Abstract
MVB12-associated β-prism (MABP) domains are predicted to occur in a diverse set of membrane-associated bacterial and eukaryotic proteins, but their existence, structure, and biochemical properties have not been characterized experimentally. Here, we find that the MABP domains of the MVB12A and B subunits of ESCRT-I are functional modules that bind in vitro to liposomes containing acidic lipids depending on negative charge density. The MABP domain is capable of autonomously localizing to subcellular puncta and to the plasma membrane. The 1.3-Å atomic resolution crystal structure of the MVB12B MABP domain reveals a β-prism fold, a hydrophobic membrane-anchoring loop, and an electropositive phosphoinositide-binding patch. The basic patch is open, which explains how it senses negative charge density but lacks stereoselectivity. These observations show how ESCRT-I could act as a coincidence detector for acidic phospholipids and protein ligands, enabling it to function both in protein transport at endosomes and in cytokinesis and viral budding at the plasma membrane.
Keywords: HIV, protein structure, protein-membrane interactions, X-ray crystallography, subcellular localization
The concept of modular membrane-targeting domains is an important theme in the architecture of regulatory proteins and central to our current understanding of the subcellular localization of proteins (1–3). The best known of these modules include the PH, PX, FYVE, C1, C2, and ENTH domains (1–3). Each of these domains was initially proposed on the basis of bioinformatics analysis and subsequently confirmed by structural, biochemical, and cell biological approaches.
A recent bioinformatics analysis (4) led to the identification of a putative membrane-targeted domain, termed the “MVB12-associated β-prism” (MABP) domain (4). MABP domains are found at the N termini of MVB12A and B, which are subunits of the human ESCRT-I complex (5). ESCRT complexes are responsible for the endosomal sorting of ubiquitinated membrane proteins into multivesicular bodies (MVBs) en route to their lysosomal degradation (6, 7). ESCRT-I in particular is also involved in plasma membrane functions in cytokinesis and the release of HIV-1 and other viruses (8). MABP domains are also found in many membrane-associated bacterial proteins and in the N-terminal region of human DENND4 isoforms. DENND4 proteins are Rab10 guanine nucleotide exchange factors that localize to a tubular Golgi-proximal membrane compartment (9).
We set out to test the inference that MABP domains interact with membranes and found that the MVB12A and B MABP domains did. The crystal structure, solved at atomic resolution, showed that the MABP domain has a β-prism fold, consistent with the bioinformatics analysis. The structure identified a potential membrane-binding site, which we confirmed experimentally in vitro and in vivo. We found that the domain bound to liposomes according to the negative charge density of the membrane but with little specificity for particular lipid head groups. We went on to characterize the subcellular localization of fluorescent MVB12 MABP domain fusions and found that under appropriate conditions they localize to both punctate structures and to the plasma membrane, consistent with the unique properties of ESCRT-I.
Results
The MABP Domain Is Necessary and Sufficient for ESCRT-I Binding to Liposomes.
