Abstract
This paper addresses how to calculate and interpret the time-delayed mutual information (TDMI) for a complex, diversely and sparsely measured, possibly non-stationary population of time-series of unknown composition and origin. The primary vehicle used for this analysis is a comparison between the time-delayed mutual information averaged over the population and the time-delayed mutual information of an aggregated population (here, aggregation implies the population is conjoined before any statistical estimates are implemented). Through the use of information theoretic tools, a sequence of practically implementable calculations are detailed that allow for the average and aggregate time-delayed mutual information to be interpreted. Moreover, these calculations can also be used to understand the degree of homo or heterogeneity present in the population. To demonstrate that the proposed methods can be used in nearly any situation, the methods are applied and demonstrated on the time series of glucose measurements from two different subpopulations of individuals from the Columbia University Medical Center electronic health record repository, revealing a picture of the composition of the population as well as physiological features.
In this paper, we show how to apply time-delayed mutual information (TDMI) to a sparse, irregularly measured, complicated population of time-dependent data. At a fundamental level, the technical problem is a probability density function (PDF) estimation problem; specifically, one can average PDF estimates or one can aggregate the data set before estimating the PDF. To understand and interpret these two means of coping with a population of time-series, one must address four issues: (1) estimator bias; (2) normalization or distribution support-based effects; (3) deviations from the single source case for average and aggregate; and (4) practical interpretation. Scientifically, this paper works to develop an infrastructure, and demonstrates how to use it, by studying the time-dependent correlation structure in physiological variables of humans—in a population of glucose time-series. In the end, we not only provide a practically actionable set of information theoretic computations that yield insight into the population composition and the time-dependent correlation structure but we also detail the time-dependent correlation structure and the degree of homogeneity within a broad population of humans via their glucose measurements.
INTRODUCTION
It is no surprise that aggregating collections of elements or data streams can allow for a productive analysis and understanding of the individual elements that make up the aggregated population. In fact, the aggregation of many elements into a measurable population can be pivotal in providing a means to study systems where the individual elements are difficult, expensive, or dangerous to measure. (Note that by aggregation, we mean combining sets of measurements in such a way that they can be treated as a single set of measurements that can be analyzed.) That aggregation provides a basis for analysis lies in the fact that the application of most statistical methods, such as statistical averages, probability density estimates, and techniques based on such fundamental methods (i.e., information theory, ergodic theory, etc.), require large numbers of data points. While some fields have gained much from the analysis of aggregated populations of elements—such as advances made in the physical sciences with the advent of statistical mechanics—many fields have not been so fortunate. A primary source of difficulty with aggregation in these less fortunate contexts lies in the fact that fortune or ruin often depends on the ability to aggregate measured elements such that statistical averages can be taken. Usually, this means that one must have a population of elements whose statistical properties being quantified are drawn from the same distributions. This requirement presents two inextricable problems, verifying that a population is homogeneous enough to produce representative statistics when aggregated, and determining whether a statistical analysis technique will yield the same outcome for the average over the population and for the aggregated population.
With these broad issues in mind, here, we focus on applying time-delayed mutual information to a population in an attempt to understand the time-dependent nonlinear correlation between measurements or the degree of predictability of measurements for members of a population. We wish to apply this, however, to a system whose members may: (1) have differing numbers of measurements; (2) have too few measurements for probability densities (or any other statistical quantities) to be estimated; (3) be non-stationary; (4) have very diverse underlying probability distributions or statistical states; and (5) may be measured in a highly irregular manner in time. In short, this paper details how to apply and interpret information theoretic analysis to a diversely measured, possibly statistically diverse population that needs to be aggregated for the information theoretic quantities to be calculable. Thus, this paper complements and contrasts with the research such as is presented in Ref. 1 where dynamical reconstruction of a uniformly measured stationary systems with short time-series are the focus. The particular population we focus on in this paper is a subpopulation of human beings who received care at the Columbia University Medical Center (CUMC). The particular time-series we are focusing on are clinical chemistry measurements (measurements such as glucose, that detail physiological functioning of humans) for this population. Nevertheless, it is important to note that the analysis presented is not limited to any particular population of measurements.
A reader’s guide: The outline of this paper
Broadly, this paper can be split into two main components. The first component is primarily theoretical and includes: a background section (Sec. 3); a section about TDMI-specific estimator bias (Sec. 4); a section focusing on how the TDMI for a population can deviate from the TDMI of an individual stationary source (Sec. 5); and finally, a section explaining how to use the TDMI population calculations to characterize diversity in a population (Sec. 6). Second, following the more theoretical sections are the computational sections including: a section explaining how to use the TDMI population calculations to characterize diversity in a population (Sec. 6); a section proposing some non-TDMI based metrics for evaluating population diversity that help verify the TDMI-based methods (Sec. 7); a section summarizing the TDMI methodology explicitly (Sec. 8); and finally the data-based Sec. 9 demonstrating the methodology. Regardless of intent, readers will need to read the introduction Secs. 1, 2, 3 and the discussion and summary.
MOTIVATING EXAMPLES
The theory-based motivation for this work is to devise a way to calculate and interpret the TDMI (Refs. 2, 3) in the context of a population of time-series that are both sampled irregularly and are from (possibly) statistically distinct sources. More concretely, the motivation for this work comes from the desire to understand human health dynamics (i.e., physiology, complex phenotype definitions such as diseases, basic biology, etc) based on the constrains of real data present in the electronic health record (EHR) repository at CUMC (note, CUMC is affiliated with NewYork-Presbyterian Hospital (NYP)).
The CUMC EHR contains all the information collected regarding the 3 million patients over 20 years at NYP and contains graphical images, laboratory data, drug data, doctor and nurse notes, billing data, and demographic data, most of which is highly dependent on time; moreover, the amount of data is growing exponentially. Despite the quantity of data, EHR data can be difficult to use; in particular, EHR data is characterized by: diverse irregular sampling, measurements correlated to statistical state, nonstationarity, statistically diverse population, very large populations with few measurements, and very diverse data types. Nevertheless, if these data prove to be useful for understanding human dynamics, a subject that is not completely without controversy,4, 5, 6 it may be possible: to define complex diseases and other phenotypes based on population scale data; to understand how disease and treatment of disease evolve in complex and interconnected ways;7, 8 correlate drugs to side effects and benefits; and to carry out many other practical applications that can be gained from understanding population-wide human health and biology. The approach taken in this paper using nonlinear physics methodology represents a radical departure from the standard utilization of biomedical data and has been termed by some9 as the physics of living things.
The advantage of motivating this paper with a data set with complex properties is that it allows for the generalization of the results to many other circumstances. Therefore, while we apply our analysis in the context of human health and physiology, our methods can be easily generalized to nearly all time-dependent contexts; e.g., astronomy,10 geology,11 climatology,12 and genetics.13
INFORMATION THEORY BACKGROUND
Begin with time-series, of real numbers. Next, denote all of the pairs of points in X separated by a either index time, τ = i − j (where i > j are the indices of ti and tj respectively), or real time, δt = ti − tj (again assume ti > tj), by X[τ] or X[δt], respectively. Note that τ is always an integer while δt can take continuous real values. For this section, we will limit the discussion to X[τ], but note that the X[δt] case follows identically. Note that in this circumstance, X[τ] can be used to approximate a joint (two-dimensional) PDF; further, note that the marginal distributions of X[τ] are approximated by X[τ](1) = X(i) and X[τ](2) = X(i − τ), respectively.
To estimate either the information entropy, or the TDMI for this time-series,2, 3 one must first estimate various PDFs.14 In order to specify a PDF, one needs to both specify the support of the PDF, S, and the PDF itself, p(X). Moreover, intuitively, the support of the PDF is the interval over which the xi’s lie, or, the support of the PDF of X is S = [min(X), max(X)]. However, when estimating a PDF from data, the support will always be collected in a series of bins; thus, there also exists an abstract support, S, which consists of the explicit bins of the data used to estimate the PDF disconnected from the values the bins are assigned externally. Thus, S does not explicitly represent numbers in X; while this may seem like a strange point to make, the difference between S and S will be critical later in this paper. Finally, note, we will always assume that PDFs in this paper have compact support.15
Now, given the random variable X and its associated PDF, p(X), the information entropy of a time series generated by X is defined by
| (1) |
Similarly, the TDMI is defined by
| (2) |
Thus, the TDMI can be thought of as an auto-information measure that depends on a delay (e.g., τ or δt).
Given this infrastructure, fundamentally, there are two ways of conjoining a population: (1) averaging the TDMI for each member of the population and (2) aggregating the population before the PDFs are estimated without intermixing the members of the population. As we will see, in the context of a heterogeneous population, these two approaches will yield both differing numerical results and differing interpretations.
Computationally, it is important to note that we will employ both a kernel density estimator (KDE) estimator16, 17, 18 and a standard histogram estimator for all PDF calculations. We explicitly use the estimator developed in Ref. 16 with a Gaussian kernel and a bandwidth of 100; the histogram estimator is of our own design and has a bandwidth of 20. The results detailed in this paper are relatively insensitive to these parameter settings (e.g., a 10% change in the bandwidth will not produce a qualitatively different result). Moreover, in this paper, we will estimate the bias using the fixed point bias estimation technique,19 which amounts to various random permutations of temporal ordering of the time-series used to generate the PDFs and will be introduced in more detail in Sec. 4B. Finally, while this paper only addresses the continuous case, the discrete case follows more or less identically with integrals replaced by sums.
Average TDMI
To formulate the average TDMI for a population, we begin by arguing that the average mutual information of a vector of individuals (a population) is the same as the average of the mutual informations of each individual, if the individuals are independent. These cases represent conjoining a population after the PDFs have been estimated; in essence, we are just arguing that taking an average before or after the TDMI integration is performed does not affect the resultant TDMI.
Assume all processes are stationary. Define a vector-valued process X, where ; this leads to a the following definition of multivariate mutual information
| (3) |
noting that p(·) is the probability density associated with the given random variable and X(·) and dX(·) are both vectors. We want the following statement to be true:
| (4) |
We claim that the sufficient condition for 4 to hold is for the Xi processes to be non-interacting or statistically independent. It is important to note that it is not necessary that the Xi’s be non-interacting copies of the same process—the processes only have to be statistically independent. It is not too difficult to verify our claim algebraically, one merely applies the chain rule for mutual information to Eq. 4; moreover, conceptually understanding why our claim is correct is rather straightforward. Begin by noting that if the Xi’s are independent, they form an orthogonal set of probability densities or a product measure on N-dimensional Euclidean space. Thus, the integral of each variable will be independent of the others simply because the variables are orthogonal and thus not functions of one another (cf. Fubini’s theorem20).
The conclusion is that, the average TDMI for the population is simply the canonically calculated TDMI for the individuals of the population, averaged.
Aggregate TDMI
To understand the construction where the population is aggregated before the PDFs are estimated, assume, as we did in Sec. 3A, a stationary, vector-valued process X, where X(t) = [X1(t), X2(t),…XN(t)], where N denotes the number of individuals in the population. Next, assume that each element emits a time-series of length ni; without loss of generality, in this section, assume that ni = n.
Aggregating the population into a time-series for which the PDFs can be estimated can be done in one of two ways. The first method involves concatenating the entire set of time-series into one scalar time-series of length Nn and then treating this concatenated time-series like a time-series from a single source; denote this aggregation method as inter-source aggregation. We will not study this as this calculation needlessly adds noise via the intermixing of elements and is hard to rectify with mathematics. The second method, denoted the intra-source aggregation because sources are not intermixed within pairs of points, involves explicitly collecting pairs of points restricted to individuals. Specifically, the pairs of points are chosen such that the individual pairs of points always originate from the same individual, and then these sets of pairs of points are conjoined such that the PDFs can be estimated. Thus, this method mixes individuals by including pairs of points from many individuals, but does not mix individuals by pairing points from differing individuals.
To concretely specify what intra-source aggregation means, begin with the time series
| (5) |
where, given an xij, i specifies the individual, j specifies the time, and a time-delay of τ for which the TDMI is to be calculated. The intra-source pairs that will be aggregated and used for estimating the PDF are then
| (6) |
Thus, denote the left column by and the right column by . Moreover, denote the TDMI calculated between these two columns as .
Much of the rest of this paper is dedicated to quantifying the implications and interpretations for when, and conditions under which the average and aggregate TDMIs differ. However, by comparing average to aggregate TDMI, we will also see that, very often (but not always), the aggregate TDMI will form an upper bound on the TDMI of an individual.
