Abstract
Next-generation sequencing is a valuable tool in our growing understanding of the genetic diversity of viral populations. Using this technology, we have investigated the RNA content of a purified nonenveloped single-stranded RNA virus, flock house virus (FHV). We have also investigated the RNA content of virus-like particles (VLPs) of FHV and the related Nudaurelia capensis omega virus. VLPs predominantly package ribosomal RNA and transcripts of their baculoviral expression vectors. In addition, we find that 5.3% of the packaged RNAs are transposable elements derived from the Sf21 genome. This observation may be important when considering the therapeutic use of VLPs. We find that authentic FHV virions also package a variety of host RNAs, accounting for 1% of the packaged nucleic acid. Significant quantities of host messenger RNAs, ribosomal RNA, noncoding RNAs, and transposable elements are readily detected. The packaging of these host RNAs elicits the possibility of horizontal gene transfer between eukaryotic hosts that share a viral pathogen. We conclude that the genetic content of nonenveloped RNA viruses is variable, not just by genome mutation, but also in the diversity of RNA transcripts that are packaged.
Keywords: deep-sequencing, virus evolution, virus assembly
Next-generation sequencing (NGS) is a powerful tool in the investigation of viral diversity from many different perspectives. NGS has been applied in directed approaches to investigate the prevalence of polymorphism or quasispecies present in a population of known viruses within a host, e.g., foot-and-mouth disease virus (1) or to follow the evolution of a viral genome and the frequency of known polymorphisms during the course of an infection, e.g., human rhinovirus (2) and HIV (3). The total genetic content of biological or clinical samples have been sequenced to uncover viruses present even at very low titers, for example, the presence of porcine circovirus-1 in Rotarix samples (4) and the identification of influenza virus subtypes and norovirus from nasopharyngeal and fecal samples collected during outbreaks (5).
NGS has also been applied in unbiased approaches aimed at the discovery of new viral species without any advance genetic information. For example, the Merkel cell polyomavirus was discovered to be the causative agent of its eponymous carcinoma by deep sequencing the transcriptome of tumor cells and analyzing nonhost transcripts (6). Similarly, it has been possible to uncover the history of viral pathogens that have plagued an organism through the de novo assembly of the sequenced siRNAs that would have been generated to tackle viral infections. This approach has revealed the viral genomes of familiar miscreants as well as of new viruses, for example, in fruit flies, mosquitoes, and nematode worms (7).
In this report, we employ NGS to investigate the total RNA encapsidated by authentic flock house virus (FHV) virions and by virus-like particles (VLPs) of FHV and of the related tetravirus, Nudaurelia capensis omega virus (NωV). Our analysis employs an unbiased approach, allowing us not just to detect the viral genome, but all packaged RNA including any host RNAs.
FHV and NωV have provided powerful systems for the study of nonenveloped viruses and constitute an important model for studying many pathogenic human viruses (reviewed in refs. 8 and 9). FHV is the prototypic member of the nodavirus family. It is a small, nonenveloped, icosahedral T = 3 insect virus containing a positive sense single-stranded RNA (ssRNA) genome. FHV provides an important model system for the study of nonenveloped viruses because of its small size and genetic tractability (10). Tetraviruses are a family of positive sense, ssRNA, nonenveloped T = 4 icosahedral viruses. Structural comparison of the nodavirus and tetravirus capsids reveals that these viruses share structural similarities despite a lack of significant homology between their capsid protein sequences and the difference in quasiequivalence. Both FHV and NωV have a bipartite genome (11, 12). In each, RNA 1 encodes the RNA-dependent RNA polymerase (RdRp). In FHV, RNA 1 also contains a frame-shifted subgenomic RNA 3 that encodes protein B2, responsible for inhibition of RNAi pathways. RNA 2 encodes the capsid precursor, α. The viral capsid of FHV consists of 180 copies of α, whereas the viral capsid of NωV has 240. Upon maturation, α undergoes an autocatalytic cleavage in its C-terminal region to form β, comprising the main structural capsid component, and γ, a short hydrophobic peptide required for endosome penetration that remains associated with the viral capsid (13–15). VLPs of FHV and NωV spontaneously form in Spodoptera frugiperda cell lines (e.g., Sf21) when RNA 2 is expressed from a baculovirus vector (16, 17). As no RdRp is present during the generation of these VLPs, there is no amplification of the viral genome. However, these VLPs have the same RNA complement (A260∶A280 ratio) as is found for authentic virions (16, 17) and therefore must package host RNAs.
Using NGS, we sequenced the RNA encapsidated by VLPs of FHV and NωV and find that they predominantly package ribosomal RNA and the transcripts from their baculovirus transfection vector. However, we also find that a number of transposable elements are packaged. Many VLPs, are structurally and biochemically indistinguishable from their parent virus (13, 16, 17). Consequently, it is likely that they can deliver their genomic cargo in the same way as an authentic virion. Such a consideration is not trivial as synthetic VLPs generated in a variety of different cell lines have therapeutic applications and transposable elements have long been known to have pathogenic roles via insertional mutagenesis (18). It is therefore important to understand what RNAs are packaged by VLPs in case they are inadvertently used to deliver foreign RNA to host cells.