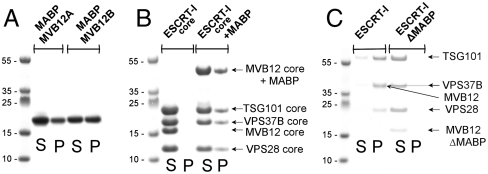
In order to test whether the MABP domain could bind to membranes, recombinant MABP domains of MVB12A and MVB12B were purified to homogeneity and liposome binding was tested. Both MABP domains bound in a sedimentation assay to liposomes prepared from Folch fraction I (Fig. 1A). Folch fraction I consists of a 1∶5∶4 ratio of phosphoinositols, phosphatidylserine, and cerebrosides isolated by organic extraction of brain. At protein concentrations of 10 μM, approximately 35% of the MVB12B-MABP bound and a slightly lesser fraction of MVB12A-MABP. To probe the possible involvement of other domains of ESCRT-I in liposome binding, a minimal ESCRT-I core was designed on the basis of homology with the crystallized yeast ESCRT-I core (10). The heterotetrameric core construct was produced as a soluble recombinant protein from Escherichia coli. The core region remained completely in the soluble fraction in sedimentation assays with Folch liposomes (Fig. 1B). A construct in which full-length MVB12 including the MABP domain was incorporated into the otherwise minimal core regained binding (Fig. 1B). We next asked whether fully assembled human ESCRT-I, consisting of the subunits TSG101:VPS28:VPS37B:MVB12A, bound to liposomes. At a concentration of 1 μM, the majority of ESCRT-I sedimented with Folch liposomes (Fig. 1C). Thus, some additional binding energy on the order of approximately 2 kcal/mol might be provided in the fully assembled complex, presumably through additional interactions with the liposomes. Alternatively, oligomerization of ESCRT-I on the membrane is possible and would also serve to increase affinity. In a variant in which MVB12A was replaced by a version in which the MABP domain was deleted (MVB12A-ΔMABP), only a trace level of pelleting was observed. To the extent that regions of ESCRT-I other than the MABP domain (Fig. S1) might interact weakly with liposomes, these interactions are insufficient for membrane association in the absence of the MABP domain. This observation demonstrates that the MABP domain is necessary for liposome binding by ESCRT-I. Taken together, these data show that the MABP domain is both necessary and sufficient for ESCRT-I to bind liposomes in vitro.
Fig. 1.
The MABP domain is necessary and sufficient for liposome binding by ESCRT-I. (A) The MABP domain binds to Folch fraction liposomes. (B) The MABP domain is sufficient to confer lipid binding on ESCRT-I. (C) The MABP is necessary for lipid binding by ESCRT-I. “S” indicates supernatant, and “P” indicates pellet in vesicle sedimentation assays.
The MABP Binds to Anionic Lipids Nonspecifically.
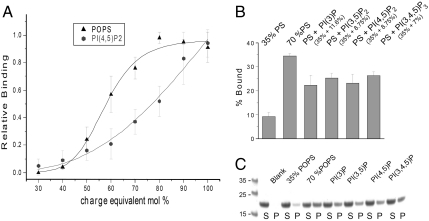
In order to determine the lipid specificity of the MABP domain, liposomes made up of a defined composition of synthetic lipids were tested. Initial screens showed that the monoanionic lipid phosphatidylserine and the polyanionic plasma membrane lipid phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2] were among the lipids that bound robustly. These two lipids were selected for a detailed binding analysis. Phosphatidylserine (PS) binds half-maximally to the MABP domain at a concentration of 55 mol % (Fig. 2A). The binding has high apparent cooperativity with respect to PS, with a Hill coefficient of 8.2 ± 1.3. In order to control for equivalent electrostatic contributions, binding of PI(4,5)P2 was compared to that of PS on the basis of a charge of -4 at the pH 7.4 of the experiment (11). Thus 40 mol % PS is compared to 10 mol % PI(4,5)P2 (Fig. 2A). Half-maximal binding was observed at 20 mol % PI(4,5)P2, which corresponds to the same negative charge density as 80 mol % of an anionic lipid (Fig. 2A). Thus whereas both lipids bound, on a charge-equivalent basis, PS has a marginally higher affinity.
Fig. 2.
The MABP domain binds to highly acidic liposomes with little head group specificity. (A) Binding of MVB12B-MABP to liposomes composed of the indicated mole fraction of PS or to the charge-equivalent amount of PI(4,5)P2, assuming charges of -1 and -4, respectively, per molecule. (B and C) Binding of MVB12B-MABP to the indicated mixtures of PS and phosphoinositides. All phosphoinositide:PS mixtures are equivalent in net charge to 70% PS. In A and B, PC is used as the background lipid in all cases. “S” indicates supernatant, “P” indicates pellet, and “blank” indicates a control containing no liposomes.