TDMI-SPECIFIC ESTIMATOR BIASES
All statistical estimates have bias associated with them. Here, we focus on three sources of bias that are particular to the estimation of the TDMI for a population: (1) sample-size-dependent estimator bias effects for the average versus the aggregate TDMI; (2) the basic methodology we use for numerically estimating the bias for the TDMI calculation; and (3) a source of non-estimator bias that is particular to the TDMI aggregation case—a sort of filtering bias.
Sample size dependent estimator bias effects
A practical reason why the order of aggregation matters for estimating probability densities lies in the fact that most probability density estimation techniques have estimator bias that is, to first order, proportional to one over the number of points to a power of at least one. Thus, because we are interested in coping with populations of poorly measured individuals and because we are comparing two methods of conjoining those individuals, it is important to understand how the number of data points will broadly affect estimator bias in the average and aggregate TDMI calculations.
Begin with a more computationally minded definition of the TDMI for a single time-series from a single source with n points
| (7) |
where is the estimated TDMI for the n pairs of points of X and BE(n) is the total estimator bias of the calculation with n pairs of points. Note that while explicit bias calculations for the entropy and TDMI calculations can be found in Refs. 19, 21, and 22, it will suffice to notice that for most PDF estimators (i.e., for kernel density estimators or histogram style estimators), the bias estimates will follow:
| (8) |
Nevertheless, it is worth noting that there is also a estimator-specific, bandwidth-specific factor on BE(n) that is dependent on the proportion of support (e.g., number of bins) for which there exist no data points, and this factor can be important when n is small (cf. (Ref. 22) where this effect is carefully quantified for the histogram estimator). To see how the bias of averaging TDMI over the population versus the bias of the TDMI for the pre-PDF-estimation aggregated populations differ, partition the time-series of length n into m pieces, where is a positive integer (thus, m divides n evenly and n ≥ m). Now, consider the difference between I[Xi(t), Xi(t − j)] calculated on a single time-series of length n, and I[Xi(t), Xi(t − j)] calculated on m disjoint time-series of length and then averaged. More specifically, consider
| (9) |
versus
| (10) |
Now, if the bias, BE, scaled linearly in the number of points, n, then, the bias contribution of Eq. 9 will be the same as the bias contribution of Eq. 10. However, we know the bias obeys a power-law in the number of points, n, so we get the difference between bias estimates to at least be
| (11) |
| (12) |
where δBE > 0 for all m > 1. Or, said differently,
| (13) |
where equality is satisfied only when m is one, or when the population consists of a single element. Note that when the population is particularly poorly sampled, say one or two measurements per element of the population, then m ≈ n and thus the difference in the bias of the population average versus the aggregated population will be will be order one. More importantly, averaging the mutual information (MI) of many poorly sampled individuals will not help the MI converge to its bias-free, high cardinality estimate.
Aside from the overall effect of n, there are other small sample size effects and these effects can have profoundly different outcomes depending on the estimator. For instance, in the presence of few points, a KDE estimator will often, in the name of smoothing, over-estimate the probability for empty portions of the support, resulting in a PDF estimate that is closer to a uniform random variable. Thus, a KDE-PDF based TDMI calculation will likely underestimate the TDMI. In contrast, a histogram estimator will underestimate the probability for empty portions of the support, thus yielding a more sharply peaked distribution that will yield an over-estimate of the TDMI. Because of these opposing effects, it is possible to verify the existence of finite-size effects by simply observing the difference between the KDE and histogram estimated TDMI estimates for the same data set.
In the end, because we are working to understand how to estimate the TDMI in the context of large, poorly measured populations, there will be a significant advantage to aggregating populations before estimating the PDFs necessary to carry out the TDMI calculations from the perspective of estimator bias minimization.
Fixed point bias estimate for average and aggregate populations
The fixed point TDMI bias estimation method19 attempts to estimate the τ = ∞ TDMI by randomly permuting the time-ordering of one of the sets of pairs used to estimate the distributions for a given δt or τ. Fundamentally, there are two different methods for estimating the TDMI fixed point (if it exists), random permutation within the individuals (i.e., not mixing individuals), and random permutation over the entire population, thus intermixing individuals.
The first method, individual-wise random permutation (IRP), involves randomly permuting the temporal ordering of one column (without replacement) of the data set used to estimate the distributions without intermixing individuals, or
| (14) |
where is the ith random permutation (without replacement) of the left index of the column vector (i.e., do not permute the first index of xi,j from Eq. 6). The IRP-method random permutation occurs only within an individual and not across the population, thus destroying information about only time-based correlations while preserving inter-individual information. Finally, there will exist a IRP bias estimate for both the average and aggregate TDMI cases, where Eq. 14 is specified for a single individual and then averaged over the population, and which is specified exactly as per Eq. 14.
The second method, population-wide random permutation (PRP), which exists only in the aggregated population context, involves randomly permuting, without regard to the individual, one column of the entire populations’ data set used to estimate the PDFs or,
| (15) |
where is the ith, random permutation (without replacement) of the both indices of column vector . Because the PRP estimate intermixes both the population and time, the PRP destroys information about both intra-individual time correlations and inter-individual information (i.e., information about differences in normalization or the supports). In the context of a single source, . Similarly, when the population is both relatively uniform over both the PDFs and the support of the PDFs, then the PRP bias estimate will be equivalent to the bias estimate of the IRP, and thus can be thought of as an estimate of the estimator bias. However, if the support of the PDFs over the population is not uniform (i.e., if the support of any of the individuals of the population differs from the support of the population), then the PRP bias estimate will differ from the IRP bias estimate (we will discuss this explicitly in Sec. 6A). Note that , , and are dependent on both τ or δt (because of the filtering effect discussed in Sec. 4C) and n, the number of points used in the estimate. In general, we will drop the n from the notation, and when there is not a τ or δt dependence, we will not include it in the notation (in general, for the data sets and δt’s, we consider in this paper, there is not a strong δt dependence).
Non-estimator bias: How the TDMI calculation can act as a population filter
While it is clear that the TDMI calculation only applies to the data used to estimate the PDFs, it is less obvious that the act of constructing the data sets used to estimate the PDFs can filter out substantial portions of the overall population. Specifically, because construction of the data sets for the PDF estimation involves collecting all pairs of points separated by some time τ or δt, if some individuals do not have pairs of points separated by τ or δt, those individuals will be filtered out of, or excluded from, the data set used to estimate the PDFs and thus the TDMI. In this sense, the TDMI calculation implicitly filters the population by measurement frequency; this is not an externally imposed data constraint, it is simply a result of calculating the TDMI in the context of population whose elements do not have identical measuring frequencies.
To understand how this filtering bias can affect the results, consider a polarized example population made up of two differently measured subsets of individuals. Specifically, the first subset of the population has individuals sampled once an hour for a month and the second subset of the population has individuals sampled once a month for 20 years. These two populations represent patients with acute and chronic conditions, respectively. If the TDMI of the population is calculated for any δt less than a month, only data set one will be represented. Similarly, if the TDMI is calculated for δt of a month or greater, only data set two will be represented. When plotting the TDMI graph versus δt, the graph has, in a sense, a bias. Namely, two the graph represents two disjoint populations for δt > one month.
Of course, for real EHR data, even more complicated problems can appear when the same individual is sampled at different rates depending on the statistical state of the individual (e.g., a patient with a chronic and acute condition). This problem is particularly acute for health care data because health correlates with presence of measurement—healthy patients are not measured often while sick patients are—thus leading to the possibility of having different subpopulations or statistical states being filtered out when calculating the TDMI for some δt values.
Thus, when estimating a TDMI for a population, it is important to quantify both who is populating the data set explicitly used to estimate the PDFs and how the proportionality of the subpopulations changes in the set used to estimate the PDFs as the delay is changed. If the population and proportionality of subpopulations in all the δt or τ TDMI estimates does not change, then the bias estimates are independent of the delay.
Methods for assessing δt bin compositions
To quantify the composition of the data set, begin with the following notation: (1) bi(τ) represents the number of pairs of points in the τ time bin contributed by individual i; (2) bmax(τ) = Nmax and bmin(τ) = Nmin correspond to the maximum and minimum number of pairs of points, over all individuals, present in the data set; N* represents the sum of bi(τ) or the total number of pairs of points in the data set; (3) N represents the total number of individuals in the population; and (4), ς(τ) represents the set of indices of individuals monotonically ordered by increasing bi. Based on these quantities, define the following functions:
| (16) |
| (17) |
noting that (Ref. 27) is Θ(τ) normalized to lie on the unit square. Next, define the following integral that quantifies the population composition of the data set:
| (18) |
When the time series of the members of the population are both uniformly sampled and of the same length, will be equal to one; thus, the closer is to one, the more composition of the data set includes the entire population uniformly, while the closer is to zero, the more composition of the data set represents a small subset of the population (possibly only an individual). A second, more gross quantification of how the population is represented in TDMI data set at a fixed δt is the percentage of individuals that contribute at least one pair to the data set, or
| (19) |
Note that an alternative, highly related quantity we have found useful is the cumulative distribution function (CDF) of the bi’s.
Finally, while it is tempting to think of the population makeup of the τ data set as a measure of homogeneity within a population, this interpretation is sometimes, but not always, correct. What , , or any other like-minded metric really detail is how a population is measured and thus represented in a given τ or δt bin. Specifically, when measurement frequency is correlated with statistical state or dynamics, then it is likely that τ bins will filter a population and make it more homogeneous. However, it is easy to think of examples where measurement frequency is random, or uncoupled from a statistical state or dynamics, and in this case, all the diversity of the population will be present in any given τ time bin.
POPULATION-BASED DEVIATIONS FROM THE INDIVIDUAL TDMI ESTIMATES
Heterogeneity-based deviations from the individual: Average TDMI case
To understand how representative the average MI over the population is of an individual in the population, begin by setting p1 as the PDF that most resembles the average (choosing p1 to be the median among the pi’s would work as well) among the set of pi’s relative to the abstract support, S; note that the average PDF is defined by
| (20) |
Note that in this situation, every pi has the same abstract support (by definition), which we will denote as . Further, note that it is possible to have a set of pi’s such that no pi resembles the mean graph of the pi’s. Next, relative to p1, we can now relate each pi to p1 as follows:
| (21) |
where is distance between the graphs of p1 and pi at a given value in . Recalling the definition of the TDMI, we get
| (22) |
Now, because integration is a linear operation, focus on the integrand instead, or more specifically, focus on
| (23) |
where, is given by
| (24) |
(for a more explicit calculation of , cf. Appendix A). As each goes to zero, goes to zero; thus, the more support independent variance (recall is relative to the abstract support ) there is within the population, the larger will be, and the less will represent the TDMI of an individual element within the population. Written explicitly, represents the “average” individual plus the sum of the deviations from that individual.
Entropy of the averaged population
While the primary topic in this paper is the TDMI, we will contend briefly with the TDMI for τ = 0 or the auto information. Based on an identical means of calculation, the information entropy of a time series for a population can be defined as follows:
| (25) |
Thus, when , the hI for the population relative to the abstract support tends toward the information contained in an individual.
Heterogeneity-based deviations from the individual: Aggregate TDMI case
To understand how the diversity in the population is rendered via the TDMI of the aggregated population begin by recalling that the TDMI for the aggregated set is defined by
| (26) |
where, under ideal (single, stationary source) circumstances the PDF of the aggregated density obeys
| (27) |
where and represent the PDF restricted to individual i. Intuitively, Eq. 27 just says that we are creating the aggregate PDF by summing the graphs of all the individuals relative to the union of the supports of all the individuals, that is, relative to .
To choose a PDF that most closely resembles a centroid, it is helpful to have a concept of abstract support; however, because is defined relative to the actual support of the population, the individual population PDFs do not separate as naturally as in the case. Nevertheless, conceptually, to define an abstract support in the aggregate circumstance, one needs to, in spirit, construct a situation where all the PDFs have roughly the same range or support. There are several ways one can imagine achieving such goal; here will define the abstract support, , such that every patient has been renormalized to have the identical support—the unit interval (i.e., [0,1]). It is important to realize that relative to the aggregate case, there can be a very severe difference between the TDMI of an aggregated population defined on support of the versus the abstract support . To allow for quantifying these potential differences, define the TDMI for an aggregated population relative to the abstract support, . Now, using the abstract support, select p1 in the same way we selected p1 in Sec. 5A, by selecting the PDF that most closely represents the mean over the population of PDFs relative to the abstract support. This definition implies an important difference in how pi is specified in the aggregate case versus the average case because, despite the fact that we use an abstract support to select a p1, is not calculated relative to the abstract support, and thus, the differences between p1 and pi are instead defined by
| (28) |
where is distance between the graphs of p1 and pi at a given value in total support, . Next, focusing on the integrand, , and substituting Eq. 28 into Eq. 27 and recalculating , we arrive at (dropping the subscript on p1)
| (29) |
where is explicitly given by
| (30) |
(the calculation of and follows a similar path to that of and as seen in Appendix A). Thus, as the average of the’s go to zero, will go to zero; moreover, when both the width of the band of PDFs decreases and when the supports of the distributions overlap (i.e., when ), the TDMI of the aggregate population will represent an individual within a homogeneous population (because the individuals within the population are similar). Similarly, when either the width of the band of PDFs increases or the supports of the distributions becomes disjoint, (i.e., when ), will represent the TDMI within the diverse population. Or, said differently, the TDMI for the aggregated population will represent the TDMI of the population plus the sum of the individual based differences from the population. As we will see in the sections that follow, this second circumstance can lead to subtle difficulties in interpretation. Finally, note that the calculation that yielded does not explicitly depend on the support; the explicit ’s will differ between and , but the explicit form of will not.