We also characterized the RNA content of authentic FHV virions. We find that a variety of host RNAs are packaged, accounting for 1% of the total encapsidated RNA. Much of this RNA is messenger RNA, ribosomal RNA, and noncoding RNA. Like the VLPs, a large number of transposable elements are also packaged. The presence of these in authentic viruses elicits the possibility of horizontal transfer of genes between organisms that share a common viral pathogen and may help explain the distribution of closely related transposons among unrelated eukaryotic organisms. The packaging of transposon transcripts, reverse transcriptases in particular, may also impact the evolution of new retroviruses. We conclude that the genetic content of nonenveloped RNA viruses is variable, not just by mutation of the viral genome, but also in the diversity of RNA transcripts that are packaged.
Results
RNA Extraction and RNAseq.
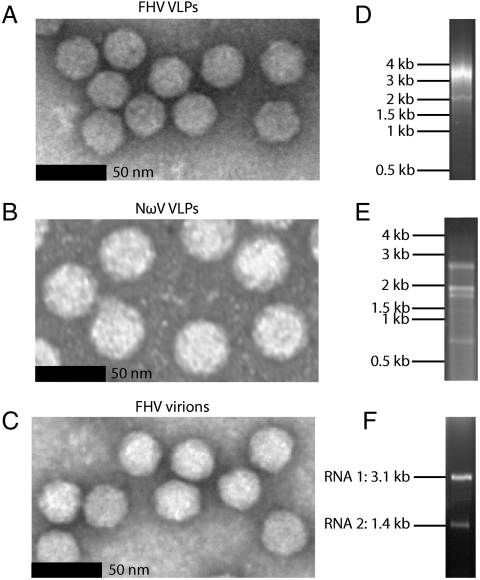
Authentic FHV virions and VLPs of FHV and NωV were generated in cell culture using established protocols and the particles were isolated over a series of sucrose gradients to exceptional purity, as evaluated by SDS-PAGE gel and native agarose gel electrophoresis (Fig. S1, Fig. S2). The samples were viewed by conventional negative-stain electron microscopy to ensure that the particles were intact and the encapsidated RNA was extracted and examined by agarose gel electrophoresis (Fig. 1). Whereas some dominant bands are evident, RNA purified from the FHV and NωV VLPs contain RNAs with a range of molecular weights (Fig. 1 D and E) reflecting the diversity of host RNAs that are packaged. The RNA extracted from authentic FHV virions consists of RNA 1 (3.1 kb) and RNA 2 (1.4 kb) (Fig. 1F). RNA was fragmented and analyzed on an Illumina platform for 40 base single-read sequencing. We thus obtained three datasets containing millions of ≤ 40 nucleotide reads corresponding to encapsidated RNAs, hereafter referred to as the FHVVLP-RNAseq, NωVVLP-RNAseq, and FHVvirion-RNAseq datasets. After processing and quality filtering, the datasets contained a total of 110, 30.7, and 40.7 M reads, respectively. The reads from each were aligned to reference sequences as appropriate for each cell line: Drosophila melanogaster in the case of FHV virions and Spodoptera frugiperda in the case of the VLPs. Such an approach allowed us to determine what genetic information is encapsidated by each sample and in what relative quantities.
Fig. 1.
Extraction of RNA from purified authentic FHV virions and VLPs of FHV and NωV. Particles of (A) FHV VLPs, (B) NωV VLPs, and (C) authentic FHV virions were visualized by negative-stain electron microscopy. Scale bars correspond to 50 nm. FHV VLPs form particles 30 nm in diameter that are indistinguishable from authentic FHV virions. NωV VLPs form 40 nm particles. RNA extracted from (D) FHV VLPs, (E) NωV VLPs, and (F) authentic FHV virions was visualized on a 0.8% nondenaturing agarose gel stained with ethidium bromide. FHV virions package two viral RNAs (RNA 1 = 3.1 kb, RNA 2 = 1.4 kb) as indicated. Molecular weights of VLP RNAs are indicated next to the gels.
FHVVLP-RNAseq.
We first analyzed the RNA content of FHV VLPs. In the first round of alignments, the FHVVLP-RNAseq reads were aligned to the FHV genome (RNA 1 and RNA 2), the Baculovirus genome (AcMNPV), and a collection of expression sequence tags (ESTs) generated from Sf21 and Sf9 cell lines collated in the SpodoBase (19). (There is not yet a publicly available entire genome sequence for the fall army worm, S. frugiperda). After this first round of alignments, over 18 M FHVVLP-RNAseq reads aligned to the ESTs, 3.45 M to the AcMNPV genome, 1.89 M to FHV RNA 2, and (unexpectedly) 3,850 to FHV RNA 1.