In cells, lipids are presented to proteins as a mixture, and the mole fractions of the anionic lipids examined are lower than the saturating values seen in Fig. 2A. Mixtures of PS and phosphoinositides were evaluated for binding. The mole fraction of PS was set at 35%, which binds weakly on its own, and supplemented with phosphoinositides on a charge-equivalent basis to match a total PS mole fraction of 70%. In other words, 35% PS was replaced with 8.25 mol % PI(4,5)P2, one-fourth the amount on a molar basis. The uncharged lipid phosphatidylcholine (PC) was used as the background in all cases. Several phosphoinositides of known or potential relevance to ESCRT-I biology at endosomes [PI(3)P and PI(3,5)P2] or the plasma membrane [PI(4,5)P2 and PI(3,4,5)P3] were tested. Each phosphoinositide:PS combination bound to the MABP domain with very similar affinity, though in no case was the binding fully equal to that of the charge-equivalent quantity of PS. In conclusion, we have detected no indication that the MABP domain has significant specificity for any of the phosphoinositides of significance in the ESCRT pathway. If anything, the MABP domain shows a preference for PS, which is broadly distributed on the plasma membrane, trans-Golgi, and endosomal network (12). This preference is marginal, however, and, in conjunction with the open nature of the lipid-binding site, described below, lipid recognition by the MABP domain seems best characterized as charge-dependent but otherwise nonspecific.
The MABP Domain Targets Punctate Structures and the Plasma Membrane.
In order to determine the subcellular targeting properties of the MABP domain, a construct expressing the MVB12B MABP domain fused to mCherry was transfected into HeLa cells for confocal microscopy analysis in live cells. MVB12B-MABP was found in the nucleus, bulk cytosol, and, to a lesser degree, punctate structures within the cytosol (Fig. 3A). The punctate structures appear to correspond to late endosomes, because most of them are positive for the late endosomal marker Rab7, but few are labeled by the early endosomal marker EEA1 (Fig. S2). Given the ability of MVB12B-MABP to bind to multiple anionic lipids, including PIPs characteristic of both endosomes and the plasma membrane, we were initially surprised to notice substantial diffuse nuclear and cytosolic localization, as well as an apparent absence of plasma membrane localization. It is important to note in this connection that the in vitro binding of MVB12B-MABP to liposomes is of moderate affinity, with K1/2 estimated at 10 μM (for liposomes of 60% PS) on the basis of the data described above. Thus MVB12B-MABP has lower affinity than widely used PI(4,5)P2 markers such as the phospholipase C-δ PH domain. The FYVE domain of Hrs is also predominantly cytosolic (13), in spite of its estimated 2.5-μM binding constant for PI(3)P-containing liposomes (14). We adopted the strategy previously used to construct a cellular PI(3)P probe from a tandem Hrs-FYVE fusion, which was shown to localize efficiently to early endosomes (13). The tandem MABP construct (“2xMABP”) showed strong plasma membrane and punctate localization while having a much reduced level of diffuse localization (Fig. 3B).
Fig. 3.
Subcellular targeting by the MABP domain. (A) mCherry-MVB12B-MABP is predominantly cytosolic with some punctate localization (arrows). (B) The tandem construct mCherry-(MVB12B-MABP)2 localizes predominantly to the plasma membrane and punctate structures (arrows).
Crystal Structure of the MVB12 MABP Domain.
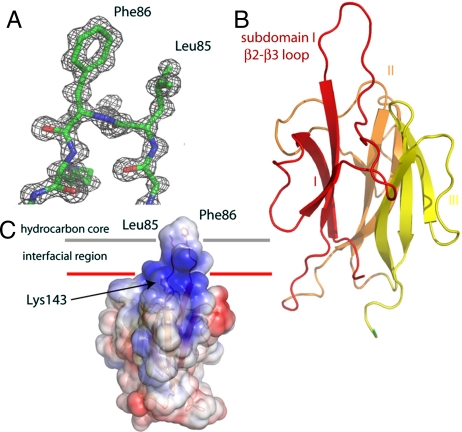
The crystal structure of the MVB12B-MABP domain was determined by multiwavelength anomalous dispersion (MAD) from selenomethionyl protein, and the structure was refined to 1.3-Å resolution (Fig. 4A and Table 1). The structure confirms the prediction of a β-prism fold (4). Consistent with the bioinformatics analysis, which identified the β-prism fragment of the Photorhabdus luminescens membrane attack complex/perforin (MACPF) protein as the only crystallized homolog, Protein Data Bank ID code 2QP2 (15) was the closest structural homolog found in a search of the Protein Data Bank (16). The β-prism domain of P. luminescens MACPF aligns with the MVB12B MABP domain with a Cα rmsd value of 2.6 Å over 130 residues, and a Z score of 13.6 (Fig. S3). The structure is organized into three β-sheet subdomains, which are arranged about a pseudothreefold axis with respect to one another. Each of the β-sheets is antiparallel and has the strands arranged in the sheet in the order 1–3–2.
