆drs2∆ strain using the plasmid shuffling assay described above. In each case, the chimeras with pairs of full TM segments exchanged (Dnf1[TM1-2], Dnf1[TM3-4], Dnf1[TM5-6], Dnf1[TM7-10], and Dnf1[TM1-10]) failed to support growth of a dnf1,2,3∆drs2∆ strain (SI Appendix, Fig. S7A). Dnf1[TM3] Dnf1[LL3-4], Dnf1[TM4] and Dnf1[WVAV-LTFW] also failed to support growth of a dnf1,2,3∆drs2∆ strain (SI Appendix, Fig. S7A). Of this group, the inability of Dnf1[TM4] to support viability was most surprising because it retained a fully functional flippase activity (Fig. 2A). Dnf1[YQS → FSN] only partially supported growth (SI Appendix, Fig. S7A), even though its uptake activity was only slightly reduced relative to WT Dnf1 (Fig. 3B). In contrast, Dnf1[YIS → FVT] and each of the point mutants (Dnf1 Y618F, Dnf1 I619V, and Dnf1 S620T) fully supported growth of a dnf1,2,3∆drs2∆ strain (SI Appendix, Fig. S7A). Seemingly, neither an increased uptake activity (as seen in Dnf1[YIS → FVT]) nor an altered substrate specificity appears detrimental to the ability of these chimeras to support growth of a dnf1,2,3∆drs2∆ strain. These data imply that flippase activity is important for in vivo function, but in the case of Dnf1[TM4], the Drs2 sequences are unable to support some other critical function of Dnf1 (compare Fig. 3B, SI Appendix, Fig. S7A). However, it is important to note that the essential function of the P4-ATPases is exerted within the Golgi and endosomal system, not the plasma membrane where activity was measured (33).
Lem3 Is Required for Dnf1[Drs2] Chimera ER Export.
WT Dnf1 interacts with the Cdc50 family protein Lem3, and this interaction is required for their mutual export from the ER (18). Cdc50 family proteins may contribute to substrate recognition and transport by P4-ATPases (19, 22) and it was possible that the Dnf1[Drs2] chimeras gained the ability to interact with Cdc50, thereby conferring specificity for PS. Therefore, we expressed the GFP-tagged chimeras in a lem3∆ strain and tested if overexpression of CDC50 could support their transport to the plasma membrane. GFP-Dnf1 and all of the chimeras tested were retained in the ER of the lem3∆ cells, even when CDC50 was overexpressed (Fig. 7A). In contrast, LEM3 overexpression rescued the polarized plasma membrane localization pattern for each chimera. To test the possibility that Cdc50 was responsible for the changes in substrate selection we assayed the uptake activity of several Dnf1 chimeras in the absence of Cdc50 or Lem3 (Fig. 7B). In each case, the Dnf1 chimeras strictly required Lem3, not Cdc50, for activity at the plasma membrane. Thus, there was no change in the Lem3 requirement for proper ER export, localization, or flippase activity of the Dnf1 chimeras tested.
Fig. 7.
GFP tagged Dnf1[Drs2] chimeras retain a Lem3 requirement for ER export and flippase activity. (A) GFP tagged chimeras were expressed in lem3Δ cells (SCY119) overexpressing CDC50 or LEM3. Each Dnf1[Drs2] chimera tested exhibited a characteristic ER localization pattern in lem3∆ cells overexpressing CDC50. In contrast, overexpression of LEM3 in a lem3Δ strain rescues each Dnf1[Drs2] chimera polarized localization pattern. In each row, the fluorescence intensity of each image is scaled equivalently to show relative expression levels of each chimera. (Scale bar: 10 μm.) (B) NBD-PL uptake by Dnf1[Drs2] chimeras expressed in dnf1,2Δcdc50Δ or dnf1,2Δlem3Δ strains.
Potential P4-ATPase Substrate Transport Pathway.