BLASTn searches indicated that the majority of the S. frugiperda ESTs corresponded to 45S ribosomal RNA, in particular, that of a related moth, Attacus ricini. To find further ribosomal sequences that may be represented among the FHVVLP-RNAseq dataset, the alignment was repeated including a 45S ribosomal DNA cassette of Attacus ricini as a reference sequence. We also included a 5S RNA gene from the genome of the silkworm, Bombyx mori. When the FHVVLP-RNAseq database was aligned to these, greater than 17 M reads aligned to ribosomal RNA.
After this second alignment, 8 M reads remained unaligned. These were assembled de novo into short contigs using Velvet (20) followed by contig alignment and finishing using SeqMan Pro (DNAStar Lasergene). Thus, a library of contigs ranging from 200 bp up to 5,730 bp was generated (Dataset S1). The identity of these contigs was established through BLASTn and BLASTx searches. Strikingly, many of the contigs bore strong homology to transposable elements found in other lepidopteran species. In addition to nucleotide sequence homology, many contained uninterrupted ORFs encoding proteins including reverse transcriptases, endonucleases, integrases, and other transposase proteins. In some striking cases (e.g., Contig_2836, Dataset S1) retrotransposons had divergent nucleotide sequences (BLASTn score < 30) whereas the protein sequences of predicted ORFs were highly conserved (BLASTx score > 30). The assignment of these as transposable elements was thus dependent upon building long enough contigs to reveal ORFs. The identity of some contigs could not be established by homology to known sequences.
A number of the contigs were generated that had very high sequence identity (< 90%) to members of a library of microsatellite sequence markers generated from wild fall army worms (21). Some of these contained fragments of transposable elements. Subsequently, the contig assembly library (Dataset S1) and the library of microsatellite sequence markers generated from wild fall army worms (21) were included as reference sequences in a final round of alignments to establish the coverage of these using the same parameters that were used for the other reference sequences.
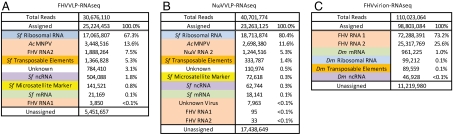
The final alignment revealed that the FHV VLPs package a large proportion of transposable elements -1.37 M reads in total equaling 5.3% of the total encapsidated RNAs. These included Class II DNA transposons and both LTR and non-LTR Class I retrotransposons (Table 1). The coverage of reads over these contigs was continuous and with overlapping single reads, confirming the validity of the contig assembly. The remaining unaligned reads could not be further assembled into meaningful contigs. A summary of the final alignments grouped into functional RNA classes is shown in Fig. 2A. (A detailed report of the alignments of reads to individual annotated transcripts is in Dataset S2.)
Table 1.
Transposable elements packaged by FHV and NωV VLPs
| Transposable element | Type | FHV VLPs (n = 1,366,828) | Directionality | NωV VLPs (n = 333,787) | Directionality |
| R1 Retrotransposon | non-LTR Class I | 998,378 | 0.99 | 9,053 | 0.99 |
| Mag Retrotransposon | LTR Class I | 188,041 | 0.95 | 1,173 | 0.97 |
| Taguchi | non-LTR Class I | 92,059 | 0.63 | 2,870 | 0.63 |
| Mariner | Class II | 29,658 | 0.96 | 318,591 | 1.0 |
| Kabuki | LTR Class I | 15,512 | 0.8 | 1,367 | 0.88 |
| RTE | non-LTR Class I | 7,905 | 0.68 | ND | — |
| Jockey | non-LTR Class I | 5,831 | 0.58 | ND | — |
| Pao | LTR Class I | 5,520 | 0.83 | 733 | 1.0 |
| Gypsy | LTR Class I | 4,134 | 0.84 | ND | — |
| Piggy-Bac | Class II | 3,772 | 1.0 | ND | — |
| Others unassigned* | — | 16,018 | — | — | — |
FHV and NωV VLPs package a range of transposable elements encompassing all three classes of transposon. The numbers of aligned reads from the FHVVLP-RNAseq and NωVVLP-RNAseq datasets are indicated for each transposable element. ND, not detected.
*See Dataset S2 and Dataset S3 for further details.
Fig. 2.
Summaries of the alignments of reads from each dataset, grouped into functional RNA classes. (A) FHVVLP-RNAseq, (B) NωVVLP-RNAseq, and (C) FHVvirion-RNAseq. Color-coded: peach, viral RNA; green, mRNA; blue, rRNA; gold, transposon RNA; purple, ncRNA; and yellow, microsatellite markers from ref. 21. In A and B, assembled contigs that did not have significant homology to other known sequences are indicated as unknown. Dm, Drosophila melanogaster; FHV, flock house virus; Sf, Spodoptera frugiperda; AcMNPV, Autographa californica multiple nuclear polyhedrosis virus.
NωVVLP-RNAseq.
Using the same methodology as for the FHV VLPs, we next analyzed the RNA content of VLPs of NωV. In the first round of alignments, the NωVVLP-RNAseq dataset was aligned to all of the references used in the alignment for the FHVVLP-RNAseq dataset. The unaligned reads were then assembled into contigs (Dataset S1) as before and a final round of alignments was performed including these. This analysis revealed that the NωV VLPs packaged a similar distribution of RNA transcripts as did the FHV VLPs (Fig. 2B). (A detailed report of the alignments of reads to individual annotated transcripts is in Dataset S3.) From a total of 23.3 M aligned reads, 18.7 M corresponded to ribosomal RNA, 2.70 M to the baculoviral expression vector, and 1.24 M to the NωV RNA 2. Importantly, NωV VLPs also packaged transposable elements demonstrating that their encapsidation is not a unique property of the FHV capsid protein.