Fig. 4.
Structure and membrane-binding mechanism of the MABP domain. (A) Electron density (2Fo - Fc) synthesis highlighting the tip of the β2–β3 loop of subdomain I. (B) Overall fold of the MABP domain colored by subdomain. (C) Model for membrane binding colored according to electrostatic potential, with blue electropositive and red electronegative. Hydrophobic residues at the tip of the β2–β3 loop are highlighted and their potential role in membrane insertion shown.
Table 1.
Statistics of crystallographic data collection and refinement
| Data collection | ||
| Construct | MVB12B47–151His6× | SeMet MVB12B47–151His6× |
| Space group | P212121 | P212121 |
| Cell dimension | a = 32.4 Å, b = 52.6 Å, c = 71.4 Å | a = 32.4 Å, b = 52.4 Å, c = 71.6 Å |
| X-ray source | SER-CAT 22 ID | SER-CAT 22 ID |
| Wavelength, Å | 1.0000 | edge = 0.9792 peak = 0.9798 remote = 0.9546 |
| Resolution, Å | 1.34 (1.39 − 1.34) | 2.08 (2.19 − 2.08) |
| No. of unique reflections | 27073 | 7675 7650 7 718 |
| I/σ(I) | 23.2 (2.0) | 19.3 (12.7) 15.7 (10.0) 19. 9 (12.9) |
| Rmerge | 7.9 (52.0) | 6.1 (9.4) 7.6 (10.1) 5.9 (10.1) |
| Data completeness, % | 96.3 (85.0) | 99.1 (94.2) 99.1 (97.5) 99.5 (97.4) |
| Multiplicity | 7.2 (3.2) | 5.8 (5.3) 5.8 (5.3) 5.8 (5.3) |
| Refinement | |
| Rwork, % | 16.5 |
| Rfree, % | 19.3 |
| rms bond angle deviation, ° | 1.5 |
| rms bond length deviation, Å | 0.014 |
| Ramachadran (outliers/favored) | 0%/99.3% |
| Number of non-hydrogen atoms | protein 1,223, water 225 |
Numbers in parentheses refer to the highest resolution shell of the respective dataset.
The structure contains two prominent features that were suggestive of roles in membrane binding. First, inserted between β-strands 2 and 3 of subdomain I is a loop (highlighted in Fig. 4B) which contains at its tip a β-turn. The residues at the II and III positions of the turn are Leu85 and Phe86, which by virtue of their placement on the isolated turn find themselves hyperexposed. In the crystal, the hyperexposed conformation is stabilized by lattice contacts (Fig. S4). In solution, it would be available for membrane binding. We speculated that these residues comprised either a docking site for another protein or, by analogy to other membrane-binding domains (3, 17), a hydrophobic membrane anchor. Such anchor regions occurs on FYVE, C1, PX, and ENTH domains and others and increase affinity for membrane-embedded lipids by allowing the protein itself to embed into the membrane (3, 17). Second, a conserved (Fig. 4C and Figs. S5 and S6) electropositive surface occurs close in three dimensions to where the above-mentioned loop exits β-sheet I. This patch is centered around Lys143 and Lys144. The juxtaposition of the electropositive patch and a potential membrane-anchoring loop suggested to us a potential mode of membrane docking (Fig. 4C).
Validation of the MABP Domain Membrane-Binding Site.