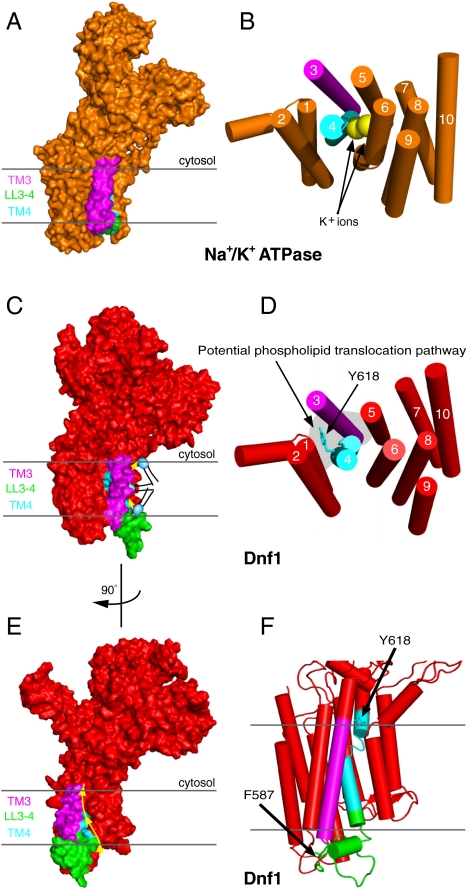
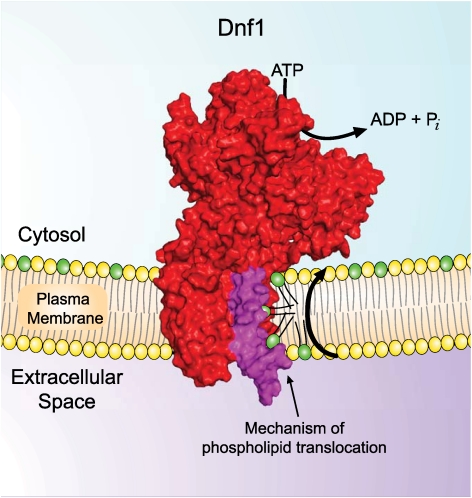
Based upon the functional data we have presented, a single amino acid substitution (Y618F) in TM4 of Dnf1, representing the loss of a single hydroxyl group, allows this protein to recognize and translocate NBD-PS. To better understand how such a subtle sequence change could lead to a dramatic change in the substrate preference, we generated a structural model of Dnf1 based upon the crystal structure of the Na+/K+ ATPase (Fig. 8A) (PDB 2ZXE) (3). We used the SWISS-MODEL workspace (34, 35) for the initial modeling and relaxed the model in Rosetta (36, 37) to obtain the final structural model (Fig. 8C). The complete conservation of the helix-breaking Pro from the PEGL motif in cation transporting ATPases (PISL in P4-ATPases), provides a concrete anchoring point for the modeling of TM segment 4, particularly for Y618 in the proline plus 4 position (PISLY) (SI Appendix, Fig. S8A). The glutamate residue from the PEGL motif in TM4 of cation transporting ATPases has a critical role in binding substrate and is oriented towards the K+ binding sites formed by residues in TM 4,5,6, and 8 (Fig. 8B). In contrast, the proline plus 4 residue in each available P-type ATPase crystal structure is rotated 180° around TM4 and is oriented in a triangular cleft between TM segments 1, 3, and 4 (3, 38–40), in the opposite direction from the canonical cation binding sites (SI Appendix, Fig. S8 B and C). The orientation of Dnf1 Tyr618 in the modeled structure is equivalently positioned at the apex of this triangular cleft (Fig. 8 D and F, SI Appendix, Fig. S8C). Tyr618 is also near the membrane interface on the cytosolic side, suggesting this residue comprises (part of) the exit site for the phospholipid substrate. LL3-4 is much longer in the P4-ATPases than in other P-type ATPase subfamilies, and is therefore not threaded onto an experimentally determined structure. This loop likely forms the base of the triangular cleft as modeled, but the orientation of F587, a residue important for PC selection, is unclear. However, the residues important for phospholipid substrate selection in TM3, LL3-4, and the proline plus 4 position of TM4 are very unlikely to lie within the canonical substrate binding pocket defined in cation transporters. Therefore, we propose a unique transport pathway for P4-ATPase flippases where the phospholipid headgroup is translocated along a groove between TM1, TM3, and TM4 while the fatty acyl chains reorient within the lipid environment of the membrane (Fig. 8 C–E).