The NωV VLPs package a smaller proportion of transposable elements (1.4%) than the FHV VLPs do (5.3%). Moreover, the distribution of packaged transposable elements differs (Table 1). The most highly packaged retrotransposon in FHV VLPs had strong homology to the R1 family of non-LTR Class I retrotransposons (22). These insert into the 28S gene of the ribosomal DNA cassette and (including R2-type insertions) can account for up to 77% of the ribosomal DNA cassettes in Drosophila cell lines (23), though many of these are transcriptionally inert as a result (24, 25). As the R1 insertions are cotranslated with the 45S ribosome transcripts, one might speculate that their abundance in FHV VLPs is linked to the packaging of ribosomal RNAs. However, the NωV VLPs do not package the R1 retrotransposons to nearly the same degree, despite packaging a similar proportion of ribosomal RNA. In contrast, the NωV VLPs primarily package the Mariner Class II DNA transposons. Consequently, there must be some specific selection mechanism favoring packaging of R1 retrotransposon in FHV VLPs and Mariner in NωV VLPs. We may conclude, therefore, that the distribution of transposable elements packaged by VLPs, or indeed by native viruses, is likely to vary on a case by case basis.
The small amount (< 0.1%, thus possibly copurified contaminant RNA) of FHV RNA1 found in both the FHVVLP-RNAseq and NωVVLP-RNAseq datasets was unexpected. We thus suspect that our Sf21 cell line has a low-level persistent infection with FHV, as has been demonstrated for other cell lines (26). We also built contigs from the NωVVLP-RNAseq reads that bore weak protein homology to other Rhabdoviridae and so may reflect a low-level infection with an unknown insect virus (Fig. 2B).
FHVvirion-RNAseq.
To see whether native ssRNA viruses also package host RNAs and in particular whether these included transposable elements, we next analyzed the RNA content of authentic FHV virions. The FHVvirion-RNAseq dataset was aligned to the FHV genome and the D. melanogaster genome (r5.22). A summary of the aligned reads grouped into functional RNA classes is summarized in Fig. 2C. (A detailed report of the alignments of reads to individual annotated transcripts is in Dataset S4.)
From a total of 110 M reads, 97.6 M aligned to the FHV genome: 72.3 M to RNA 1 and 25.3 M to RNA 2 giving a ratio of 2.86∶1, approximating the expected ratio of 2.22∶1. The deviation here may be due to uneven read coverage (Fig. S3). Strikingly, 1.20 M reads from the FHVvirion-RNAseq dataset aligned to the D. melanogaster genome (Fig. 2C). These reads account for approximately 1% of the original dataset. Therefore, authentic FHV virions encapsidate a considerable amount of nonviral genetic information. A total of 961,225 reads aligned to host mRNA, of which 89.4% aligned to exonic sequences, 6.8% to intronic regions, and 3.7% to intergenic regions. In addition, 99,212 reads mapped to rRNA, comprising the entire 45S cassette and 5S rRNA.
Strikingly, 89,559 reads mapped to transposable elements, which corresponds to 0.1% of the total encapsidated RNA. Most of these are Class I LTR retrotransposons (Table 2), which are the most abundant transposons in the D. melanogaster genome (27, 28). Interestingly, unlike the mRNAs, a number of the transposable elements are present in both positive and negative sense directions (e.g., Gypsy, Springer, and Diver). This bidirectionality suggests that multiple copies of these transposable elements are inserted in both sense and antisense directions of transcribed genes. Class II DNA transposons were also detected, but have very low sequence coverage (e.g., ′1360′ transposons with only 132 reads, Dataset S4).
Table 2.
Transposable elements packaged by FHV virions with > 1,000 reads
| Transposable element | Type | No. reads (n = 89,559) | Directionality | Copies in Dm Genome* |
| ′1731′ | LTR Class I | 56,740 | 0.99 | 2† |
| Gypsy | LTR Class I | 4,894 | 0.52 | 6 |
| Springer | LTR Class I | 4,540 | 0.71 | 11 |
| Copia | LTR Class I | 3,853 | 0.98 | 30 |
| 3S18 | LTR Class I | 3,388 | 0.88 | 6 |
| Max-element | LTR Class I | 1,871 | 0.97 | — |
| Diver | LTR Class I | 1,826 | 0.50 | 9 |
| 297 | LTR Class I | 1,341 | 0.81 | 57 |
| < 1,000 reads‡ | — | 11,106 | — | — |
FHV virions package a range of transposable elements. The number of aligned reads from the FHVvirion-RNAseq dataset is indicated for each transposable element.
*Taken from ref. 27.
†Transposon ′1731′ is known to be highly duplicated in cell lines (28).