Entropy of the aggregated population
Again, while the TDMI is the primary topic of this paper, in both the interest of completeness and later analysis, we define hI for the aggregated population, which was calculated in analog with , as follows:
| (31) |
In contrast to the situation where the information entropy is averaged over the population, when the average , the information entropy for the aggregated population, , relative to the real support of the population tends toward the information contained in an individual who has the most data pairs in the PDF estimate.
HOW TO INTERPRET THE TDMI FOR A POPULATION, OR, TDMI-BASED METHODS FOR INTERPRETING POPULATION DIVERSITY
To achieve a practical understanding of the meaning of the TDMI in the context of a population, we have to combine information from Sec. 5B1 to construct an explicitly numerically computable means of interpreting and . Practically speaking, there are two broad situations: (1) is practically calculable (when is calculable, always will be); and (2), is not calculable (usually to estimate , there need to be at least 100 pairs of points per representative element) leaving us only with -related quantities. Relative to the first situation, define the difference between and , or
| (32) |
This allows for the following conjecture which we will not prove in this paper:
Conjecture 1:In the circumstance, where can be accurately estimated, δI(τ) ∼ 0 if and only if the population used to estimate and is statistically homogeneous temporally (i.e., the PDFs representing the individuals in the population are identical, as are the PDFs under temporal evolution).
The forward direction of the if and only if statement, that δI(τ) ≠ 0 implies a heterogeneous population will be briefly discussed in Sec. 6B; this direction is more complicated to prove. The reverse direction of the if statement in this conjecture claims that if the population represents a single, stationary, homogeneous distribution then δI(τ) ∼ 0; this claim relies on the fact that in this circumstance, all ε’s are zero and thus (Eq. 22 and (Eq. 26) represent a homogeneous source and are equivalent up to bias. Essentially, when one can estimate δI(τ), one can interpret the population make-up without delving deeply into the detailed sources of the TDMI. In contrast, when only is practically calculable, the interpretation of can only be understood by understanding the source of the TDMI. Nevertheless, in general, it is insightful to understand the sources of the TDMI, and the sources of the TDMI are tied to the make-up of the population.
From a detailed perspective, the make-up of the population is important because the deviation of the TDMI from the homogeneous case is due to non-zero ε’s, and the source of non-zero ’s can differ from the source of non-zero ’s. Specifically, can only be non-zero because of differences between the graphs of the pi’s. This is because all the pi’s for the average TDMI have the same support. In contrast, the source of non-zero ’s is due to a heterogeneous population can be split into three broad categories: (1) differences in the TDMI estimates due to differences in the supports independent of the graphs of the PDFs; (2) differences in the TDMI estimates due to differences in the graphs independent of the supports; and (3) differences in the TDMI estimates due to the supports’ effect on the graphs.
Support dependent, graph independent, effects on the population TDMI
To understand and quantify the differences in the TDMI estimates due to differences in the supports independent of the graphs of the PDFs, consider the difference between the random permutation bias estimates defined in Sec. 4B.
First, recall that the population-wide random permutation bias estimate will be roughly equivalent to the estimator bias, or BPRP(τ) ≈ BE(τ) regardless of the supports or densities of the elements (cf. (Ref. 19) for small sample size qualifications of this statement). Next, note that the individual-wise random permutation bias estimate, represents the bias due to heterogeneity in the supports plus the estimator bias. Thus, the contribution to the bias due to the diversity in population normalization is approximated by the difference between the individual-wise and population-wise random permutation bias estimates
| (33) |
There are two reasons why BRP(τ) can be non-zero. First, the number of points used to calculate the two can differ by orders of magnitude (say, a population of 1000 with 10 points each); in this case, BRP(τ) represents the 1/n effect on the bias estimates. In the case where the number of pairs used to estimate and are relatively similar (e.g., more than 100 and within an order of magnitude; to control for the number of points, it is easy reduce the cardinality of the set used to calculate ) Fig. 1b shows visually how these bias estimates would render differently. In this context, would be identical to I, where as randomly permuting the entire population, such as is done to estimate , will result in one of the marginal distributions becoming —a uniform distribution instead of three Gaussians with distinct means—thus greatly changing the amount of mutual information. These effects are primarily support-driven effects; note that while it is possible that differences in the underlying distribution function can be rendered through BRP(τ), differences in the support of those distributions will always be rendered through BRP(τ). As we will see in a moment, is not enough to imply that δI(τ) ≈ 0, but is enough to imply that the variance in the boundaries of the supports will all be relatively small. Nevertheless, while in some circumstances, it may be difficult to use the bias estimates to detect a difference in the average versus aggregate TDMI, we can use the bias estimates to interpret the average and aggregate TDMI signal. In particular, when BRP(τ) ≤ BE(τ), intermixing individuals’ measurements has no effect on the random permutation bias estimate, implying that there is very little population selection information in the TDMI estimate. Thus, BRP(τ) ≤ BE(τ) at least implies overlapping distribution supports. Similarly, when , intermixing elements has a profound effect on the random permutation bias estimates; in this instance, BRP(τ) reveals a bias whose source is the diversity of the supports among the elements. This leads us to the measure of homogeneity of supports that is very computable even for poorly measured populations (e.g., when only is calculable); the TDMI homogeneity of support is defined by the following equation:
| (34) |
The closer is to one, the less the diversity of the supports over the population; similarly, the closer is to zero, the greater the diversity of the supports over the population. (Again, note one must control for the dependence on the number of pairs used to estimate the above quantities.)
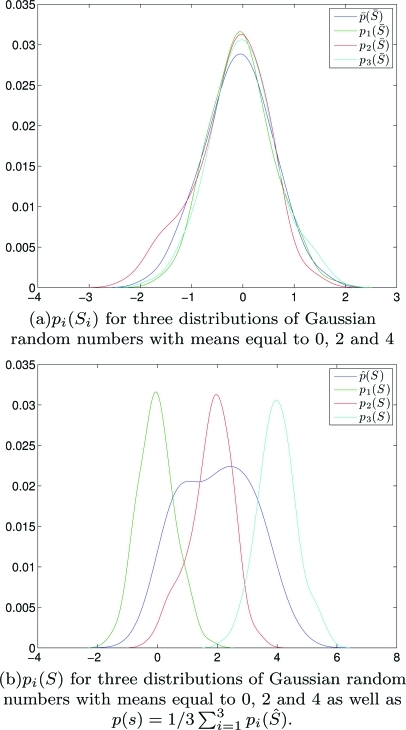
Figure 1.
(Color) Graphically comparing (average PDF) and (PDF of the aggregate) for a collection of three collections of Gaussian random numbers whose distributions have means 0, 2, and 4 respectively.
It is worth noting that a similar analysis can be done by comparing to , as their difference will reveal support based effects. The principles behind a and are similar in that they both address normalization of support based effects, only depends on quantities that represent distributions— and can both be estimated many times—and thus are likely more robust.
Graph dependent, support independent, effects on the population TDMI
To understand in detail how differences in the graphs independent of the supports can affect the and , begin by assuming that all the pi’s have the same support, or that . In this circumstance, the for all i. Thus, the contribution of the diversity of PDFs within the population to I, or the deviation from the mean at a particular x ∈ S value, is captured by and as defined in Eqs. 24, 30. Consequently, the only way that can be different from up to the estimator bias is for the variation in the collections of PDFs to be due to the order of averaging as rendered through the G’s.
Based on the aforementioned intuition, we claim (e.g., conjecture 1) that δI(τ) is equal to zero if and only if all the ε’s are zero. While we will not present a qualified proof of this claim here, we can offer an intuitive argument as to why our claim is justified. First, note that by inspection, if for all i, . Now, what remains is to understand what happens to the G’s when there are non-zero ε’s; to do this, note that we reduce the G’s to the terms they do not have in common
| (35) |
| (36) |
| (37) |
| (38) |
where , , , , , , , and ; then, define the difference in these quantities
| (39) |
Now, further noting that is convex (or concave, depending on the p’s) and applying standard convexity arguments, δg will not equal zero unless for all i. Intuitively, the difference lies in when the summation of the differences between the p’s and the ε’s is taken. Specifically, the difference between and the ’s is taken after the ’s are averaged, whereas the difference between and the ’s is itself averaged (the summation is applied over the differences between and the ’s). Thus, while it is possible that, through the act of integrating the G’s, symmetries will allow for the G’s to be equal, δg equaling zero is extremely unlikely. This is the primary reason why be believe that conjecture 1 is true. Nevertheless, because the convexity or concavity of depends on the nature of the p’s, it is non-trivial to specify whether will be, in general, greater or less than . Nevertheless, it appears in computational experiments that is often less than . In any event, it is now more clear how diversity amongst the distribution of p’s over the same support can (and likely will) force δI(τ) ≠ 0.
In the situation, where is not accessible, it may not be possible to fully understand the meaning of . While can help identify support based effects, pure graph-based temporally dependent effects may be difficult to estimate. In particular, if the sample size for some of the individuals is small, then it will be difficult to determine the contribution to due to purely graphic diversity simply because there will be such high variance in the graphical PDF estimates due to small sample sizes.28 In this case, the best that can be done is to estimate more static measures of graphic diversity such as those presented in Sec. 7.
Support dependent, graph-based effects on the population TDMI
There are two potential contributors to support dependent, graph-based effects on δI(τ), δG(τ), and δρ(τ).
The contribution to δI(τ) due to δρ(τ) is entirely due to the limits of integration; the integrand for the average and aggregate ρ component of the TDMI are identical. Thus, intuitively, δρ > 0 because of the relative location of the support of p1 in reference to the total support of the population; p1 will represent a more peaked distribution when defined on compared to . Note that while δρ is, in general, computable, it has similar characteristics to with more severe bias issues.
The contribution due to is not as intuitive; to understand how diversity in the supports contributes to via the induced differences in the ε’s, consider Figs. 1a, 1b. Relative to Fig. 1a, begin by defining as the average of the PDFs relative to the abstract support, or ; here, all the ’s will be small and independent of the support. This is how variation in the population is rendered when calculating , and thus how will render. In contrast, define the average of the PDFs relative to the total support, or ; this is the aggregate scenario. Here, it is clear that both the averaged PDF will not resemble any of the PDFs and relative to a selected p1. Moreover, all ’s will be relatively large and on the order of the various ’s over a non-trivial portion of the population support . Because of this, when the supports of the individuals differ, the largest term in , , will be accounting primarily for variation within the distribution of the supports of the population, rather than support-independent variation within the population. Moreover, when the supports of the individuals are relatively invariant, will be independent of time even when the I of an individual varies with τ. In any event, the point is, variation in the supports of otherwise identical distributions affects how the distributions are rendered through the TDMI calculation.
Finally, when only is available, which implies the presence of individuals with too few pairs of points to accurately estimate a PDF and thus the TDMI, and when there are support-dependent graph-based effects in the TDMI, it will likely be difficult to separate the support dependent, graph-based effects from the support independent graph-based effects on the TDMI (e.g., on the structure of the temporal correlation).
NON-TDMI-BASED METHODS FOR INTERPRETING POPULATION DIVERSITY
In this paper, we claim that the TDMI-based analysis can be used to both detail nonlinear correlation in time and interpret the composition of the population to which that correlation pertains to (i.e., whether the TDMI reflects and individual/homogeneous population or a diverse population). To verify this claim, we require a set of methods for establishing a baseline that are independent of information-theoretic machinery and can be used to interpret the make-up of the population. We propose three different quantifications of homogeneity of a population: (1) homogeneity in measurement representation, which addresses the variance in the distribution of the number of measurements per element of the population; (2) homogeneity in support, which addresses variation in the supports of each elements’ distribution; and (3) homogeneity in density, which addresses variation in the PDFs (or the graphs of the PDFs) over the population. Note that all but one of the methods for quantifying homogeneity are independent of time, and all are independent of any time-based correlation structure existent within the data set. Moreover, the homogeneity qualification methods we propose here are neither exhaustive nor particularly innovative; rather they are simple intuitive methods devised to interpret and confirm the TDMI-based results. Nevertheless, many of these methods are useful in their own right; moreover, at least one of the quantities we define here is required to supplement the TDMI analysis when very few measurements exist per individual. Finally, Table TABLE I. contains a summary of the twelve TDMI-independent quantities are we use to verify the TDMI methodology.