Fig. 8.
Model of the Dnf1 structure showing the orientation of Y618 and a proposed pathway for phospholipid translocation. (A) Na+/K+ ATPase crystal structure in the E2(2K+)-Pi conformation [PDB 2ZXE (3)]. Dark gray lines represent the membrane boundaries. (B) View of the Na+/K+ ATPase TM domain from the cytosolic side. (C) Structural model of Dnf1 highlighting segments involved in phospholipid selection and the groove between TM1, TM3, and TM4 forming a potential pathway for phospholipid flip. (D) View of the Dnf1 TM domain from cytosolic side of bilayer. Tyr618 is oriented on the opposite side of TM4 relative to the cation binding pocket of ion-transporting P-type ATPases. (E) Dnf1 model rotated 90° compared to (C). The yellow arrow represents the potential phospholipid transport pathway at the protein/lipid interface between TM1,TM3, and TM4. (F) Triangular cleft formed from TM1, TM3, LL3-4, and TM4. Arrows indicate F587 and Y618 of Dnf1, residues involved in PC and PS selection, respectively.
Discussion
Cation transporting P-type ATPases utilize a spatially restrictive ion binding pocket in the center of the TM domain. In this work, we have identified residues that contribute to phospholipid substrate specificity in S. cerevisiae P4-ATPases that lie outside of the canonical binding pocket. Based on the orientation of a key tyrosine residue in TM4 and the involvement of TM3 and LL3-4 residues in substrate selection, we suggest a pathway for phospholipid translocation along the protein/lipid interface that would accommodate transport of this large substrate.
We generated a series of chimeras between Dnf1 and Drs2, two P4-ATPases that differ in substrate specificity, to map amino acid residues involved in substrate selection. We sought to identify Drs2 sequences required for PS recognition by construction of chimeras that convert Dnf1 from a PC/PE flippase to a PS flippase. We have partially succeeded in this endeavor as Dnf1[TM3-4] gained the ability to flip NBD-PS and shows a preference for NBD-PS over NBD-PC. While we have significantly altered the substrate specificity of Dnf1 through incorporation of these Drs2 sequences, it appears that we have not fully recapitulated the substrate specificity of Drs2 because Drs2 appears to have negligible activity towards NBD-PC (5, 25). Substitution of Dnf1 TM3-4 with analogous Drs2 sequences significantly attenuates uptake of NBD-PC and NBD-PE, even though the protein localizes normally to the plasma membrane. The striking increase in PC and PE activity of the Dnf1[TM4] chimera relative to Dnf1[TM3-4] also implies that TM3 and LL3-4 from Dnf1 contain sequences that recognize these substrates much better than the analogous Drs2 segments. In fact, we have identified substitutions of a single residue (Phe587) within LL3-4 that reduce PC recognition without affecting recognition of PE. These are original observations as TM3-LL3-4 residues have not been implicated in substrate recognition by other P-type ATPases. LL3-4 is much larger in the P4-ATPases relative to the cation transporters and no structural information on this loop is available, but it is likely to sit at the membrane surface where phospholipid substrate would initially be selected for translocation.
The most surprising observation of this work is that the substitution of a single tyrosine residue for phenylalanine in TM4 (Y618F) conferred an NBD-PS transport activity to Dnf1 without altering the ability of this protein to flip NBD-PC or NB-PE. TM4 is also important for substrate interaction by cation transporting P-type ATPases and contains a highly conserved, helix-breaking Pro followed by a glutamate residue critical for cation binding by SERCA (Glu309) and the Na+/K+ ATPase (Glu334) (SI Appendix, Fig. S8A). The PEGLX motif in the cation transporters is replaced by PISLY (Dnf1) and PISLF (Drs2) in these P4-ATPases. Conservation of Pro implies that the P4-ATPases maintain the same break in the TM4 helix observed in the heavy metal (P1), cation (P2), and proton (P3) transporter crystal structures and that structural features of this TM segment would be similar. The critical Tyr/Phe identified in this study lies only four residues from the conserved proline, and in each P-type ATPase crystal structure, the residue analogous to Dnf1 Tyr618 is oriented on the opposite face of TM4 relative to the cation binding site (Fig. 8, SI Appendix, Fig. S8). This Tyr/Phe residue in the Dnf1 structural model juts into the apex of a triangular cleft formed by TM segments 1, 3, 4, and LL3-4 that lies at the protein/lipid interface (Fig. 8F, SI Appendix, Fig. S8C).