‡See Dataset S4 for further details.
We detected 46,928 reads that corresponded to noncoding RNAs (ncRNAs). We also found many ncRNAs in the VLPs. Some of these elements have previously been demonstrated to be packaged by retroviruses, e.g., U6 snRNA in Rous–Sarcoma virus (29) and 7SL signal recognition particle RNA in HIV (30). The packaging of ncRNAs is interesting as some of these sequences (e.g., the Alu element widely found in the human genome that is derived from 7SL RNA) are known to be highly mobile elements that constitute members of the short interspersed element (SINE) class of nonautonomous retrotransposons that rely on reverse transcriptases encoded by other transposons for their genomic mobility. Thus their detection here could either indicate the packaging of ncRNA transcripts or of SINEs.
There is a striking difference between the distribution of host RNA packaged by authentic FHV virions and FHV VLPs. Authentic FHV virions predominantly package mRNAs (Fig. 2C). During an FHV infection, mitochondria are morphed into particle-creating factories that assemble the capsids and replicate the viral RNA genome in a semiclosed environment (31). Any host RNA that can reach this site of assembly must come from the cytoplasm. In this environment, ribosomal RNAs would not be available for encapsidation as they are in complex with ribosomal proteins and other rRNAs. In contrast, VLPs predominantly package ribosomal RNA (Fig. 2 A and B). Consequently, the VLP particles are likely assembled in the nucleus where the rRNA is more highly transcribed compared to mRNA and remains unbound to other rRNAs and ribosomal proteins.
RT-PCR Analysis.
Because of the low coverage of some of the transcripts present in the FHVvirion-RNAseq dataset, it was important to confirm these did not correspond to copurified contaminant RNA or DNA and that these were indeed packaged inside the virion capsids. To this end, FHV virions were treated overnight at room temperature with DNase I and RNase A to remove contaminating genomic DNA or RNA. Packaged transcripts would be protected from digestion by the FHV capsid. RNA was then extracted as before, and RNAs from each functional class were probed by RT-PCR to confirm the packaging of full-length transcripts within the capsid of the FHV (Fig. 3).
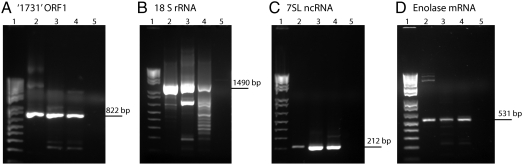
Fig. 3.
RT-PCR analysis confirms that full-length RNA transcripts are packaged inside authentic FHV virions. RNAs from each functional class were amplified and analyzed by gel electrophoresis: (A) ′1731′ retrotransposon ORF 1; (B) 18S rRNA; (C) 7SL ncRNA; and (D) enolase mRNA. Lane 1, molecular weight standard 1 Kb plus DNA ladder (Invitrogen); lane 2, amplified from DL-1 DNA (without reverse transcription); lane 3, amplified from DL-1 RNA; lane 4, amplified from RNA encapsidated by FHV virions; lane 5, same as lane 4 but without a reverse transcription step. The expected product lengths are indicated on the left of each gel.
We assayed for retrotransposon ′1731′ ORF 1, 18S rRNA, 7SL ncRNA, and Enolase mRNA. Transcripts were amplified from DNA extracted from DL-1 cells (without reverse transcription step) (lane 2), RNA extracted from DL-1 cells (lane 3), and RNA extracted from FHV virions (lane 4). In all cases, the transcripts were successfully amplified producing bands of the expected lengths. Following gel extraction, the identities of the bands were confirmed by sequencing. We also attempted to amplify the transcripts from FHV-extracted RNA without prior reverse transcription (lane 5). There were no detectable PCR products, demonstrating that bands obtained in lane 4 were not amplified from DNA. Only 529 reads mapped to the 7SL ncRNA and 1,492 to Enolase mRNA (sum of four variants) (Dataset S4). These values correspond to 17.9 and 6.86 reads per kilobase per million mapped sequence reads, respectively. Yet, we are still able to RT-PCR-amplify these transcripts from RNA extracted from the RNase A/DNase I-treated FHV virions. This analysis is important because it demonstrates that full-length RNA transcripts are present, rather than fragments thereof, and that they are encapsidated within authentic FHV virions.
Virus Genome Mutation Frequency.