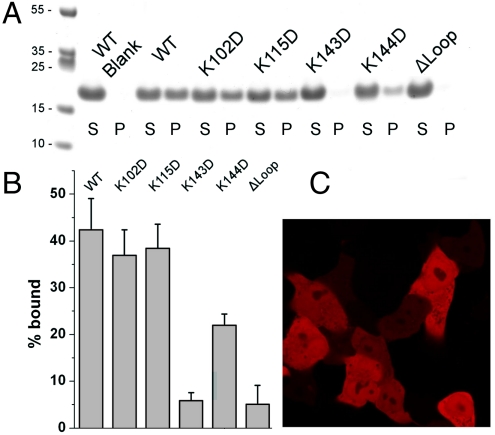
In order to test the putative role of the electropositive patch in membrane binding, conserved patch residues Lys143 and Lys144 were mutated to Asp (Fig. 5 A and B). Nonpatch residues Lys102 and Lys115 were also mutated, as controls for structurally nonspecific net charge effects. MVB12B-MABP bearing the mutations K102D and K115D bound to Folch liposomes to the same extent as the wild type (Fig. 5 A and B). The K143D mutation blocked binding completely, whereas K144D reduced binding to about half of the wild type (Fig. 5B). To probe the role of the putative membrane-anchoring loop, a construct MVB12B-MABP-Δloop was made in which the loop K82DGLFKSK89 was replaced by the linker Gly-Ser-Ser-Gly. MVB12B-MABP-Δloop completely lost the ability to bind liposomes (Fig. 5 A and B). From these data, we concluded that the membrane-binding model shown in Fig. 4C is a reasonable representation of the mode of binding to liposomes in vitro. To determine whether this mechanism extended to the situation within cells, the localization of an mCherry-MVB12B-MABP-Δloop construct was compared to wild-type mCherry-MABP. The mCherry-MVB12B-MABP-Δloop construct was found to be exclusively diffuse in localization, with no puncta observed (Fig. 5C). We conclude that MABP most likely binds to cellular membranes by using the K82DGLFKSK89 as a membrane anchor in the mode shown in Fig. 4C.
Fig. 5.
Mechanism of membrane binding by the MABP domain. (A and B) Binding of MVB12B-MABP mutants to Folch liposomes. (C) Subcellular localization of mCherry-MVB12B-MABP hydrophobic loop mutant is diffuse with no detectable punctate localization.
Discussion
Here we have established that the MABP domain is a bona fide membrane-targeting module. The MABP domain binds to acidic lipids more or less nonspecifically, though with a modest preference for phosphatidylserine. In these respects, it resembles another recently defined membrane-targeting module, the KA1 domain (18). The presence of this domain in MVB12 helps rationalize a confusing aspect of human ESCRT-I biology, namely, its ability to function at both endosomes and the plasma membrane, with their differing lipid compositions. The MABP domain of MVB12 can bind to mixtures of anionic lipids that are present both at late endosomes and at the plasma membrane, consistent with the cellular functions of ESCRT-I. The properties of other MABP domains, such as those of DENND4 isoforms whose membrane-binding determinants are partially conserved with MVB12, remain to be further explored.
The moderate affinity of the interaction is a critical feature of the MABP domain. Estimated to be in the tens of micromolar, the affinity is too low to target to membranes in the absence of a second factor. Membrane affinity is an order of magnitude higher in the context of the full-length ESCRT-I complex. Nevertheless, the specificity of subcellular localization cannot be accounted for by membrane binding alone. Thus, human ESCRT-I appears to fit the paradigm of a coincidence detector, which prevents ESCRT-I from binding inappropriately to acidic internal membranes other than late endosomes and the plasma membrane. It also prevents ESCRT-I from binding late endosomes and the plasma membrane until its activity is needed there. At the plasma membrane, CEP55 (19–21) and the P(S/T)AP motifs of HIV-1 Gag (22–25) and other viral proteins provide the additional increment in affinity to drive recruitment. In the case of endosomes, the P(S/T)AP of the Hrs subunit of ESCRT-0 and the proteins Tom1L1 and GGA3 could serve as additional factors providing specificity (26–31).