We propose that the cleft bordered by TM3, LL3-4, and Y618 in TM4 provides a pathway for the flip of a phospholipid headgroup from the extracellular to the cytosolic leaflet. Alternatively, the groove between TM3, 4, and 5 could also provide a route for substrate, but unfortunately none of the chimeras we have created bearing TM5-10 from Drs2 were functional. Thus, the influence of TM5-10 on substrate selection is not yet known and cannot be ruled out as potentially contributing to phospholipid transport. By providing a substrate pathway at the protein/lipid interface, the acyl chains can simply reorient within the lipid environment and do not have to be accommodated in a proteinaceous binding pocket. This mechanism could explain why phospholipids with acyl tags such as spin labels (41) or the NBD moiety (42) are transported by P4-ATPases. If these tagged phospholipids were transported using a tight binding pocket similar to the mechanism of cation transporting P-type ATPases, the bulky tag would likely prevent transport. A transport mechanism that allows the acyl chains to remain in the lipid environment significantly alleviates the giant substrate problem.
The position of the substrate-selective Tyr/Phe residue near the cytosolic side of TM4 in the membrane domain was also surprising. We assumed that residues conferring substrate specificity would define a binding site near the extracellular leaflet where substrate would load into the pump. The role of LL3-4 in PC selection, particularly Phe587, fit this expectation. The proximity of Tyr618 to the cytosolic leaflet suggests this residue forms part of the exit pathway for the phospholipid headgroup. Dnf1 can weakly recognize PS and likely retains other residues involved in loading PS into the pump, but perhaps Y618 prevents PS exit into the cytosolic leaflet. Conversely, Drs2 F511Y was specifically deficient in recognizing PS. Therefore, the Y618 hydroxyl group may either generate a steric clash with the PS carboxyl group, thereby preventing PS transport, or the hydroxyl alters the packing of TM segments in this region in a manner that specifically restricts PS transport. Interestingly, substitution of the proline plus 4 Tyr/Phe for Leu, the Atp8a1 residue, reduces the activity of Dnf1 or Drs2 without a noticeable change of substrate specificity. Perhaps the Leu is incompatible with the surrounding yeast residues that form this restriction point. However, the phospholipid is likely selected at multiple points during its transport across the membrane because (i) we identified at least two residues (on opposing membrane faces) which specifically affect substrate specificity and (ii) substitution of TM3, LL3-4, and TM4 with Drs2 sequences was required to confer the greatest preference for PS.
Assuming that P4-ATPase TM segments undergo conformational changes during the catalytic cycle comparable to the Ca2+ ATPase, the remarkable pumping motions of TM segments 1–4 could provide the physical motion needed to flip a phospholipid substrate (43). Dnf1 is modeled in the E2 conformation and phospholipid substrate is thought to be pumped during the E2 ∼ P → E1 transition. During this power stroke, TM1 starts in the 90° bent conformation with residues in the TM1 kink modeled in close proximity to Tyr618 in TM4. As the pump relaxes back to the E1 conformation, TM1 straightens and slides down in the membrane (towards the exofacial leaflet) while TM4 moves upward. These movements place Tyr618 in proximity to the cytosolic end of the TM1 helix in the E1 conformation, such that a phospholipid headgroup docked near Tyr618 would then have access to a large opening between TM1 and TM2, perhaps representing an exit site.
Cdc50 family members have been suggested to provide the substrate specificity of their P4-ATPase counterpart by loading it with phospholipid (19). Even though we have altered the phospholipid specificity of Dnf1 by introduction of Drs2 sequences, the chimeras retain a Lem3 requirement. In no case was Cdc50 necessary or sufficient to facilitate the ER export, localization, and activity of the chimeras (Fig. 7). Our data suggest that, like Arabidopsis thaliana P4-ATPases (22), S. cerevisiae P4-ATPases do not rely on a Cdc50 family member to determine the substrate specificity of the heterodimer complex (Fig. 3B and 7).