From the reads in each dataset that aligned to the FHV genome, we could calculate the genomic mutation frequency (Table 3). The exact error inherent in the Illumina sequencing cannot be directly quantified. However, the mutation frequency of FHV RNA 2 from the FHVVLP-RNAseq was 7.4 nucleotides for every 10,000 aligned nucleotides (expressed, 7.4 × 10-4). In the FHVvirion-RNAseq dataset, the mutation frequency was 8.7 × 10-4 for FHV RNA 1 and 9.8 × 10-4 for FHV RNA 2. These values are within the range of the estimated error rate for the related poliovirus RdRp (32). In the FHVvirion-RNAseq dataset, mutation frequency across the ORFs of the FHV genome is imbalanced with over twice the frequency of mutation at the third nucleotide position of each codon than at the first and second positions [as was also noticed for wild foot-and-mouth disease virus samples (1)]. This imbalance is not observed for the FHVVLP-RNAseq alignment to FHV RNA 2, which is transcribed by Sf21 RNA polymerase II. This observation implies that, even during the limited passaging of FHV in cell culture, there is negative selection of mutational error over the first two positions of each codon, which is stronger than that for the third nucleotide position (which frequently encodes a wobble base). Interestingly, the imbalance of mismatch frequency over different codon positions is not observed in the region of FHV RNA 1 where the subgenomic RNA 3 is expressed. RNA 3 is expressed from a frame-shifted ORF. Thus, any evolutionary conservation of the first codon position from RNA 3 would result in conservation of the third codon position in RNA 1 (and vice versa) eliminating the observed imbalance. This observation may be useful in general when attempting to detect the expression of unknown subgenomic RNAs in other viruses whose mismatch frequency has been mapped in a similar way.
Table 3.
Mutation frequency of the FHV genome
| Sample | Average | First position | Second position | Third position |
| FHV virion RNA 1 | 8.7 × 10-4 | 8.5 × 10-4 | 5.8 × 10-4 | 11.9 × 10-4 |
| FHV virion RNA 1 + RNA 3 | 7.0 × 10-4 | 7.1 × 10-4 | 7.3 × 10-4 | 6.6 × 10-4 |
| FHV virion RNA 2 | 9.8 × 10-4 | 6.1 × 10-4 | 5.8 × 10-4 | 18.9 × 10-4 |
| FHV VLP RNA 2 | 7.4 × 10-4 | 8.4 × 10-4 | 7.9 × 10-4 | 8.4 × 10-4 |
Mutation frequency is expressed as the average number of mismatched nucleotides per 10,000 aligned nucleotides for both authentic FHV virions and FHV VLPs. The averages over the whole gene and at each codon position for the ORFs of both RNA 1 and RNA 2 are shown.
Discussion
In this study, we employed NGS to investigate the RNA content of authentic FHV virions and VLPs of FHV and NωV. We demonstrated that both the FHV VLPs and NωV VLPs package transposable elements. VLPs expressed in cell lines are presumed to be inert because of the lack of viral genome. However, if VLPs are capable of delivering their genomic cargo in the same way as would an authentic virion, then the demonstration that VLPs carry a variety of transposons suggests that VLPs may not be as innocuous as assumed. We thus advocate the use of NGS as a quality control tool for therapeutic VLPs. Using NGS technology, the presence of transposable elements in VLPs could be validated on a case by case basis. Additionally, the activity and cross-species tropism of an identified transposable element can be investigated. Comparing the total genetic content of a given VLP to the RNA and DNA complement of the cell that the VLP may transfect should be sufficient to detect the delivery of any transposable elements.
The extensive mutation found throughout viral genomes due to the error-rate of viral polymerases is essential for the rapid adaptation of viruses to new hosts and for the evasion of immune responses. Here, we have shown that approximately 1% of the RNA encapsidated by purified authentic FHV virions is host RNA. Therefore, the genetic content of nonenveloped RNA viruses is variable, not just by mutation of the viral genome, but also in the diversity of RNA transcripts that are packaged. This observation may have an impact upon the evolution of new viruses.
Of particular significance is the presence of components of endogenous viruses within the genetic cloud of FHV. If these components were to acquire the ability to interact with the viral capsid proteins and if such an interaction were to prove evolutionarily beneficial, then one might witness the evolution of a new virus. For example, our observation that transcripts that encode reverse transcriptase are packaged within FHV may essentially capture a stepping-stone in the evolution of a new retrovirus. Indeed, the reverse situation has already been observed, where a gypsy LTR retrotransposon has picked up a viral envelope protein from an insect baculovirus, thus enabling it to package retrotransposon transcripts into virus-like particles forming a rudimentary, yet infectious, retrovirus (33). Similarly, should a retrotransposon transcript acquire an RNA sequence recognized by a viral capsid as a packaging motif (as is present in a number of ssRNA nonenveloped viruses), this element may learn to parasitize a prevalent RNA virus. In this scenario, the retrotransposon will become a passenger to the viral infection which may provide it with the opportunity to jump between host cells and even cross species barriers.
The horizontal transfer of functional genes between eukaryotic organisms has been well-documented (for a review, see ref. 34). Of particular note is the apparent proliferation of transposable elements among organisms that have no vertical lineage, e.g., between Drosophila species (35), among higher plants (36), and among distantly related tetrapods (37, 38). These transposable elements show remarkable sequence identity despite host genome variation arising from divergent evolution. However, the mechanism of such transfer is unknown. In rare cases, transposons encode their own gag proteins that can assemble into virus-like particles (39) and so may autonomously mediate horizontal transfer. Alternatively, many parasitic and symbiotic organisms have been suggested to be suitable vectors for eukaryotic gene transfer, e.g., parasitic mites, fungi, trypanosomes, bacteria, as well as viruses. Indeed, the genomes of poxviruses (40) and granuloviruses (41, 42) carry transposons. Such viruses have very large genomes (> 130 kbp) and so are suitable candidates for the accommodation of additional viral sequences. In this study, we open up the range of potential vectors for horizontal gene transfer to include small nonenveloped RNA viruses, which are ubiquitous through all of life.