Although ESCRT-0 does not colocalize completely with ESCRT-I in cells, there is abundant evidence that ESCRT-0 has an important functional role in ESCRT-I recruitment. One way to resolve this apparent contradiction is to postulate that the negative charge density of the endosome surface might increase as the endosome matures from early to late. The phosphorylation of PI(3)P to PI(3,5)P2 is essential for maturation of late endosomes (32, 33). The addition of a charge on the 5-phosphate suggests one mechanism for an increase in negative charge density. It is unclear at this time if enough PI(3,5)P2 is produced to increase the charge density by the necessary amount. Endosomes are enriched in PS (12). PI(3)P, a lipid that is intimately associated with the ESCRTs, has in common with PS that both lipids are incorporated into the intralumenal vesicles (ILVs) of MVBs (12, 13). By taking the observation that PS is incorporated into ILVs in cells in combination with the in vitro preference of human ESCRT-I for PS, a functional connection between the ESCRTs and PS in ILV budding (34) seems plausible. The cooperativity of the PS binding suggests a mechanism to amplify the response to small changes in charge, as seen in the example of cooperative PI(4,5)P2 binding by the neuronal Wiskott–Aldrich Syndrome Protein (35). Thus, high levels of PS in endosomes could provide a baseline on top of which small increments in membrane surface charge density could trigger a switch-like response in ESCRT-I recruitment.
How does MVB12-MABP act as an almost pure charge-density-dependent membrane binder? The electropositive membrane-binding patch is open and contrasts to the well-defined pocket seen in highly specific PIP binding pockets of the PH (36) or FYVE domains (37). The MABP domain membrane-binding site seems more reminiscent of that of the KA1 domain (18), which also lacks a stereoselective binding pocket. Membrane targeting via nonspecific binding to acidic lipids is most commonly carried out by polylysine tracts and basic amphipathic helices (38). Indeed, the ESCRT pathway itself provides some examples, including basic putative helices at the N termini of Vps37 of yeast ESCRT-I (10) and Vps22 of yeast and human ESCRT-II (39). It is intriguing that sometimes specialized domains, such as MABP and KA1, use well-defined three-dimensional folds to carry out the same function.
In addition to MVB12A-B, there are at least three additional human genes coding for MVB12-like proteins that, based on sequence homology, appear to be capable of forming alternative ESCRT-I complexes in place of MVB12 (4). One of these, UBAP1, forms an alternative ESCRT-I complex that localizes to endosomes and functions in epidermal growth factor receptor down-regulation (40). UBAP1 lacks a MABP domain but instead has three potentially ubiquitin-binding UBA domains at its C terminus (see Fig. S1). This structural arrangement is predicted to place them close to the ubiquitin-binding UEV domain of TSG101 in three dimensions. The incorporation of UBAP1 could provide an alternative means to recruit ESCRT-I to endosomes or other sites where high concentrations of ubiquitinated proteins are found. Thus, ubiquitin or acidic lipids appear able to drive the recruitment of distinct ESCRT-I complexes to different sites in the cell, providing some insight into the diversity of ESCRT-I isocomplexes and cellular activities.
Materials and Methods
Plasmid Construction.
Plasmids were constructed by restriction cloning. All mutants were constructed by a Phusion® Site-Directed Mutagenesis Kit (Finnzymes) according to the manufacturer’s instructions. All plasmids were verified by sequencing. Plasmids designed for this study are listed in Table S1. VPS23/pCAG, VPS37B/pCAG, VPS28/pCAG, and MVB12A/pCAG were kindly provided by Wesley Sundquist (University of Utah, Salt Lake City, Utah).
Protein Expression and Purification.
