Abstract
Objective
Renal transplantation has dramatically improved the survival rate of hemodialysis patients. However, with a growing proportion of marginal organs and improved immunosuppression, it is necessary to verify that the established allocation system, mostly based on human leukocyte antigen matching, still meets today's needs. The authors turn to machine-learning techniques to predict, from donor–recipient data, the estimated glomerular filtration rate (eGFR) of the recipient 1 year after transplantation.
Design
The patient's eGFR was predicted using donor–recipient characteristics available at the time of transplantation. Donors' data were obtained from Eurotransplant's database, while recipients' details were retrieved from Charité Campus Virchow-Klinikum's database. A total of 707 renal transplantations from cadaveric donors were included.
Measurements
Two separate datasets were created, taking features with <10% missing values for one and <50% missing values for the other. Four established regressors were run on both datasets, with and without feature selection.
Results
The authors obtained a Pearson correlation coefficient between predicted and real eGFR (COR) of 0.48. The best model for the dataset was a Gaussian support vector machine with recursive feature elimination on the more inclusive dataset. All results are available at http://transplant.molgen.mpg.de/.
Limitations
For now, missing values in the data must be predicted and filled in. The performance is not as high as hoped, but the dataset seems to be the main cause.
Conclusions
Predicting the outcome is possible with the dataset at hand (COR=0.48). Valuable features include age and creatinine levels of the donor, as well as sex and weight of the recipient.
Keywords: Machine learning, statistics, bioinformatics, promoter detection, transplantation outcome, nephrology
Introduction
Compared with hemodialysis, renal transplantation has dramatically improved the survival rate of patients with end-stage renal disease (ESRD). Due to demographic changes, ESRD's incidence has increased in most Western countries, and a growing number of grafts are required. In Austria, Belgium, Croatia, Germany, Luxembourg, The Netherlands and Slovenia, the organization Eurotransplant is responsible for organ procurement.1 Donor–recipient allocation is performed via Eurotransplant's kidney allocation system (ETKAS2), a scoring system based on the number of human leukocyte antigen (HLA) mismatches, the time on dialysis, the distance from the explant site to the transplantation center and additional factors that favor balanced national import/export rates. Particular groups of patients, such as children and patients in a critical condition or with a low chance of finding an adequate organ, receive extra points. Specific allocation procedures were further added for highly sensitized patients (Acceptable Mismatch Program) and for the elderly (Eurotransplant Senior Program). The latter has led to a remarkable increase in available donors,3 but the number of marginal organs has grown equally.
A broad range of factors are known to influence the allograft survival, such as the number of HLA mismatches,4 the cold ischemia time (CIT),5 the number of previously received transplants,6 the age of the donor7 and a history of hypertension,8 diabetes,9 or obesity10 in the recipient. However, there is still no technique to predict an allograft outcome reliably, and the clinical decision of accepting an organ is in the physician's hands.
The future challenge for organ procurement is to predict the outcome of transplantation with high accuracy using the donor–recipient data available in order to allocate an adequate organ to each patient.
Background
A number of authors, generally with the same mathematical methods, have tried to predict the outcome of transplantation and have reported very different performances, indicating a strong variability in the quality of the datasets. Most literature predicts a binary outcome using logistic regression and neural networks (NNs) on the same core of features: donor's and recipient's clinical details (age, sex, weight, medical/viral history, etc), CIT, and HLA mismatches.11 12 The origin of the data, the number of samples, a few extra features and the chosen outcome make up most of the difference. As an example, Shoskes et al13 had 100 training transplantations with extensive medical data from donors and recipients, as well as CIT and HLA matching to predict delayed graft function (DGF). They tested their NN on 20 transplantations, and achieved 80% accuracy. However, the number of parameters seems far too high for the model to be trained properly. Brier et al14 also predicted DGF using 198 transplantations with the following variables: donor's sex–race, recipient's age–sex–height–weight–body surface area–race, CIT and HLA matching. They tested their NN on 106 transplantations, and achieved 30% sensitivity and 70% specificity. Shadabi et al15 used 896 transplantations and 23 variables: extensive medical data from donor and recipient, as well as CIT and HLA matching, to predict rejection after two years. They tested their NN on 448 transplantations, and achieved about 62% accuracy. Lin et al16 predicted graft survival after 1 year with 57 389 transplantations using 10-fold cross-validation and 71 variables: extensive medical data from donor and recipient, as well as CIT and HLA matching. They achieved an area under the curve (AUC) of 0.73. This is probably the largest database used in this kind of application. Krikov et al17 predicted the allograft survival after a different number of years using decision trees trained on about 60 000 transplantations. Based on 30 000 test transplantations, they achieved an AUC of 0.64 for 3-year graft survival. Akl et al18 predicted 5-year graft survival of living-donor kidney transplants using NNs trained on 1500 transplantations with 11 features including donors' and recipients' ages, HLA haplotype, and number of acute rejections. On 319 test transplantations, they achieved an AUC of 0.88. Note that most studies found in the literature predict a binary variable, denoting success or failure of the transplant.