When considering that an individual FHV virion packages just two small genes, the packaging of an additional transposable element seems implausible. However, small RNA viruses such as FHV cannot achieve an infection when delivered to a host cell individually. In the case of FHV, 300 virus particles are required per cell for an infection. We observed that 1% of the RNA content of authentic FHV virions was host RNA. Consequently, at a multiplicity of infection of only 1, an average of three particles are each delivering up to 4.5 kb of host genetic material to another cell. Therefore, by considering the genetic makeup of a population of viruses rather than of just a single virion, there is plenty of scope for the addition of passenger sequences and their delivery to a new host cell.
Materials and Methods
See also SI Materials and Methods. All virions and VLPs were expressed and isolated to purity using a series of ultracentrifugation steps in sucrose using well-established procedures (10, 43). Particles were imaged by conventional negative-stain electron microscopy (10). Particles were disrupted with 0.1% SDS and 100 mM NaCl and RNA was phenol/chloroform extracted.
Directional RNAseq.
One microgram of RNA was prepared for NGS using a modified version of the Illumina protocol where 12 cycles of PCR were performed and standard TruSeq adapters and TruSeq barcoded primers were used. A final size selection was performed by native agarose gel electrophoresis to yield a library of inserts 50–150 bases in length suitable for 40 base single read sequencing. The library was extracted from the agarose gel using standard oligo purification columns. The prepared library was then loaded onto an Illumina HiSeq v2 single read flow cell, standard cluster generation was performed on a Cbot and sequenced for 40 bases of the insert and 7 bases of the index read using standard HiSeq sequencing reagents. Reads were processed using CASAVA 1.8 and demultiplexed based on index sequences.
Alignment of RNAseq Reads.
After assessment of read quality, alignment of the short reads to reference genomes was performed using the CLC Genomics Workbench 5.1. Alignments of reads to exonic, intronic, and intergenic regions were determined using SAMMate (44). Reads that did not align to the reference genomes were assembled into contigs using the Velvet package (20). Contig finishing was carried out using SeqMan Pro (DNAStar Lasergene).
RT-PCR.
One hundred micrograms FHV virions in 100 μL were treated overnight at room temperature with 4 U DNase I (NEB) and 500 ng RNase A (Roche) in NEB DNase I buffer (10 mM Tris pH 7.6, 2.5 mM MgCl2, 0.5 mM CaCl2) to remove any traces of nonencapsidated DNA or RNA. The particles were then extensively washed with 50 mM Hepes pH 7.0 using a standard 100 kDa molecular weight cutoff centrifugal concentrator to remove the enzymes. Extracted RNA was reversed transcribed using RT Superscript III (Invitrogen) and amplified using standard PCR techniques using the primers indicated in Table S1.
Supplementary Material
Acknowledgments.
We thank Mark Young and Anette Schneemann for advice and stimulating discussions. We thank Megan Guelker, Mandy Janssen, and Jeff Speir for providing laboratory reagents and technical support. We thank Lana Schaffer and Steven Head for support and advice with next-generation sequencing technology. A.R. is supported by a European Molecular Biology Organization Long-Term Fellowship, ALTF 573-2010. T.D. is supported by a Pew Fellowship and Conselho Nacional de Desenvolvimento e Pesquisa (CNPq-Brazil). This work was funded by National Institutes of Health Grant R37-GM034220 (to J.E.J).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116168109/-/DCSupplemental.