TABLE I.
Summary of all the non-TDMI based metrics used to assess homogeneity in a population (both among the graphs and the supports) used to verify the TDMI-type analysis.
| non-TDMI-based quantities for characterizing a population | ||
|---|---|---|
| difference between the population and individual element means | ∼0 implies either (1) most elements have a similar number of measurements or (2) the individuals come from distributions with similar means; implies the converse | |
| V(f(n)) | variance of the PDF of the number of measurements per individual | (1) V ∼ 0, imply elements were measured similarly; , implies elements measured at different rates; , implies elements measured at different rates with differing source distributions |
| E[smin(i)] | lower support boundary mean | |
| E[smax(i)] | upper support boundary mean | |
| Var(smin) | lower support boundary variance | |
| Var(smax) | upper support boundary variance | |
| length of support mean. | ||
| length of support variance | ||
| HRA | area between the (point-wise) least and greatest PDF graph | quantifies variance between the PDFs of the population; ∼0 implies element PDFs are homogeneous; very sensitive |
| VS(p) | , variance of the PDFs relative to a specified support, S | ∼0 implies homogeneity in PDFs; larger VarS(f) implies greater heterogeneity in the PDFs. |
| VS(p) calculated relative to the support of the aggregate population; ; note that there does exist an aggregate normalized support, , but we will not use this quantity here. | has the same interpretation as VS(p) in general, but has the potential to include support-based effects. | |
| VS(p) calculated relative to the abstract support of the population, | has the same interpretation as VS(p) in general, but excludes support-based effects. | |
Homogeneity in measurement composition
To quantify homogeneity in measurement composition, begin with the following two quantities. First, consider the difference between the mean of the raw measurements over the population versus the mean of the individual-wise measurement means, or
| (40) |
where nk is the number of points contributed by individual k, N is the number of individuals in the population, and n0 = 0. Now, under two circumstances: (1) the distribution of nk’s has zero or small variance, regardless of the collection of individual distributions; or (2) each individual comes from an identical distribution. Second, consider the variance of the probability density of the number of measurements per individual
| (41) |
where f(n) denotes the density of measurements per individual. Combining these two quantities, we arrive at three cases: (1) Vf(n) ∼ 0 implies that , together implying that the elements were measured similarly—no insight into the original distributions can be made; (2) and together imply that the elements were measured at different rates regardless of their source distributions (which can be identical); and (3) and together implies that the elements were measured at different rates and likely have differing source distributions. Note, that in general, both of these metrics are rather sensitive to diversity in a population.
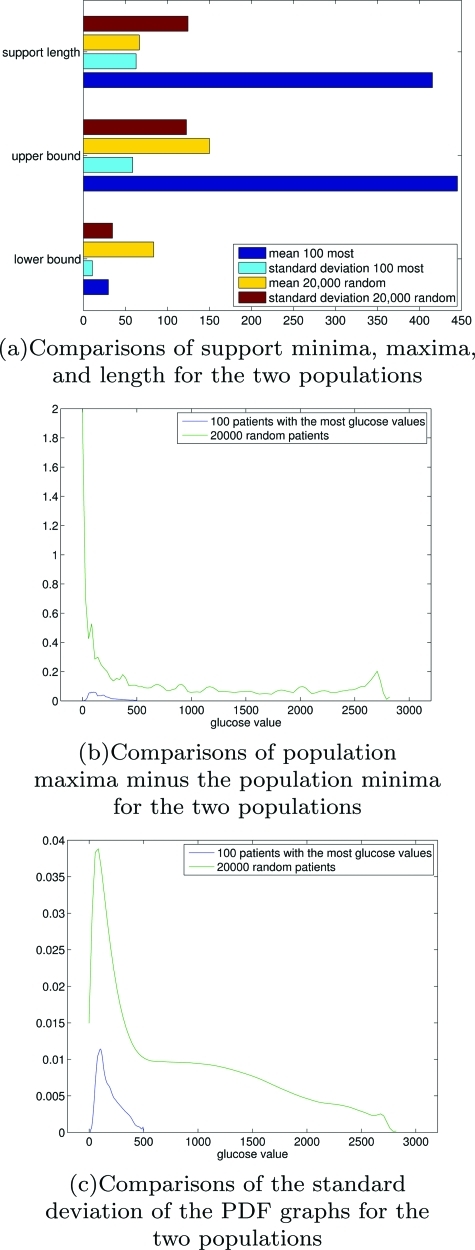
Homogeneity in measurement distribution supports
To characterize homogeneity in distribution support, we rely on a brute force homogeneity characterization technique. Begin by recalling that the support for element i’s distribution as Si = [smin(i), smax(i)]. Given these sets, which are defined by the individuals’ measurements, define the mean and variance of the support minima, maxima, and length by
| (42) |
| (43) |
| (44) |
| (45) |
| (46) |
| (47) |
These quantities afford relatively simple representations. For instance, when the minima, maxima, and lengths for the population have small variance, the intersection of the supports will not differ significantly from the union of the support—meaning the supports overlap. While a large variance in any either the minima, maxima, or lengths implies that the supports differ significantly over the population.
Homogeneity in the distribution of the graphs of the measurement PDFs
To specify homogeneity in the PDF of the population, we will use two methods. Intuitively, all of the methods characterize, in one way or another, the width of the maximum and minimum band of PDFs of the population over the support of the entire population. Begin by defining the PDF for an individual by pi(x), the supremum of the PDFs of the population by maxi(p(x)) = pM(x) and the infimum of PDFs of the population by mini(p(x)) = pm(x), over the union of the supports, . First, using the L1 (pseudo) distance,29 we can define the relative area of the width of the band of PDFs by
| (48) |
The relative area, HRA is literally the proportion of the supremum of the collection of PDFs that coincides with the infimum of the collection of PDFs. When HRA is close to one, the maximum distance between PDFs over the population occupies all the volume of the population-wide PDF. In other words, the population has at least two substantially different PDFs. Similarly, when HRA is near zero, this implies that the proportion of the area between the supremum and infimum over the collection of pi’s relative to the total area occupied by the supremum of the pi’s over the population is very small. Thus, the implication of HRA being near zero is that the pi’s are all nearly identical. However, this method is very sensitive to heterogeneity; a single individual’s PDF differing from the rest of the population can maximize HRA at one. In contrast, the second method for evaluating the diversity in PDFs over the population quantifies diversity from a mean within the population by estimating the variance of the PDFs at a given at a given x integrated over a given support (S), or
| (49) |
Note, VS(p) can be estimated relative to two different supports, the union of the supports, or the abstract support. This is an L2 flavored representation of the variation in PDFs; the variance of the pi’s at a given x is maximized when pi’s are maximally orthogonal (in the sense of an inner product between the pi’s) to one another, and minimized when the pi’s are minimally orthogonal (meaning they coincide). Thus, VS(p) has the potential to capture both support- and graph-based variation, depending on whether V is calculated relative to , which will include support-based effects, or , which will not include support-based effects.
ASSEMBLING THE PIECES: AN EXPLICIT PRESCRIPTION FOR TDMI ANALYSIS AND INTERPRETATION FOR A POPULATION OF TIME SERIES FOR A FIXED TIME SEPARATION δt
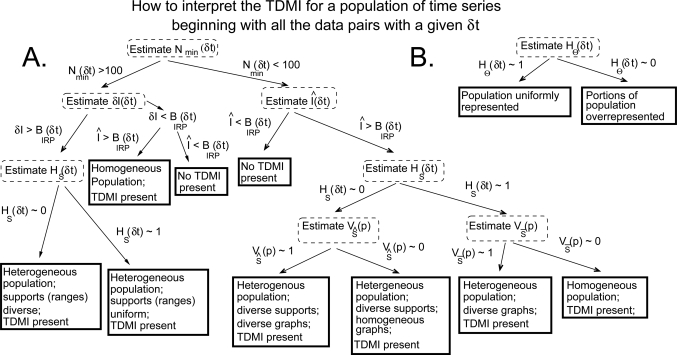
The interpretation of the TDMI and entropy for a complex, diversely measured population can be split into three broad steps: (1) performing a preliminary interpretation of and ; (2) performing an interpretation of δI(δt) or for the population; and (3) understanding the make-up of the data explicitly used to estimate the PDFs, yielding an understanding of what proportion of the population as used in the calculation. All the TDMI quantities used for the TDMI-based analysis are shown in Table TABLE II., a graphical schematic for applying this infrastructure is shown in Fig. 2, and a detailed algorithmic schematic for applying the TDMI infrastructure to a population is depicted via pseudocode in Appendix B.
TABLE II.
Summary of all the TDMI-based metrics used to interpret the TDMI and determine the population composition.
| TDMI-based analysis quantities | ||
|---|---|---|
| Quantity | What it signifies | What it quantifies |
| population averaged TDMI | quantifies average TDMI of a population | |
| aggregated population TDMI | quantifies TDMI of an aggregated population | |
| aggregated population calculated relative to the abstract support | support independent TDMI of an aggregated population | |
| δI(δt) | ; difference between the average and aggregate TDMI | ∼0 implies homogeneity, <0 implies heterogeneity |
| BE(δt) | PDF estimator bias; usually, BE(δt) ∼ BPRP(δt); BE(δt) can be estimated in a variety of ways | the number above which the I is considered to be positive |
| individual permutation bias averaged over a population | bias estimate that preserves information about the relative ranges of individuals | |
| individual permutation bias | bias estimate that preserves information about the relative ranges of individuals | |
| population permutation bias | bias estimate that destroys information about the relative ranges of individuals | |
| ; quantifies diversity of supports | ∼1 implies homogeneous supports; ∼0 implies diverse supports | |
| BRP(δt) | ; quantifies diversity of supports; quantifies cardinality of individual data sets | can imply diverse supports or cardinality per-element data sets; ∼0 can imply homogeneity in supports |
| δG(δt) | difference in the difference between how population diversity renders in and | >0 implies population diversity |
| δρ(δt) | ; quantifies diversity in supports | >0 implies population diversity. |
| HΘ(δt) | how representative the population used to estimate I at δt is of the time-independent (e.g., the entire) population | ∼0 implies the entire population is well represented; ∼1 implies portions of the population are overrepresented |
| Nmin(δt) | minimum number of pairs of points contributed by any one individual | a lower bound on the representation of an individual; 1/Nmin(δt) is a rough estimate of BE(δt) for the individual with the fewest pairs |
Figure 2.
The graphical schematic for the TDMI analysis of a population; note that by TDMI Present, we mean that the relevant TDMI measure (e.g., ) is greater than bias.
Step one: Determining the computability of
To begin, one must determine whether and are calculable for a given (or set of) δt(s). In general, to estimate , every representative individual must (under most circumstances) have at least 100 pairs of points available for the TDMI calculation. Similarly, to estimate , there must be at least 100 pairs of points gathered over the entire population—this is why is so useful in the context of a population. It is important to note that the rough number, 100 pairs of points per PDF estimate, is based on how PDF estimation technique converge in practice and is discussed in detail in Ref. 19. Nevertheless, the number of pairs of points needed is process dependent as is true for all quantities that depend on PDF-estimate schemes (very wide supports can require many more than 100 points whereas some distributions can require fewer than 100 points). Moreover, for a given δt, the number of pairs of points separated by a given δt depends both on length of the time series and the sampling rate; hence, the reason why we specify the number of pairs of points instead of the length of the time series. Because of these reasons, it is important to perform sensitivity analysis, estimate the bias, observe the PDF estimates for all quantities and their variation under perturbation, and generally use care to insure the robustness of the results.
Assuming that is calculable, because the calculation of I for an individual is independent of the support of the distribution, the variance in the distribution of is due to differences in the graphs of the PDFs representing each patient at a given δt. Further, because is made of individuals who have been averaged, the interpretation of the statistical moments of (i.e., the mean, variance, etc), is a scientific problem that depends on the particular circumstances.
The interpretation of is more difficult because can be composed of purely graphical, purely support, and intermixed support and graphical components, Thus, because is a population-dependent quantity where the individual contributions cannot be separated, it will be treated in Sec. 8B with δI(δt).