These studies provide the first glimpse into the mechanism of substrate recognition and translocation by phospholipid flippases, a distinct clade in the P-type ATPase phylogeny. The residues reported here are not likely to encompass all residues involved in phospholipid selection and many more studies will be required to fully solve the giant substrate problem. It remains to be determined whether the TM4 “proline plus 4” position is universally used by all members of this family for substrate recognition in a manner similar to the “proline plus 1” position of the cation transporters. In addition, tests for the validity of structural model will be required to support or refute the proposed substrate pathway along the TM1,3-4 protein/lipid interfacial region we propose herein. Regardless of the precise orientation of Tyr618 in Dnf1, models of substrate translocation by the P4-ATPases will need to accommodate the observation that the presence or absence of a hydroxyl group at this position substantially impacts substrate selectivity.
Materials and Methods
Reagents.
All lipids were purchased from Avanti Polar Lipids, Inc.. 5-fluoroorotic acid (5-FOA) was purchased from Zymo Research. Edelfosine was purchased from Tocris Biosciences. Papuamide B was a gift from Raymond Andersen (University of British Columbia). Duramycin was purchased from Sigma Aldrich.
Strains and Culture.
Strains used in this study are listed in SI Appendix, Table S1. Yeast were grown in standard rich medium or synthetic minimal glucose medium (SD) (44) and transformations were performed using the lithium acetate method (45). For complementation tests, 50,000 cells were spotted with 10-fold serial dilutions onto synthetic media or synthetic media containing 5-FOA. Plates were grown for 3 d before imaging.
Escherichia coli strain DH5α was used for plasmid amplification and construction. Dnf1[Drs2] chimeras were generated by either site-directed mutagenesis (for point mutations) or gene splicing overlap extension (46) (for > 4 amino acid changes). Dnf1[Drs2] chimeras were generated by gap repair through homologous recombination of a gapped yeast expression vector and PCR product, followed by recovery of the plasmid from yeast. All plasmids used in this study are listed in SI Appendix, Table S2 and primers are listed in SI Appendix, Table S3.
Edelfosine Screen.
In order to generate random mutations targeted to TM3-4 of Dnf1, we used pRS313-DNF1 as template with primers Dnf1(forw)@1500 and Dnf1(rev)@2297 using PCR conditions described previously (47). We cotransformed ZHY704 with the PCR products and pRS313-DNF1 gapped with restriction enzymes MfeI and NruI to allow for gap repair through homologous recombination. Transformations were plated onto synthetic dropout plates and allowed to grow for 3 d, after which colonies were picked into 96-well plates and spotted onto synthetic dropout plates containing 20 μg/mL edelfosine or 5-FOA. We sequenced DNF1 in strains which grew on both edelfosine and 5-FOA.
Viability Assay.
For papuamide B and duramycin treatments, midlog phase yeast were seeded in 96-well plates at 0.1 OD600/mL with or without drug. Plates were incubated at 30 °C for 20 h and the OD600/mL was measured with a Multimode Plate Reader Synergy HT (Bio-Tek). Relative growth was compared to mock-treated yeast in the assay.
Fluorescence Microscopy.
Images were collected with an Axioplan microscope (Carl Zeiss) with a CCD camera and processed with MetaMorph 4.5 software (Molecular Devices). To observe GFP-tagged proteins, cells were grown to midlogarithmic phase, pelleted, and resuspended in imaging buffer [10 mM Tris-HCl, pH 7.4, 2% glucose (wt/vol)]. An aliquot of cells was distributed onto slides, and visualized using a GFP filter set.
NBD-Lipid Uptake.
Lipid uptake was performed essentially as described previously (48). Briefly, overnight cultures were diluted to 0.15 OD/mL and allowed to grow to early-midlog phase. 500 μL of cells were harvested, resuspended in ice-cold SD media containing 2 μg/mL NBD-lipid (approximately 2.5 μM), and incubated on ice for 60 min. Cells were washed twice with SA media [SD media + 2% sorbitol (wt/vol), +20 mM NaN3]+ 4% BSA (wt/vol), once with SA media, and resuspended in SA media prior to analysis by flow cytometry.