Human MABP domain (MVB12A9–151 and MVB12B47–192) and all its mutants were expressed with a C terminal 6× histidine tag in E. coli at 30 °C O/N after induction with 0.5 mM isopropyl thiogalactoside (IPTG) at an optical density (OD 600 nm) of 0.8. The proteins were affinity purified by using Ni-NTA resin (QIAGEN) and further purified by size exclusion chromatography at Superdex 75 column (GE Healthcare) in 20 mM Tris, pH = 7.4, 50 mM NaCl, 3 mM DTT, 1 mM EDTA buffer. Human full-length ESCRT-I was purified as described in ref. 5. Briefly, the four subunits were coexpressed in a mammalian expression system with a Strep tag attached to the VPS23 subunit. ESCRT-I complex was affinity purified by using StrepTrap HP column (GE Healthcare) followed by size exclusion chromatography at Superdex 200 column (GE Healthcare) in 20 mM Tris, pH = 7.4, 150 mM NaCl, 3 mM DTT, 1 mM EDTA buffer. The ΔMABP form of ESCRT-I was prepared in the same manner except that full-length MVB12A was replaced by MVB12A152–273. To prepare the core ESCRT-I complex, all four subunits were coexpressed in E. coli. VPS23218–388, VPS37B6–164, and VPS2816–113 were expressed from the pST-39 vector, and MVB12B254–303 was expressed from pRSFD with a N-terminal 6× His and GB1 fused in frame. Expression was carried out in E. coli at 30 °C overnight after induction with 0.5 mM IPTG at an optical density (OD 600 nm) of 0.8. The minimal core with the MABP domain attached was made in the same manner but with MVB12B47–303 replacing the smaller MVB12B fragment. The complex was affinity purified by using Ni-NTA resin (QIAGEN) and further purified by size exclusion chromatography at Superdex 200 column (GE Healthcare) in 20 mM Tris, pH = 7.4, 150 mM NaCl, 3 mM DTT, 1 mM EDTA buffer. Proteins were flash frozen in liquid nitrogen and stored at -80 °C until use.
Crystallization and Crystallographic Analysis.
For crystallization the MABP domain was concentrated to approximately 10 mg/mL. Crystals grew in 2–3 d at 291 K in hanging drops consisting of a 1∶1 mixture of the protein and a well solution (100 mM Tris, pH 8.5, 33% PEG 400). SeMet crystals were grown in the same condition by microseeding using the native crystals as seeds. Both native and Se-MAD datasets were collected from a single frozen crystal by using a MAR CCD detector at beamline 22-ID, Advanced Photon Source. All data were processed and scaled by using the HKL2000 suite. All five expected SeMet sites were found by SHELX (41). A single copy of the MABP domain occurs per asymmetric unit. A polyalanine model obtained from anomalous data was used for molecular replacement against the native dataset. Model building and refinement was done by using Coot (42) and Phenix (43). The model was refined to Rwork = 16.5% and Rfree = 19.3%. Structural figures were generated by using PyMol (44).
Confocal Microscopy.
HeLa cells were grown in Dulbecco’s modification of Eagle’s medium (DMEM) + 10% fetal bovine serum (FBS) (Invitrogen). For live cell imaging, the cells were grown at an eight well chambered coverglass (Lab-Tek) to 70% confluency. The medium was changed to OptiMEM I reduced serum medium (Invitrogen) and cells were transfected by 200 ng of each plasmid DNA by Lipofectamine (Invitrogen) according to manufacturer’s instructions. Briefly, the cells were incubated for 4 h with the transfection mixture and washed with 1× PBS, and the medium was exchanged back to DMEM + 10% FBS. The cells were cultured for an additional 12–16 h and visualized by using a Zeiss LSM780 confocal microscope equipped with a heated stage at 37 °C. The 561-nm laser was used for mCherry illumination. The LSM780 array detector was set to collect in windows of ± 20 nm around the emission maximum (600–620 nm for mCherry). In each experiment, at least 200 cells were observed. Results are presented are based on at least three replicate experiments.
Liposome Pulldown Assays.