Step two (A in Fig. 2): Interpreting δI(δt) or
Step two has two courses of action depending on whether it is possible to calculate or not: (1) and are calculable and thus δI(δt) can be computed and (2) only , BRP(δt), and are calculable (when is calculable, this will always be the case). When δI(δt) is available, it, as estimated by both a KDE and histogram estimator, is all we need know: the closer δI(δt) is to zero, the more homogeneous the population is and the more represents a single, statistically singular source and the larger in magnitude δI(δt) is, the more statistically heterogeneous the population is and the more represents the population. Of course, if the histogram and KDE TDMI estimates differ substantially, it is likely that there are significant small sample size effects present in , and this needs to be taken into consideration when interpreting δI(δt), and . Moreover, in this circumstance, calculation of either BRP(δt) = |BIRP(δt) − BPRP(δt)| or can be used to further qualify the small sample size effects on the variation in the supports versus the graphs. Finally, when δI(δt) is positive, and shows no diversity due to the supports, then all the diversity in the population is due to the graph-based diversity.
When is not calculable, one is left with only , , and BRP(δt) or . In this case, one can still use BRP(δt) or to detect the homo- or heterogeneity in the supports. If there is no support-based variation, then pure graph-based heterogeneity maybe difficult to determine; in this circumstance, we recommend using a non-TDMI metric such as VS(p), which will have greater statistical power while sacrificing temporal dependence, to help determine the graphical composition of the population. In general, if there is support-based variation, it will likely be difficult to separate support-based, versus graph-based, contributions; it will be even more difficult to specify the proportion of diversity contributed by the support- versus graph-based effects.
Step three (B in Fig. 2): Assessing population representation
Finally, it is extremely important to understand what portions of the population actually have points in a given δt bin. Recall that the make-up of the population used to estimate I at a specific δt is a concern because of the filtering effect (cf. Sec. 4C); specifically, it is possible to have entire portions of the population excluded from the data set as well as a highly nonuniform distribution of the population represented in the data set used to estimate the PDFs. Written differently, it is important to always remember that δI is always calculated relative to a fixed δt which will have a particular bin population—when studying the evolution of I as δt is varied, the representative population can change as δt changes. Thus, it is important to at least calculate HΘ(δt) or an HΘ-like quantity to verify what proportion of the population is being included in the PDF estimate. Moreover, we also find it convenient to keep track of the minimum (and sometimes maximum) number of pairs of points contributed by an element represented in the data set used to estimate the PDFs; we denote this number by Nmin(δt) as a measure of the least representative individual.
QUANTITATIVE EXAMPLES FOR TDMI INTERPRETATION AND POPULATION HOMOGENEITY EVALUATION
Simulated data examples: The quadratic map and the Gauss map
To explicitly demonstrate how to interpret and in the presence of a diverse population in a variety of circumstances, consider two sources of simulated data, the quadratic map
| (50) |
where a is set to 4 and the Gauss map
| (51) |
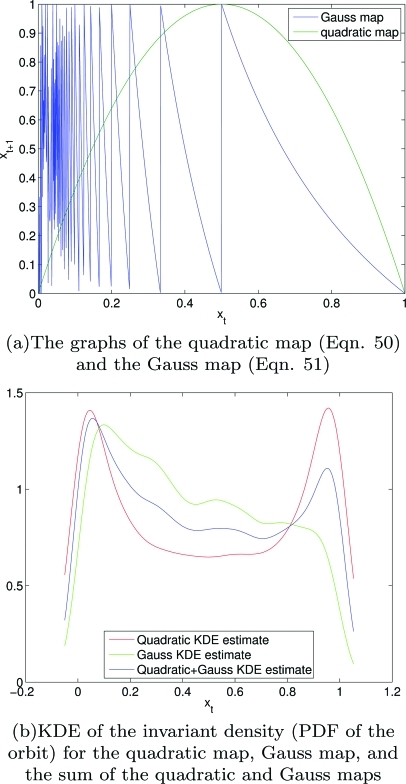
These sources were chosen because their statistical structures are well understood,2, 23, 24 they are chaotic, they are both 1-dimensional maps defined over the unit interval (meaning, they have the same support), and they have relatively different invariant densities. Figure 3 shows the graphs of the quadratic and Gauss maps, their individual invariant densities (PDFs of the orbit), and the sum of their invariant densities. Thus, in this context, the difference between pf and pg, , is both large enough such that the G’s will be non-zero and is non-uniform over the domain or nonlinearly dependent on x. The data sets we will use, based on the maps above, include:
Figure 3.
(Color) The graphs of the quadratic map (Eq. 50) and the Gauss map (Eq. 51)—note the significant difference between the graphs of the mappings, and invariant density (PDF of the orbit) for the quadratic map, Gauss map, and the sum of the quadratic and Gauss maps—note the significant differences between the relative p’s.
Dataset1:Quadratic map time-series with 1000 points; this is one of the data sets meant as a baseline from which all the other cases can be compared.
Dataset2:Gauss map time-series with 1000 points; this is one of the data sets meant as a baseline from which all the other cases can be compared.
Dataset3:Data sets 1 and 2 concatenated into a single data set with 2000 data points; this data set is used primarily to test the effects of differing PDFs within a population on ι, G, and thus, versus .
Dataset4:50 independent, concatenated quadratic map time-series with 20 points each totally 1000 points; this data set is meant to highlight the effect of the estimator bias when calculating versus .
Dataset5:10 independent, concatenated quadratic map time-series with 100 points each totaling 1000 points; this data set is meant to form a baseline for data set 6.
Dataset6:10 independent, concatenated quadratic map time-series with 100 points with disjoint supports with increasing means totaling 1000 points; this data set is used to demonstrate the effect of diverse supports amongst the population where the PDFs are identical on ι, G, B, and thus versus .
Each data set will be denoted by Di where i is the indexed label of the respective data; the data sets are detailed in Table TABLE III..
TABLE III.
Complete list of the simulated data sets.
| Synthetic data sets | ||
|---|---|---|
| Data set | Source | Size |
| D1 | chaotic quadratic map | 1000 pts |
| D2 | chaotic Gauss map | 1000 pts |
| D3 | D1 concatenated with D2 | 2000 pts |
| D4 | 50 independent concatenated 20 pt chaotic quadratic map time series | 1000 pts |
| D5 | 10 independent concatenated 100 pt chaotic quadratic map time series | 1000 pts |
| D6 | 10 independent concatenated 100 pt chaotic quadratic map time series each with non-overlapping, monotonically increasing support | 1000 pts |
Finally, to save space, we will demonstrate the TDMI and non-TDMI-based computations on all the simulated data sets at one time. We will adhere to the algorithm shown in Fig. 2 when analyzing the real data sets.
TDMI-based analysis of the simulated data
Base cases: testing the TDMI-based metrics on individuals—In Table TABLE IV., one can see that both the quadratic and Gauss maps have distinctly different I(τ = 1) values. Note that the Gauss map has a faster decay in correlations; for both maps, all correlations in time decay by τ = 6. Further notice that all bias estimation schemes are essentially identical as expected. This also implies that support-variation detecting quantities such as register no variation in supports.
TABLE IV.
TDMI results and homogeneity metrics for the simulated data sets one through six.
| TDMI-based quantities | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source | BRP(τ = 1) | δρ(τ = 1) | δG(τ = 1) | δI(τ = 1) | ||||||
| D1 | 0.72 | — | 0.008 | 0.008 | 0.008 | 0 | 0.99 | 0 | 0 | 0 |
| D2 | 0.31 | — | 0.012 | 0.012 | 0.012 | 0 | 0.96 | 0 | 0 | 0 |
| D3 | 0.52 | 0.37 | 0.01 | 0.008 | 0.007 | 0.001 | 0.98 | 0 | 0.15 | 0.15 |
| D4 | 0.34 ± 0.07 | 0.71 | 0.18 ± 0.03 | 0.013 | 0.011 | 0.002 | 0.98 | 0 | δI | 0.37 ± 0.07 |
| D5 | 0.48 ± 0.01 | 0.71 | 0.04 ± 0.01 | 0.006 | 0.007 | 0.001 | 0.99 | 0 | δI | 0.24 ± 0.01 |
| D6 | 0.48 ± 0.01 | 1.12 | 0.04 ± 0.01 | 1.12 | 0.011 | 1.11 | 0 | unknown | unknown | 0.55 ± 0.01 |
Support dependent, graph independent analysis—To see how diverse supports are rendered, consider the contrast between D5 and D6, whose only difference is in the location of the supports. Both of the support-based TDMI based metrics, BRP and , produced dramatic representations of the disjoint nature of the supports of data set six (cf. Table TABLE IV.). Notably, the difference between both BRP and on D5 and D6 are near their respective maxima.
Graph dependent, support independent analysis—Data set three, the quadratic-Gauss aggregated data set, has homogeneity in support in all support-based metrics (BRP and ) as can be seen in Table TABLE IV.. In particular, both and all the random permutation bias estimates are totally unaffected by the existence of . Furthermore, δI ≠ 0, meaning that the population averaged TDMI and the TDMI of the aggregated population were different. In particular, , thus leading to the conclusion that , which is not surprising given that when the for all i, it is reasonable that the ε’s register greater through the sum than the aggregate. In any event, all the TDMI based metrics registered the diversity in the population of PDFs.
Support dependent graph-based analysis—To begin to see how support and graph effects intermix, consider for a data set identical to D6 except where the quadratic data has been replaced with uniform random numbers, thus yielding data with purely population location information; denote this data set as . Now, , thus comparing to , we notice that the presence of intra-agent time-based correlation decreases the population scale TDMI by a small but measurable amount—here, . Therefore, while nearly all the intra-agent TDMI is subsumed by the inter-agent TDMI, when there is a presence of both strong intra-agent information as well as strong inter-agent information (i.e., highly disjoint supports), will contain both intra-agent and inter-agent components.
What the example in the previous paragraph shows is that deducing the contribution of the intra-agent and inter-agent components to will, in many cases, be non-trivial. Nevertheless, the use of metrics that detail the PDF variation can sometimes aid in the interpretation of . First, consider how the heuristic metrics of PDF variation render the variation in PDFs. Both the super sensitive HRA and more robust, less sensitive V(p), for D6 are about double their values for D5, even though D5 will yield considerably noisier PDF estimates due to the smaller sample size per element. Similarly, the TDMI metrics for PDF variation also render population diversity; δI for D6 is more than twice δI for D5. However, δI for D6 has a slightly more complicated interpretation. In particular, while δI represents the difference between the population and the individual TDMI, there is likely a non-trivial component of that is a function of sample size. Thus, δI is not purely the difference between the individual and the population TDMI for unlimited data as it was for D3. Nevertheless, because and , we know that has components of both individual and population scale TDMI. In fact, considering versus , one can see that the TDMI for D6 is due to support-based effects instead of intra-element effects; presumably if the supports for D6 were nearly overlapping instead of disjoint, would be much closer to zero (as is the case for ). While it is unusual to be able to compare identical, stationary systems with differing supports, this analysis does suggest that calculating for the raw data and for the data with normalized supports may be useful for determining the proportion of that is due the diversity of the supports.
Non-TDMI-based analysis of the simulated data
Base cases: testing the non-TDMI metrics on individuals—Begin by considering D1 and D2, both of which represent only a single source. Both data sets are well defined in p (cf. Fig. 3) and have supports whose lengths, |S|, and boundaries, smin, smax, are well resolved and within their expected ranges (cf. Table TABLE V.).
TABLE V.
Heuristic homogeneity metrics for the simulated data sets one through six.
| non-TDMI-based population diversity metrics | |||||||
|---|---|---|---|---|---|---|---|
| Source | Var(ni) | |S| ± V|S| | HRA | ||||
| D1 | 0 | 0 | 0.0001 | 0.999 | 0.9989 | 0 | 0 |
| D2 | 0 | 0 | 0.0002 | 0.9998 | 0.9997 | 0 | 0 |
| D3 | 0 | 0 | 0.0002 ± 0.0003 | 0.9989 ± 0.0015 | 0.9987 ± 0.0018 | 0.16 | 0.09 |
| D4 | 0 | 0 | 0.02 ± 0.02 | 0.996 ± 0.006 | 0.98 ± 0.03 | 0.9 | 0.39 |
| D5 | 0 | 0 | 0.001 ± 0.002 | 0.9997 ± 0.0006 | 0.998 ± 0.003 | 0.37 | 0.13 |
| D6 | 0 | 0 | 5.5 ± 3 | 6.5 ± 3 | 0.997 ± 0.004 | 0.68 | 0.32 |
Support dependent, graph independent analysis—To see how variations in the supports are rendered, consider the contrast between D5 and D6, whose only difference is in the location of the supports. In contrast to D5, for D6, the variation in support shows up in the heuristic metrics smin, smax, |S|, and especially in the variance of smin and smax.