Flow Cytometry.
Flow cytometry was performed with a BD LSRII-3 laser (BD Biosciences) using BD FACSDiva v6.1.3. NBD-lipid uptake was measured with the FITC filter set (530/30 bandpass with a 525 longpass). Prior to analysis, propidium iodide was added to the cells at a final concentration of 5 μM. At least ten thousand events were analyzed using forward/side scatter to identify single cells and propidium iodide fluorescence was used to exclude dead cells.
Data Analysis.
Within each experiment, at least three independently isolated transformants harboring the same construct were assayed. The mean of these values from at least three separate experiments were reported as ± SEM. For each experiment, the chimera-independent background uptake for each NBD-PL was measured by the average fluorescence of the dnf1,2Δ strain (PFY3275F) transformed with an empty vector and subtracted from the uptake of WT Dnf1 or the Dnf1[Drs2] chimeras. In order to compare results from independent experiments, the NBD-PC uptake by WT Dnf1 after 1 h was normalized to 100%. Each value is reported relative to WT Dnf1 PC uptake at 1 h. A student’s T-test was used to determine significance where indicated.
Modeling.
A model of Dnf1 was generated based on the crystal structure of the Na+/K+ ATPase (Protein Data Bank accession code 2ZXE). We used the SWISS-MODEL workspace (34, 35) to generate the initial model based on sequence alignment of Dnf1 and the Na+/K+ ATPase (SI Appendix, Fig. S9). The initial structural model was relaxed in Rosetta-Membrane (36, 37) to obtain the final structural model. Images were generated with PyMOL (The PyMol Molecular Graphics System, Version 1.3r1, Schrodinger, LLC) and the pdb file is available from the Vanderbilt University Center for Structural Biology at http://csb.vanderbilt.edu/research/grahamlab/baldridgePNAS2012_model.html.
Supplementary Material
Acknowledgments.
We thank Jonathan Sheehan for assistance with refining the structural model and valuable discussion. We acknowledge Patrick Williamson at Amherst College for originally articulating the “giant substrate problem.” These studies were supported by National Institutes of Health (NIH) Grant GM62367 to T.R.G. and the Cellular and Molecular Microbiology Training Program T32AI007611-11, which supported R.D.B.. The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 1822.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115725109/-/DCSupplemental.
References
- 1.Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-Type ATPase superfamily. J Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- 2.Toyoshima C. Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch Biochem Biophys. 2008;476:3–11. doi: 10.1016/j.abb.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 3.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 4.Morth JP, et al. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat Rev Mol Cell Biol. 2011;12:60–70. doi: 10.1038/nrm3031. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci USA. 2009;106:16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman JA, Kwok MCM, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem. 2009;284:32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua Z, Fatheddin P, Graham TR. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell. 2002;13:3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulusma CC, Oude Elferink RP. The type 4 subfamily of P-type ATPases, putative aminophospholipid translocases with a role in human disease. Biochim Biophys Acta. 2005;1741:11–24. doi: 10.1016/j.bbadis.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Xu P, et al. Identification of a novel mouse P4-ATPase family member highly expressed during spermatogenesis. J Cell Sci. 2009;122:2866–2876. doi: 10.1242/jcs.047423. [DOI] [PubMed] [Google Scholar]
- 10.Dhar MS, et al. Mice heterozygous for Atp10c, a putative amphipath, represent a novel model of obesity and type 2 diabetes. J Nutr. 2004;134:799–805. doi: 10.1093/jn/134.4.799. [DOI] [PubMed] [Google Scholar]