References
- 1.Wright CF, et al. Beyond the consensus: Dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J Virol. 2011;85:2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordey S, et al. Rhinovirus genome evolution during experimental human infection. PLoS One. 2010;5:e10588. doi: 10.1371/journal.pone.0010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: Application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Victoria JG, et al. Viral nucleic acids in live-attenuated vaccines: Detection of minority variants and an adventitious virus. J Virol. 2010;84:6033–6040. doi: 10.1128/JVI.02690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura S, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Q, et al. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci USA. 2010;107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai B. Penetration of nonenveloped viruses into the cytoplasm. Annu Rev Cell Dev Biol. 2007;23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- 9.Odegard A, Banerjee M, Johnson JE. Flock house virus: A model system for understanding nonenveloped virus entry and membrane penetration. Curr Top Microbiol Immunol. 2010;343:1–22. doi: 10.1007/82_2010_35. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee M, Johnson JE. Activation, exposure and penetration of virally encoded, membrane-active polypeptides during nonenveloped virus entry. Curr Protein Pept Sci. 2008;9:16–27. doi: 10.2174/138920308783565732. [DOI] [PubMed] [Google Scholar]
- 11.Krishna NK, Schneemann A. Formation of an RNA heterodimer upon heating of nodavirus particles. J Virol. 1999;73:1699–1703. doi: 10.1128/jvi.73.2.1699-1703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanzlik TN, Gordon KH. The Tetraviridae. Adv Virus Res. 1997;48:101–168. [PubMed] [Google Scholar]
- 13.Canady MA, Tihova M, Hanzlik TN, Johnson JE, Yeager M. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis omega virus (NωV) J Mol Biol. 2000;299:573–584. doi: 10.1006/jmbi.2000.3723. [DOI] [PubMed] [Google Scholar]
- 14.Fisher AJ, Johnson JE. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 15.Schneemann A, Zhong W, Gallagher TM, Rueckert RR. Maturation cleavage required for infectivity of a nodavirus. J Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneemann A, Dasgupta R, Johnson JE, Rueckert RR. Use of recombinant baculoviruses in synthesis of morphologically distinct virus-like particles of flock house virus, a nodavirus. J Virol. 1993;67:2756–2763. doi: 10.1128/jvi.67.5.2756-2763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agrawal DK, Johnson JE. Assembly of the T = 4 Nudaurelia capensis omega virus capsid protein, posttranslational cleavage, and specific encapsidation of its mRNA in a baculovirus expression system. Virology. 1995;207:89–97. doi: 10.1006/viro.1995.1054. [DOI] [PubMed] [Google Scholar]
- 18.Kazazian HH., Jr Mobile elements and disease. Curr Opin Genet Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 19.Negre V, et al. SPODOBASE: An EST database for the lepidopteran crop pest Spodoptera. BMC Bioinf. 2006;7:322. doi: 10.1186/1471-2105-7-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arias RS, Blanco CA, Portilla M, Snodgrass GL, Scheffler BE. First microsatellites from Spodoptera frugiperda (Lepidoptera: Noctuidae) and their potential use for population genetics. Ann Entomol Soc Am. 2011;104:576–587. [Google Scholar]
- 22.Xiong Y, Eickbush TH. The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol Cell Biol. 1988;8:114–123. doi: 10.1128/mcb.8.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakubczak JL, Zenni MK, Woodruff RC, Eickbush TH. Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics. 1992;131:129–142. doi: 10.1093/genetics/131.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamrich M, Miller OL., Jr The rare transcripts of interrupted rRNA genes in Drosophila melanogaster are processed or degraded during synthesis. EMBO J. 1984;3:1541–1545. doi: 10.1002/j.1460-2075.1984.tb02008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye J, Eickbush TH. Chromatin structure and transcription of the R1- and R2-inserted rRNA genes of Drosophila melanogaster. Mol Cell Biol. 2006;26:8781–8790. doi: 10.1128/MCB.01409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li TC, Scotti PD, Miyamura T, Takeda N. Latent infection of a new alphanodavirus in an insect cell line. J Virol. 2007;81:10890–10896. doi: 10.1128/JVI.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminker JS, et al. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0084. 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Franco C, Pisano C, Fourcade-Peronnet F, Echalier G, Junakovic N. Evidence for de novo rearrangements of Drosophila transposable elements induced by the passage to the cell culture. Genetica. 1992;87:65–73. doi: 10.1007/BF00120994. [DOI] [PubMed] [Google Scholar]
- 29.Giles KE, Caputi M, Beemon KL. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA. 2004;10:299–307. doi: 10.1261/rna.2150604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onafuwa-Nuga AA, Telesnitsky A, King SR. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA. 2006;12:542–546. doi: 10.1261/rna.2306306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanman J, et al. Visualizing flock house virus infection in Drosophila cells with correlated fluorescence and electron microscopy. J Struct Biol. 2008;161:439–446. doi: 10.1016/j.jsb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freistadt MS, Vaccaro JA, Eberle KE. Biochemical characterization of the fidelity of poliovirus RNA-dependent RNA polymerase. Virol J. 2007;4:44. doi: 10.1186/1743-422X-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik HS, Henikoff S, Eickbush TH. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 34.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 35.Jordan IK, Matyunina LV, McDonald JF. Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc Natl Acad Sci USA. 1999;96:12621–12625. doi: 10.1073/pnas.96.22.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol. 2006;4:e5. doi: 10.1371/journal.pbio.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464:1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pace JK, 2nd, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecher P, Bucheton A, Pelisson A. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J Gen Virol. 1997;78:2379–2388. doi: 10.1099/0022-1317-78-9-2379. [DOI] [PubMed] [Google Scholar]
- 40.Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci USA. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friesen PD, Nissen MS. Gene organization and transcription of TED, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol. 1990;10:3067–3077. doi: 10.1128/mcb.10.6.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jehle JA, Nickel A, Vlak JM, Backhaus H. Horizontal escape of the novel Tc1-like lepidopteran transposon TCp3.2 into Cydia pomonella granulovirus. J Mol Evol. 1998;46:215–224. doi: 10.1007/pl00006296. [DOI] [PubMed] [Google Scholar]
- 43.Schneemann A, Marshall D. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J Virol. 1998;72:8738–8746. doi: 10.1128/jvi.72.11.8738-8746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu G, et al. SAMMate: A GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol Med. 2011;6:2. doi: 10.1186/1751-0473-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.