Graph dependent, support independent analysis—Data set three, the quadratic-Gauss aggregated data set, has homogeneity in support in all support-based metrics (smin, smax, |S|) as can be seen in Table TABLE V.. In contrast, both of the heuristic metrics designed to detect variation in PDFs (HRA, ) registered as non-zero, meaning they detected variation in the PDFs. Moreover, the l1-like diagnostic, HRA was more sensitive than the variance based metric, , as expected.
Support dependent graph-based analysis—By design, none of the examples mix graph and support effects simultaneously.
Quantifying small sample-size effects
To form a baseline of small sample size effects for both real data applications and the support-based effects, we focus on comparing and constraining results for D4 and D5, the quadratic map data sets with 50 sets of 20 points, and 10 sets of 100 points.
Small sample size effects on non-TDMI-based support analysis metrics—The heuristic metrics of support diversity, smin, smax, and |S| show homogeneity in support for D4 and D5 in an absolute sense. However, it is important to note that the invariant density of the quadratic map has most of its mass at the end points, and thus may represent the best case scenario for support based metrics on small data sets. Moreover, differences between D4 and D5 can be observed—smin for D4 is roughly an order of magnitude larger than smin for D5.
Small sample size effects on TDMI-based support analysis metrics—The TDMI based metrics of support diversity (BRP, ) show homogeneity of support, although the individual-wise random perturbation for the random case (BIRP) is rather high, especially for the 20 point data sets, as one might expect. However, we hypothesize that the primary reason why BIRP is so high for the 20 point data sets is that, upon randomly permuting any data set, the average τ will be the length of the data set over 3, in this case, . Thus, for very short data sets, it can be difficult to approximate the estimator bias using only the random permutation method.19
Small sample size effects on non-TDMI-based graph analysis metrics—In contrast to the support-based effects, the heuristic-based PDF variability metrics (HRA, ) register substantial diversity among the PDFs D4 and D5, effects that are entirely a function of small sample sizes. These results are not surprising given that there will be great variance in the PDF estimate of a quadratic time-series with only 20 points.
Small sample size effects on TDMI-based graph analysis metrics—The small sample size situation highlights both the difference between and and also displays the motivation for why one would want to estimate . The average based TDMI results for both D4 and D5do not approximate the 1000 point analogs; and moreover, the addition of more sets of data with similar lengths will not help to converge to the higher point analog but rather decrease the variance in the mean value. Thus, the meaning of is, in a sense, a precision/accuracy type issue; adding more 20 point data sets will make the estimate of more precise, but not necessarily more accurate. That said, accuracy is always defined relative to a target; there is likely less TDMI in the 20 point data set because there is considerably less time-based information in a 20 or 100 point data set than in a 1000 point data set. Therefore, while adding more data sets will not aid in convergence to the infinite point analog, the infinite point analog may not be right target to be aiming for with 20 point data sets. In contrast, the aggregated data sets produce a TDMI equivalent to the 1000 point analog, thus inducing a δI. Moreover, adding points to the aggregated data set will help with convergence to I(τ = 1) for infinitely long data strings.
Interpreting δI when individual elements have few pairs of points—The existence of δI for D4 and D5 introduces a form of divergence from I(τ = 1, N = ∞) that is not quite a bias (either estimator or non-estimator); the “true” amount of information in a data string of length 20 is fundamentally different from the “true” amount of information in a data string of length N = ∞—thus δI can also exist due to finite sample size effects. Or, said more quantitatively, , even for an unlimited collection of 100 point data strings, will never be within estimator bias or any other kind of bias, of I(τ = 1, N = ∞) because I(τ = 1, N = ∞) ∼ 0.72 while I(τ = 1, N = 20) ≈ 0.48 ± 0.1. What this means for is that, unless the aggregated data sets are homogeneous enough in their time-dependent correlation structure, will likely represent population distribution information, as would represent the upper bound on time-correlation based information present in each data string. Often the composition of most real world data streams can be difficult to infer; and moreover, it can be a non-trivial problem to discern whether or most faithfully represent a population or individual effects. For instance, in Ref. 25, the authors claim both the presence of time-correlation information and population-based time-correlation being simultaneously present. Usually, a careful analysis of the population composition of the δt bins will help rectify this difficulty.
Real data examples: Glucose values for 100 densely sampled individuals versus 20,000 random individuals
We now move on to applying the insights and techniques of the previous sections to real data. In particular, we will consider two data sets that contain different populations of patients from the CUMC data repository. More specifically, the data sets include
Data set 7:A collection of the 100 patients with the most glucose measurements in the database, ranging from ∼4000 to ∼1500 measurements per patient.
Data set 8 : A collection of 20 000 random patients with at least 2 glucose measurements from among the 800 000 patients with glucose values.
Each data set will be denoted Di where i is the indexed label of the respective data; the data sets are detailed in Table TABLE VI..
TABLE VI.
Complete list of the real patient data sets.
| EHR-based data sets | ||
|---|---|---|
| Data set | Source | Size |
| D7 | glucose time series from the 100 patients with the most glucose values | length of time series and sampling rate varies with patient |
| D8 | glucose time series from 20 000 randomly selected patients | length of time series and sampling rate varies with patient |
To visualize these populations, consider Fig. 4 where the normalized PDFs for each individual for each population and the PDF of the overall populations are plotted. While the population-wide PDFs, shown in Fig. 4c are not wildly different, the relative diversity within the two populations, as shown in Figs. 4a, 4b, is dramatic. The motivation for choosing D7 is that, for this set, each patient has at least 1000 lab values, both and are calculable. Moreover, the authors hypothesize that patients with so many glucose values are more likely to represent a more homogeneous population compared with the population at large. Given the makeup of D7, D8 represents not only a contrast to D7 in that D8 is a snapshot of the entire population, but D8 also represents a pathologically difficult situation data-wise—very few patients have more than 100 glucose values, and the set of possible causes for the existence of a glucose measurement is extremely large (or broad). Thus, not only will be difficult to calculate for D8 (most patients would not have enough data to generate a PDF estimate), but there is likely tremendous and differing diversity amongst the patients actually included in the estimates of and .
Figure 4.
(Color) PDFs of glucose measurements for individuals within a population and for a population for two data sets, the 100 patients with the largest records and 20 000 random patients.
Finally, note that in contrast to the previous analysis of simulated data, we will present the TDMI results first, followed by an analysis using the non-TDMI metrics to verify the TDMI results. The point of this ordering is to demonstrate the TDMI infrastructure without hindsight knowledge.
TDMI-based analysis for data set 7, the well measured population
Analysis of the δt = 6 h time separation using the algorithm in Fig.2—First, considering Table TABLE VII., note that for D7 with a δt = 6 h, we are able to estimate , and thus δI because Nmin(6 h) > 100. Next, note that δI(6 h) is considerably above BIRP(6 h), meaning that the population on the time-scale of 6 h is heterogeneous. Moreover, both and are greater than zero, meaning that there is TDMI present in individuals and the aggregated population. To determine the nature of heterogeneity, further consider the support-based metric; points to the population having uniformity in supports or ranges (BRP(6 h) ≈ BIRP(6 h) which corroborates this conclusion). Finally, the entire population is reasonably represented for δt = 6 h as confirmed by the fact that Nmin(6 h) ∼ 500 and . Thus, the concluding interpretation is as follows: the population is heterogeneous on the δt = 6 h time scale; the heterogeneity in the population is in the graphs not the supports (or the normalizations; there is diverse but present temporal correlation among the population (i.e., the TDMI is not due to the population aggregation but exists because of the individuals); and the entire population is well represented in the TDMI-based quantities.
TABLE VII.
TDMI results and homogeneity metrics for the real patient data sets seven and eight; note all δt times are in hours.
| TDMI-based quantities for the δt = 6 h time separation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source | δI | BRP | HΘ | Nmin | ||||||
| D7 | 0.64 ± 0.03 | 0.22 | 0.42 ± 0.03 | 0.02 ± 0.01 | 0.02 ± 0.005 | 0.001 ± 0.0005 | 1 ± 0.0005 | 0.31 | 470 | |
| D8 | 0.29 ± 0.16 | 0.38 | 0.09 ± 0.37 | 0.2 ± 0.2 | 0.08 ± 0.005 | 0.006 ± 0.0005 | 1 ± 0.02 | 0.003 | 1 | |
Analysis of the δt = 24 h time separation using the algorithm in Fig.2—First, considering Table TABLE VIII., note that for D7 with a δt = 24 h, we are able to estimate , and thus δI because Nmin(24 h) > 100. Next, note that δI(24 h) is within the error bars of zero (e.g., below BIRP(24 h)), meaning that the population is on the time-scale of 24 h is homogeneous. Moreover, both and are greater than zero, meaning that there is TDMI present in individuals and the aggregated population. To determine the nature of heterogeneity, further consider the support-based metric; points to the population having uniformity in supports or ranges (BRP(24 h) ≈ BIRP(24 h) which corroborates this conclusion). Finally, the entire population is reasonably represented for δt = 24 h as confirmed by the fact that Nmin(24 h) 500 and . Thus, the concluding interpretation is as follows: the population is homogeneous on the δt = 24 h time scale; there is present temporal correlation among the population (i.e., the TDMI is not due to the population aggregation, but exists because of the individuals); and the entire population is well represented in the TDMI-based quantities.
TABLE VIII.
TDMI results and homogeneity metrics for the real patient data sets seven and eight; note all δt times are in hours.
| TDMI-based quantities for the δt = 24 hrs time separation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source | δI | BRP | HΘ | Nmin | ||||||
| D7 | 0.093 ± 0.06 | 0.077 | 0.016 ± 0.06 | 0.02 ± 0.01 | 0.02 ± 0.005 | 0.001 ± 0.0005 | 0.99 ± 0.01 | 0.33 | 479 | |
| D8 | 0.21 ± 0.15 | 0.17 | 0.04 ± 0.15 | 0.3 ± 0.2 | 0.07 ± 0.01 | 0.005 ± 0.001 | 0.97 ± 0.001 | 0.005 | 1 | |
Analysis independent of time—Considering the entropy calculations in Table TABLE IX., D7 renders some heterogeneity because the difference between and is non-zero. Nevertheless, as we will see for D8, an entropy difference of 0.73, which is about half the magnitude of , would argue that the static information theoretic interpretation of the population is of relative homogeneity.
TABLE IX.
Time independent TDMI results for the real patient data sets seven and eight.
| Time independent TDMI-based quantities | ||
|---|---|---|
| Source | ||
| D7 | 1.39 ± 0.07 | 2.12 |
| D8 | 0.8 ± 0.22 | 2.05 |
Sample size issues—There were no sample size issues with respect to either δt time separations studied; in both cases, Nmin was well over 100, and thus all PDFs and their respective biases could be accurately estimated. In fact, careful analysis of the population make-up in each δt between 6 h and 56 h revealed that the proportionally of each individual remained relatively constant. Finally, Fig. 6, where both the TDMI estimated using both KDE and histogram estimation schemes are shown, confirms the lack of any small sample size effects because both estimation schemes are essentially equal.
Figure 6.
(Color online) The TDMI for both and with δt bins of 6 h for a period of a few days for D7 and D8; note that the bias estimates can be found in Tables 8, TABLE VII.. With respect to (a), note the following: for δt ≤ 6 h, δI > 0 and for δt > 6 h, δI ≈ 0; the KDE and histogram estimates are extremely similar; the diurnal (daily) periodic variation in correlation of glucose is clearly evident in both and . With respect to (b), note the following: for all δt δI is consistent and likely zero within bias; the KDE and histogram estimates differ greatly, implying the presence of small sample size effects in the average TDMI calculation; the diurnal (daily) periodic variation in correlation of glucose is clearly evident in both and in all but the KDE estimated TDMI average.
Non-TDMI-based analysis for data set 7, the well measured population
Non-TDMI support-based analysis—To verify the TDMI-based results, begin by observing that heuristic metric that quantifies variation in the supports, , which is considered small. Thus, while there is some diversity among how the patients were measured, variation how patients are measured is small. This claim is also justified by the fact that the variance in the number of points contributed, per patient, to the δt = 6 h bin, Var(ni), is small. Finally, the variance in smin, smax and |S| is small compared to the respective values (cf. Fig. 5a). Because these are time-independent measures of the support, and because adding the temporal aspect of the analysis only makes the data set smaller, it is likely that the TDMI analysis of the homogeneity of support are correct.
Figure 5.
(Color) Comparisons of the supports and PDF graph variations for two data sets, the 100 patients with the largest records and 5000 random patients.