- 11.Bull LN, et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 12.Siggs OM, Schnabl B, Webb B, Beutler B. X-linked cholestasis in mouse due to mutations of the P4-ATPase ATP11C. Proc Natl Acad Sci USA. 2011;108:7890–7895. doi: 10.1073/pnas.1104631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stapelbroek JM, et al. ATP8B1 is essential for maintaining normal hearing. Proc Natl Acad Sci USA. 2009;106:9709–9714. doi: 10.1073/pnas.0807919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Beserra C, Garbers DL. A novel aminophospholipid transporter exclusively expressed in spermatozoa is required for membrane lipid asymmetry and normal fertilization. Dev Biol. 2004;267:203–215. doi: 10.1016/j.ydbio.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Siggs OM, et al. The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat Immunol. 2011;12:434–440. doi: 10.1038/ni.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yabas M, et al. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat Immunol. 2011;12:441–449. doi: 10.1038/ni.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muthusamy BP, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta. 2009;1791:612–619. doi: 10.1016/j.bbalip.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito K, et al. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puts CF, Holthuis JC. Mechanism and significance of P4 ATPase-catalyzed lipid transport: lessons from a Na+/K+-pump. Biochim Biophys Acta. 2009;1791:603–611. doi: 10.1016/j.bbalip.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Ding J, et al. Identification and functional expression of four isoforms of ATPase II the putative aminophospholipid translocase Effect of isoform variation on the ATPase activity and phospholipid specificity. J Biol Chem. 2000;275:23378–23386. doi: 10.1074/jbc.M910319199. [DOI] [PubMed] [Google Scholar]
- 21.Lenoir G, Williamson P, Puts CF, Holthuis JCM. Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J Biol Chem. 2009;284:17956–17967. doi: 10.1074/jbc.M109.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López-Marqués RL, et al. Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA alpha-subunit. Mol Biol Cell. 2010;21:791–801. doi: 10.1091/mbc.E09-08-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomorski T, et al. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato U, et al. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J Biol Chem. 2002;277:37855–37862. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- 25.Natarajan P, Wang J, Hua Z, Graham TR. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc Natl Acad Sci USA. 2004;101:10614–10619. doi: 10.1073/pnas.0404146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alder-Baerens N, Lisman Q, Luong L, Pomorski T, Holthuis JCM. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol Biol Cell. 2006;17:1632–1642. doi: 10.1091/mbc.E05-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens HC, Malone L, Nichols JW. The putative aminophospholipid translocases, DNF1 and DNF2, are not required for 7-nitrobenz-2-oxa-1,3-diazol-4-yl-phosphatidylserine flip across the plasma membrane of Saccharomyces cerevisiae. J Biol Chem. 2008;283:35060–35069. doi: 10.1074/jbc.M802379200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodish HF. Transport of secretory and membrane glycoproteins from the rough endoplasmic reticulum to the Golgi. A rate-limiting step in protein maturation and secretion. J Biol Chem. 1988;263:2107–2110. [PubMed] [Google Scholar]
- 29.Chen S, et al. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- 30.Parsons AB, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 31.Navarro J, et al. Interaction of duramycin with artificial and natural membranes. Biochemistry (Mosc) 1985;24:4645–4650. doi: 10.1021/bi00338a025. [DOI] [PubMed] [Google Scholar]
- 32.Hanson PK, Malone L, Birchmore JL, Nichols JW. Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J Biol Chem. 2003;278:36041–36050. doi: 10.1074/jbc.M305263200. [DOI] [PubMed] [Google Scholar]
- 33.Liu K, Hua Z, Nepute JA, Graham TR. Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol Biol Cell. 2007;18:487–500. doi: 10.1091/mbc.E06-07-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 35.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 38.Obara K, et al. Structural role of countertransport revealed in Ca(2+) pump crystal structure in the absence of Ca(2+) Proc Natl Acad Sci USA. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- 40.Gourdon P, et al. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475:59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 41.Zachowski A, Henry JP, Devaux PF. Control of transmembrane lipid asymmetry in chromaffin granules by an ATP-dependent protein. Nature. 1989;340:75–76. doi: 10.1038/340075a0. [DOI] [PubMed] [Google Scholar]
- 42.Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- 43.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 44.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 45.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 46.Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. Genome Res. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 48.Hanson PK, Nichols JW. Energy-dependent flip of fluorescence-labeled phospholipids is regulated by nutrient starvation and transcription factors, PDR1 and PDR3. J Biol Chem. 2001;276:9861–9867. doi: 10.1074/jbc.M009065200. [DOI] [PubMed] [Google Scholar]