Lipids were dissolved in chloroform (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, and Folch fraction I) or in a mixture of methanol:chloroform:water at a ratio of 1∶2∶0.8 [for PI(3)P, PI(3,5)P, PI(4,5)P2, and PI(3,4,5)P3] and were pipetted in glass tubes at the desired ratios. One percent of fluorescent dye 1,1'-dioctadecyl-3,3,3',3'-tetramethlyindodicarbocyanine was added to allow better visualization of lipids. The solvents were evaporated under a stream of dry nitrogen, and the residual organic solvents were removed by overnight incubation in vacuum. Lipids were hydrated for 5 h in binding buffer (20 mM Tris, pH = 7.4, 150 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine, 1 mM EDTA) at 4 °C, and multilamellar vesicles were prepared by vigorous vortexing. All cosedimentation assays were done with a total lipid concentration of 1 mg/mL. The protein concentration was 10 μM for MABP constructs and all its mutants and also for the ESCRT-I core and ESCRT-I core with MABP domain attached complexes. For full-length ESCRT-I and its mutant ΔMABP contruct, the protein concentration used was 1 μM.
Supplementary Material
Acknowledgments.
We thank W. Sundquist, J. Bonifacino, and Y. Ye for reagents and B. Beach and L. Saidi for excellent technical assistance. Crystallographic data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. This work was supported by the Intramural Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (J.H.H.), the Intramural AIDS Targeted Anti-viral Program of the Office of the Director, National Institutes of Health (J.H.H.), and an Intramural AIDS Research Fellowship (to E. B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the RCSB Protein Data Bank, www.pdb.org (accession code 3TOW).
See Commentary on page 1816.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117597109/-/DCSupplemental.
References
- 1.Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr Opin Cell Biol. 2001;13:146–152. doi: 10.1016/s0955-0674(00)00191-5. [DOI] [PubMed] [Google Scholar]
- 2.Cho WH, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 4.de Souza RF, Aravind L. UMA and MABP domains throw light on receptor endocytosis and selection of endosomal cargoes. Bioinformatics. 2010;26:1477–1480. doi: 10.1093/bioinformatics/btq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morita E, et al. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 7.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald B, Martin-Serrano J. No strings attached: The ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshimura S, et al. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostelansky MS, et al. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kooijman EE, King KE, Gangoda M, Gericke A. Ionization properties of phosphatidylinositol polyphosphates in mixed model membranes. Biochemistry. 2009;48:9360–9371. doi: 10.1021/bi9008616. [DOI] [PubMed] [Google Scholar]
- 12.Fairn GD, et al. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High-affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry. 2001;40:8581–8587. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 15.Rosado CJ, et al. A common fold mediates vertebrate defense and bacterial attack. Science. 2007;317:1548–1551. doi: 10.1126/science.1144706. [DOI] [PubMed] [Google Scholar]
- 16.Holm L, Sander C. Dali—A network tool for protein-structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 17.Hurley JH. Membrane binding domains. Biochim Biophys Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moravcevic K, et al. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 20.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HH, et al. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 23.Martin-Serrano J, Zang T, Bieniasz PD. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 24.Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VerPlank L, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pornillos O, et al. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Q, et al. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puertollano R. Interactions of Tom1L1 with the multivesicular body sorting machinery. J Biol Chem. 2005;280:9258–9264. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- 29.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 30.Im YJ, et al. Crystallographic and functional analysis of the ESCRT-1/HIV-1 Gag PTAP interaction. Structure. 2010;18:1536–1547. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren X, Hurley JH. Proline-rich regions and motifs in trafficking: From ESCRT interaction to viral exploitation. Traffic. 2011;12:1282–1290. doi: 10.1111/j.1600-0854.2011.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 33.Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurley JH, Boura E, Carlson LA, Rozycki B. Membrane Budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papayannopoulos V, et al. A polybasic motif allows N-WASP to act as a sensor of PIP2 density. Mol Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 36.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high-affinity complex of inositol trisphosphate with a phospholipase-C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 37.Dumas JJ, et al. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin S, Wang JY, Gambhir A, Murray D. PIP2 and proteins: Interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 39.Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14:902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stefani F, et al. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol. 2011;21:1245–1250. doi: 10.1016/j.cub.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 41.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delano W. New York: Schrödinger; 2008. PyMOL Molecular Graphics System. ver 1.1, http://pymol.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.