Non-TDMI graph-based analysis—The most sensitive PDF variation metric, HRA points to a relatively diverse population, while the less sensitive PDF variation metric , based on the standard deviation of the distribution of PDFs, points to a relatively homogeneous, yet not totally homogeneous population. Figure 5 confirms this analysis visually. The maxima minus the minima, which, when integrated is essentially HRA, shown in Fig. 5b, can be seen to be relatively large, thus making HRA render diversity. In contrast, the variance in the graphs of the PDFs, shown in Fig. 5c, is seen as relatively small for D7, thus making render relative homogeneity. It is important to note, however, that , which is independent of time, does not detail the fact that the population has diverse predictive information for time periods less than 6 h; this is an important distinction to make as it implies that prediction can vary with time despite the overall distribution of physiological variables. Finally, both the TDMI and the heuristic analysis conclude that the population is homogeneous in supports and in the long term (i.e., independent of time), the population is homogeneous; this is because δI ∼ 0 for δt > 12 h and is small.
TDMI-based analysis for data set 8, the random (less well measured) population
Analysis of the δt = 6 h time separation using the algorithm in Fig.2—First, considering Table TABLE VII., note that for D8 with a δt = 6 h, we are not really able estimate because Nmin(6 h) = 1. To interpret , we consider the support-based metric; which points to the population, which was filtered and has time points separated by 6 h, having uniformity in supports or ranges (BRP(6 h) ≈ BIRP(6 h) which corroborates this conclusion). To give intuition to the graph-based variation, consider (Table TABLE X.), which implies a somewhat diverse population. Moreover, for D8 is twice that of D7, implying that the population in D8 is more diverse than that of D7. Moving beyond the algorithm shown in Fig. 2, we did estimate and thus, δI(6 h), only including individuals with enough points to estimate I. Based on this restricted version of δI(6 h), the population appears to be homogeneous. Nevertheless, both the restricted and are greater than zero, meaning that there is TDMI present in individuals and the aggregated population. This means that there is an apparent contradiction; the restricted δI(6 h) implies a population that is somewhat homogeneous/heterogeneous while implies a heterogeneous population. This contradiction is resolved by recalling that is calculated on the entire, non-filtered population and is independent of time and will overestimate graphic diversity, while δI is overly restricted and will underestimate diversity. This interpretation will be substantiated further in Sec. 9B5. Finally, the overall population is poorly represented for δt = 6 h as confirmed by the fact that Nmin(6 h) = 1 and HΘ(6 h) ≈ 0. In fact, for D8, we know that 63% of the patients (12 763) have no points in the δt = 6 h bin, and only 12% (2400) of the patients have ten or more points in the δt = 6 h bin. Thus, the concluding interpretation is as follows: the population is homogeneous on the δt = 6 h time scale up to what is resolvable by δI(6 h); the represented population has relatively uniform supports; there is diverse but present temporal correlation among the population (i.e., the TDMI is not due to the population aggregation, but exists because of the individuals); the population has diversity relative to their time-independent graphs, but this graph diversity may not reflect the graph diversity of the represented population (i.e., the population used to estimate the TDMI-based quantities); the overall population of patients is poorly represented in the TDMI-based diagnostics; and finally, the overall population of 20 000 patients is diverse, but the patients that have enough data to estimate the TDMI on time-scales of δt ≤ 48 h (i.e., the represented population), which represents a strongly filtered subpopulation, is relatively homogeneous in predictive information regardless of δt.
TABLE X.
Heuristic homogeneity metrics for the real patient data sets seven and eight.
| non-TDMI-based analysis metrics | |||||||
|---|---|---|---|---|---|---|---|
| Source | Var(ni) | |S| ± V|S| | HRA | ||||
| D7 | 1.042 | 463.7 | 29.7 ± 10.7 | 445.0 ± 58.8 | 415.4 ± 62.7 | 0.898 | 0.432 |
| D8 | 30 | 55 | 84 ± 35 | 150 ± 122 | 66 ± 125 | 1 | 0.90 |
Analysis of the δt = 24 h time separation using the algorithm in Fig.2—Considering Table TABLE VIII. (and later, Fig. 6b), the analysis of the TDMI diagnostics for δt = 24 h is essentially identical to δt = 6 h case. Even representative population for both the δt = 6 and 24 h bins is essentially identical down to the individual proportional contributions to the aggregated data set. Thus, the key observation here is the difference between D7 and D8; D7 registered heterogeneity at δt = 6 h and homogeneity at δt = 24 h whereas D8 does not render a δt dependence in the TDMI-based diagnostics.
Analysis independent of time—Considering the entropy calculations in Table TABLE IX., D8 renders heterogeneity because the difference between and is non-zero. In particular, compared to the entropy differences for D7, the D8 has an entropy difference of ∼1.25, which is substantially larger in magnitude than . Thus the static information theoretic interpretation of the population in D8, which includes all patients (there is not filtering effect), is of heterogeneity.
Sample size issues—There are three sample size issues present in the TDMI analysis of D8, the poor representation of the overall population, the inability to estimate I for every representative member of the population, and the overall small sample size and bandwidth/normalization issues. The first issue implies that the probability mass used to estimate the PDFs comes from a very small subset of the population; e.g., only 12% of the population has 10 or more points in the δt = 6 h bin. Thus, the restricted (i.e., filtered) population is likely substantially more homogeneous than the overall population, and the TDMI analysis cannot be said to represent the overall population. Relative to the second issue, since Nmin = 1 (for both δt = 6 and 24 h), is representative of a smaller population than . Finally, the third issue, small sample size effects, can be seen in the large difference (about a factor of 2) between the KDE and histogram estimator based TDMI values seen in Fig. 6.
Non-TDMI-based analysis for data set 8, the random (less well measured) population
Non-TDMI support-based analysis—Begin by noticing that there is considerable diversity in how the 20 000 patients are measured, as can be seen in , which is 30 times larger for D7. Considering this in conjunction with Var(ni) ≈ 50 for D8, which is much smaller than Var(ni) for D7, implies that very few of the patients have many points. Said differently, the reason why Var(ni) is relatively small compared to is that ni is bounded from below by 0 and is never very large for any member of D8. That this is the fact is reflected in variance in smin, smax, and |S|, which is large (on the order of, or greater than) the values of smin, smax, and |S|, respectively (cf. Fig. 5). Heuristically, this effect can be seen by observing the range of values seen in Fig. 4b versus Fig. 4a—the population of 20 000 yields a range of glucose values roughly five times that of D7.
Non-TDMI graph-based analysis—The most sensitive PDF variation metric, HRA points to a relatively diverse population. In contrast to the results for D7, the less sensitive PDF variation metric , also points to a heterogeneous population; in particular, is just about twice the for D7.
Analysis of the TDMI under variation of δt
A central motivation for using the TDMI is to observe how nonlinear correlation evolves in time; however, in the context of a diversely measured population, one must take care to ensure the TDMI signal represents a relatively constant population. Relative to D7 and D8, we know that, for δt between 6 and at least 56 h, the representative population is roughly constant. Figure 6 details the temporal evolution of the TDMI, and with it, exhibits five notable features.
First, both data sets display diurnal peaks in predictability; a full explanation of these peaks, which is dependent the structure of meal times.26 This is scientifically interesting because it is a signal that can be used to test physiological models, it can be used to distinguish populations, it implies that outside of very local time windows, measurements separated by 24 are more informative than measurements separated by fewer hours, and finally, the diurnal peaks confirm the presence of diurnal cycles in humans that are believed to exist.
Second, relative to D7, the population appears to be heterogeneous on time scales of 6 h and less, and homogeneous on time scales longer than 6 h. This can be seen in Fig. 6a, where δI(6 h) is relatively large and drops to zero by δt = 12 h. This is an interesting result that we are still working to understand.
Third, by comparing the results for D7 and D8, we can observe a difference in the degree of homogeneity amongst the population. In particular, combining the facts that the error bars for are large for D8 compared to D7, δI is independent of δt for D8, δI for D8 is much larger than for D7, and the broad qualitative TDMI signal (i.e., the diurnal peaks) is the same for both D7 and D8, it seems clear that both data sets have somewhat homogeneous populations (i.e., homogeneous enough to resolve a similar signal), but D7 is considerably more homogeneous than D8.
Fourth, considering Fig. 6b, it is clear that the aggregate TDMI resolves the diurnal peaks considerably better than the average TDMI. This is confirms the usefulness of the aggregate TDMI in the context of a complex, diversely measured population.
And fifth, the small sample size effects are clearly evident when comparing the difference between the histogram and KDE estimates of the TDMI between Figs. 6a, 6b. In particular, the two different estimates for the aggregate TDMI on D7 are essentially identical, while the aggregated TDMI estimates on D8 differ in a nontrivial way (by more than a factor of two). The average TDMI calculations display an even stronger effect. Finally, the error bars for D8 are about ten times the magnitude of the error bars for D7.
The point is, the time evolution of the TDMI is both scientifically valuable in that it leads to insights not otherwise observed and interpretable in the context of a time dependent, complex, diversely measured population using the infrastructure presented in this paper.
DISCUSSION AND COMMENTS
Specific results of the interpretative framework relative to real data
The methods in this paper were shown to work for both a well understood computer-generated data set and for a pathologically diverse real data set. The entropy for all populations registered the populations as diverse. Nevertheless, the TDMI produced a more nuanced picture. In particular, for one set of patients, the TDMI calculation implied that a set of patients have differing predictive information up to 6 h and are homogeneous in correlation afterwards. In contrast, the same calculation on a heavily filtered general population (the population that had frequent data measurements), yielded a population that seemed homogeneous with respect to time-dependent correlation. Thus, while it is likely that these populations are different, a full explanation, which requires more clinical study, is beyond the scope of this paper. Nevertheless, the TDMI analysis yielded results that were understandable, given this pathologically difficult population of data.
Using categorical billing code data to help verify the TDMI analysis
Because the patients in D7 and D8 are real patients in the hospital, we can consider the billing codes, which can act as a proxy for population composition, assigned to the patients in the various populations. To do this, consider the fraction of patients with the two most frequent billing codes for three data sets, D7, D8, and the subset of D8 used to estimate the TDMI-based diagnostics, (members of the subpopulation have at least 10 glucose measurements separated by 6 h or less). There are two features of that are important to pay attention to: (1) the overall fraction of patients that have a given billing code and (2), the drop off between the fraction of patients with the most and second most common billing codes. For D7, 75% of the patients are covered by a single billing code and the drop between the most and second most common billing codes is around 5%—thus 70+% of these patients likely have relatively similar afflictions. In contrast, the most frequently seen billing code in D8 only covers 25% of the population, followed by a 10 point drop off. In contrast, at least 50% of is covered by a single billing code, while the second most common billing code only covers only a quarter of the population—a 25 point drop. This implies more homogeneity than D8 but less than D7. Thus, broadly speaking, the billing code analysis corroborates the conclusions drawn from the time-based information theoretic analysis in Sec. 10A. Moreover, the billing codes are largely independent of the specific lab values, and thus, can be seen as an outside test of the validity of the TDMI analysis.
How our method addresses nonstationarity
At various points in this paper, we have alluded to how nonstationarity is addressed within our framework. To be more explicit, consider three cases: (1) a single nonstationary source, (2) multiple different stationary sources, and (3) multiple different nonstationary sources. Relative to case (1), because there is diversity in the data sets, δI ≈ 0, , and —thus, there will be no distinction between stationarity and nonstationarity. Case (2) is the case we handled in Sec. 9A and does not need explanation. And case (3) will behave identically to case (2); nonstationary will be difficult to detect, but multiple different statistical states will be detectable. While it might be too much to ask to be able to distinguish nonstationarity amongst a population from a population with multiple stationary sources, we can detect nonstationarity within an individual, given enough data points. In particular, relative to case (1), the reason why all the diagnostics fail to detect multiple statistical states is that there is no concept of averaging over a population. To address this issue, one only needs to partition the single time series into multiple pieces (of sufficient length), and then apply the standard TDMI analysis from this paper to the new “population” of time series. Said differently, to detect nonstationarity in a single source, one only needs to treat the single source as multiple sources and apply our machinery; if it appears that there are multiple sources, then you know that the single source has multiple statistical states and is thus nonstationary.
Comments regarding the connection between the supports and the normalizations of the distributions
In a sense, all support-based variation amongst the population could be eliminated by normalizing all individuals to some standard support (or to a distribution with mean zero and variance one). We did not implement this because sometimes the normalization of the support matters with respect to the composition of the population, and we wanted to allow for the TDMI infrastructure to capture this type of dependence. Relative to the example in this paper, having glucose oscillate around 500 means the patient is very sick, whereas glucose oscillation around 100 means the patient is likely healthy (at least from a blood glucose perspective)—we wanted to be able to capture this type of heterogeneity. That said, if one begins with a normalized population and performs the TDMI analysis, any δI must exist because of variation in the graphs of the PDFs. However, if one has enough points per patient to estimate , one knows this anyway upon calculating and ; when there are not enough points to estimate I for every individual, then deducing temporal, graph-based variation is difficult.
Future directions regarding the use of this technique
One of the sources of motivation for performing this calculation is based on the idea of stratifying or clustering populations of individuals by their predictive information. Based on the TDMI infrastructure here, we have identified at least 3 different subpopulations based on their predictive information structure. Thus, future computational problems will involve developing and testing a more automated form of this interpretive structure that can be used for generating hypothesized sub-categories of individuals and eventually an infrastructure that can be integrated with classification and clustering schemes.
Some remaining statistical problems
In this work, we attempted to outline and show, mathematically, how to interpret the TDMI and information entropy for aggregated populations. Nevertheless, there are many problems that remain unresolved. In particular, a partial list might include full rigorous proofs regarding: the technical conditions under which our claims (i.e., δI > 0 if an only if for some i) apply; the convergence properties of various quantities we propose (i.e., δI, . etc); and the full relationships between what the information entropy and TDMI can imply about one another.
SUMMARY
We have fashioned a methodology for computing and interpreting the TDMI for a nonuniform population of time series. Within this methodology, one of the means of estimating the TDMI for a population of time series allows for a meaningful TDMI estimate to be performed on a system that has many poorly measured components, none of which support a TDMI calculation. Specifically, given a population of time-series that are: non-uniformly measured in time, of diverse lengths, from statistically diverse sources, and pathologically sparse, our methods will likely still yield interpretable results. Achieving such a methodology was the original motivation for this work. An explicit prescription for interpreting I for a fixed time separation δt for a population can be found in Fig. 2 within Sec. 8. Moreover, an algorithmic portrayal can be found in Appendix B.
The process of interpreting the TDMI for a population revealed a way to quantify the degree of diversity within a population, should the population of time series have diverse sources. Specifically, our methodology gives a way of detecting and quantifying temporal heterogeneity (or homogeneity) in a population, given enough data. Moreover, our methodology allows for the calculation of statistical quantities such as bias (which is built into the methodology) and statistical significance of the TDMI signal (which is easily estimated via standard methods such as applying a bootstrap technique).
Broadly, these results have two primary consequences. First, our methodology extends the circumstances under which temporal nonlinear correlation for real world, complicated data sets can be calculated in two broad directions: (1) populations of time series where the sources are not necessarily the same and (2) poorly sampled collections of time series from diverse or identical sources. Second, the act of interpreting the TDMI for a population with an unknown temporal makeup yields three advancements: (1) the population can be stratified by their temporal structure; (2) the population can be categorized as homo or heterogeneous when this information was previously unknown; and (3), because of (2), it is possible to reduce the number of confounding factors that generate the signal.
We have expended substantial effort to make this framework as easy to use as possible. Nevertheless, as is the case with nearly all time series techniques, one must use care and intelligence when using this methodology. Aside from the standard correlation is not causation issues that arise when using the TDMI even for an individual, there are some hazards that are worth mentioning or recalling.
First, whenever one aggregates a population that is possibly diverse, diversely measured, and potentially non-stationary, temporal signals can arise simply through the act of aggregation. Thus, one must always be mindful of this very real issue, and be vigilant in working to understand the nature of the source of any given signal that appears as δt is varied.
Second, the methodology-specific sources of confounding factors, such as effects from variation in the representative population as δt is varied, are covered in this paper. Nonetheless, it is easy to imagine a large number of application-specific confounding factors (i.e., factors that are a function of specific details of the system being studied) that could lead to signals being generated when aggregating an uncontrolled population of time series.
Third, one must have, at some fundamental level, enough data to make robust and believable PDF estimates; how much data is enough data depends on the details of the particular system being studied. We gave a rough lower bound of 100 pairs of points being necessary to estimate to . We have found that, upon bootstrapping and observing PDF estimates, 100 points appears to be the minimum number of points required to get stable PDF estimates. However, if the support of the distributions is extremely wide, 100 pairs points may be far too few. Similarly, if the distribution is bimodal where the modes are clearly separated, fewer than 100 pairs of points may be required. It is, therefore, important to observe the PDFs being generated for all the estimates, and sometimes, perform a bootstrapping analysis on the population can be used to estimate a confidence interval (in particular, for ), and the bias estimate techniques can give insight into the meaning, accuracy, and precision of the results.
And fourth, when possible, once one has a TDMI-based interpretation, it can be useful to corroborate the interpretation with other available data. For instance, we verified or crosschecked our results with other data sets (e.g., the billing codes) to help interpret the representative sets of humans.
One final note; our methodology was designed for application in complex situations with complex data sets. In these situations, it is especially important to use common sense, context, and observation of the computational process to fully interpret the numbers the framework produces.
ACKNOWLEDGMENTS
The authors would like to thank two anonymous reviewers, J. Dias, N. Elhadad, A. Perotte, and D. Varn for carefully reading this paper and providing many useful comments. D.J.A. would like to thank C. Shalizi for early discussions related to this work. Finally, the authors would like to acknowledge the financial support provided by NLM Grant No. RO1 LM06910.
APPENDIX A: DETAILED AVERAGE TDMI CALCULATION
Begin by recalling the definition of the average TDMI
| (A1) |
Next, recall that for the average TDMI, we have PDFs defined entirely with respect to the abstract support, . In this situation, we define the ith PDF relative to the “average” PDF, p1, by
| (A2) |
where is distance between the graphs of p1 and pi at a given value in . Next, for convenience, define the following: p(Xi(j), Xi(j − τ)) = p(j,τ), p(Xi(j)) = p(j), p(Xi(j − τ)) = p(τ), , , , and . With this notation, we can now re-write the integrand in Eq. A1
| (A3) |
| (A4) |
Next, factoring out of the summation term, one arrives at
| (A5) |
| (A6) |
| (A7) |
Multiplying and collecting terms under the sum, one obtains
| (A8) |
| (A9) |
| (A10) |
| (A11) |
| (A12) |
| (A13) |
| (A14) |
| (A15) |
where can be shown to have the more digestible form
| (A16) |
APPENDIX B: PSEUDOCODE FOR INTERPRETING THE TDMI FOR A POPULATION OF TIME SERIES
Algorithm 1:
How to interpret the TDMI for a population of time series
| if there are enough points to estimate (usually ∼100 pairs of points per representative individual are required) |
| then, |
| estimate δI and HΘ |
| ifδI > BIRPthen |
| the population is heterogeneous |
| ifthen |
| supports (or ranges) are diverse or disjoint |
| elseif, then |
| supports (or ranges) are uniform |
| endif |
| elseifδI ≤ BIRP, then |
| the population is homogeneous |
| endif |
| ifHΘ ∼ 0, then |
| the population is well represented |
| elseifHΘ ∼ 1, then |
| the portions of the population are overrepresented |
| endif |
| else if not enough pairs to estimate , then |
| estimate , , and HΘ |
| if, then |
| supports (or ranges) are diverse or disjoint |
| if there are enough pairs of points per patient to estimate a PDF for each patient at the specific δt,then |
| (i.e., V(p) relative to the abstract supports) |
| if, then |
| the population used to estimate has graph-based heterogeneity |
| elseif, then |
| the population used to estimate is graphically homogeneous |
| endif |
| elseif it is not possible to accurately estimate a PDF for each patient at the specific δt,then |
| it is not possible to determine the contribution of the graph-based heterogeneity to the overall heterogeneity |
| endif |
| elseif, then |
| supports (or ranges) are uniform |
| if, then |
| the population used to estimate has graph-based heterogeneity |
| elseif, then |
| the population used to estimate is homogeneous |
| endif |
| ifHΘ ∼ 0, then |
| the population is well represented |
| elseifHΘ ∼ 1, then |
| the portions of the population are overrepresented |
| endif |
| {NOTE: there are 10 possible sharp interpretations for both δI and -only cases.} |
| {All TDMI interpretations should include: I-like quantities (e.g., , δI, etc), population diversity qualification (support- and graph-based contributions to diversity; if they are unknown, this should be specified), and the make-up of the population used to estimate the I-based quantities (e.g., HΘ.}. |
| {NOTE: even under the best circumstances, it may be difficult to determine what proportion of the heterogeneity is due to support-based versus graph-based diversity.} |
References
- Komalapriya C., Thiel M., Ramano M. C., Marwan N., Schwarz U., and Kurths J., Phys. Rev. E 78, 066217 (2008). 10.1103/PhysRevE.78.066217 [DOI] [PubMed] [Google Scholar]
- Sprott J. C., Chaos and Time-series Analysis (Oxford University Press, New York, 2003). [Google Scholar]
- Kantz H. and Schreiber T., Nonlinear Time Series Analysis, 2nd ed. (Cambridge University Press, UK, 2003). [Google Scholar]
- Hogan W. and Wagner M., J. Am. Med. Inform Assoc. 5, 342 (1997). 10.1136/jamia.1997.0040342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lei J., Methods Inf. Med. 30, 79 (1991). [PubMed] [Google Scholar]
- Sagreiya H. and Altman R. B., J. Biomed. Inf. 43, 747 (2010). 10.1016/j.jbi.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. M. and Mahadevan L., Proc. Natl. Acad. Soc. U.S.A. 107, 20587 (2010). 10.1073/pnas.1012747107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shudo E., Ribeiro R. M., and Perelson A. S., J. Viral Hepat. 15, 357 (2008). 10.1111/j.1365-2893.2007.00954.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M. S., Phys. Today 62, 8 (2009). 10.1063/1.3226778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scargle J. D., Astrophys. J. 263, 835 (1982). 10.1086/160554 [DOI] [Google Scholar]
- Baisch S. and Bokelmann G. H. R., Comput. Geosci. 25, 739 (1999). 10.1016/S0098-3004(99)00026-6 [DOI] [Google Scholar]
- Schulta M. and Stattegger K., Comput. Geosci. 23, 929 (1997). 10.1016/S0098-3004(97)00087-3 [DOI] [Google Scholar]
- Liew A. W. C., Xian J., Wu S., Smith D., and Yan H., BMC Bioinf. 8, 137 (2007). 10.1186/1471-2105-8-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman L., All of Statistics: A Concise Course in Statistical Inference, (Springer, New York, 2004). [Google Scholar]
- Loéve M., Probability Theory I (Springer-Verlag, 1977). [Google Scholar]
- Gray A. G. and Moore A. W., “Very fast multivariate kernel density estimation using via computational geometry,” in Joint Stat. Meeting (August 4th, 2003).
- Moon Y.-I., Rajagopalan B., and Lall U., Phys. Rev. E 52, 2318 (1995). 10.1103/PhysRevE.52.2318 [DOI] [PubMed] [Google Scholar]
- May R. J., Dandy G. C., Maier H. R., and Fernando T. M. K. G., “Critical values of a kernel density-based mutual information estimator,” in International Joint Conference on Neural Networks (IEEE, Vancouver, BC, 2006).
- Albers D. J. and Hripcsak G., Estimation of time-delayed mutual information from sparsely sampled sources, e-print arXiv:1110.1615, 2011. [DOI] [PMC free article] [PubMed]
- Wheeden R. L. and Zygmund A., “Measure and integral,” in Monographs and Textbooks in Pure and Applied Mathematics (Marcel Dekker, Inc., New York, 1977), Vol. 43. [Google Scholar]
- Basharin G. P., Theor. Probab. Appl. 4, 333 (1959). 10.1137/1104033 [DOI] [Google Scholar]
- Roulston M. S., Physica D 125, 285 (1999). 10.1016/S0167-2789(98)00269-3 [DOI] [Google Scholar]
- Graxzyk J. and Światek G., Ann. Math. 146, 1 (1997). 10.2307/2951831 [DOI] [Google Scholar]
- Jakobson M., Commun. Math. Phys. 81, 39 (1981). 10.1007/BF01941800 [DOI] [Google Scholar]
- Albers D. J. and Hripcsak G., Phys. Lett. A 374, 1159 (2010). 10.1016/j.physleta.2009.12.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers D. J. and Hripcsak G., Using population scale EHR data to understand and test human physiological dynamics, e-print arXiv:1110.3317, 2011.
- It may seem odd to normalize indices, but this just keeps the domain of Θ̃ between zero and one.
- To see the variation in the PDF estimates due to small sample sizes, observe the PDF estimates for different sets of uniform random numbers with small cardinality.
- Note, the L1 difference is not technically a distance function or a metric because it does not satisfy the triangle inequality.