In this work, the goal is to establish a link between the data available before transplantation (measurements on the donor and the recipient) and the estimated glomerular filtration rate (eGFR) of the recipient one year after surgery, and to identify which method performs best. It is worth mentioning that the eGFR is real-valued, making the task harder, however predicting an interpretable real quantity may be more helpful than a binary success / failure prediction. A principled machine-learning approach is considered, and a thorough study is conducted. Our data comprise 707 transplantations performed at Charité-Universitätsmedizin Berlin (Campus Virchow-Klinikum) between 1998 and 2008. To predict the eGFR after one year, we use linear regression (LR), support vector machines (SVMs) with a Gaussian kernel (G-SVMs), NNs and random forests (RFs).
Methods
Datasets
Donors' data were obtained from Eurotransplant's database2 using all the features available at the time of allocation. Recipients' and outcomes' data were retrieved from Charité's transplant database. All (first or repeated) single renal transplantations performed between 1998 and 2008 were listed, but for an optimal simulation of organ-procurement procedures, grafts from living donors were excluded from the analysis.
Transplantations were labeled with the eGFR of the recipient, that is, the volume of fluid filtered by the renal glomerular capillaries into the Bowman capsule per time unit, one year after the transplant was received. The eGFR was computed using the Modification of Diet in Renal Disease (MDRD) formula.19 This quantity is real-valued so regression is performed, as opposed to classification. Since each patient's sex and age are already known before surgery, what is effectively predicted is the level of creatinine.
All the features with more than 50% missing values (except diabetes information) and all the transplantations for which the patient's eGFR was missing were removed, leaving in total 707 transplantations described by 56 features, listed in table 1. Missing values were filled in with predictions, and the completed data were subsequently standardized, that is, every feature has its mean set to 0 and its variance set to 1. Exact details can be found in appendix section 1.
Table 1.
Variables
| Variable | Type | Missing values | |
| Estimated glomerular filtration rate (outcome) | Real | 0 | |
| 1 | Donor's age | Real | 0 |
| 2 | Donor's sex | Binary | 0 |
| 3 | Donor's blood type | Discrete | 0 |
| 4 | Donor's weight (kg) | Real | 0 |
| 5 | Donor's height (cm) | Real | 1 (0.1%) |
| 6 | Donor's hepatitis B | Binary | 4 (0.6%) |
| 7 | Donor's hepatitis C | Binary | 4 (0.6%) |
| 8 | Donor's cytomegalovirus | Binary | 7 (1%) |
| 9 | Donor's sodium (mmol/l) | Real | 3 (0.4%) |
| 10 | Donor's potassium (mmol/l) | Real | 1 (0.1%) |
| 11 | Donor's glucose (mg/dl) | Real | 28 (4%) |
| 12 | Donor's creatinine (mg/dl) | Real | 4 (0.6%) |
| 13 | Donor's urea (mg/dl) | Real | 14 (2%) |
| 14 | Donor's cause of death | Discrete | 4 (0.6%) |
| 15 | Recipient's age | Real | 0 |
| 16 | Recipient's sex | Binary | 0 |
| 17 | Recipient's blood type | Discrete | 0 |
| 18 | Recipient's weight (kg) | Real | 20 (2.8%) |
| 19 | Recipient's height (cm) | Real | 10 (1.4%) |
| 20 | Recipient's hepatitis B | Binary | 2 (0.3%) |
| 21 | Recipient's hepatitis C | Binary | 3 (0.4%) |
| 22 | Recipient's cytomegalovirus | Binary | 7 (1%) |
| 23 | Recipient's previous transplants | Real | 71 (10%) |
| 24 | Recipient's previous heart transplants | Real | 71 (10%) |
| 25 | Recipient's previous liver transplants | Real | 71 (10%) |
| 26 | Recipient's previous lung transplants | Real | 71 (10%) |
| 27 | Recipient's previous kidney transplants | Real | 71 (10%) |
| 28 | Cold ischemia time | Real | 0 |
| 29 | HLA mismatches (broad) | Real | 0 |
| 30 | HLA mismatches (split) | Real | 0 |
| 31 | HLA mismatches (A broad) | Real | 0 |
| 32 | HLA mismatches (A split) | Real | 0 |
| 33 | HLA mismatches (B broad) | Real | 0 |
| 34 | HLA mismatches (B split) | Real | 0 |
| 35 | HLA mismatches (DR broad) | Real | 0 |
| 36 | HLA mismatches (DR split) | Real | 0 |
| 37 | Donor's smoking | Binary | 182 (25.7%) |
| 38 | Donor's number of packs a year | Real | 348 (49.2%) |
| 39 | Donor's diabetes | Binary | 402 (56.9%) |
| 40 | Donor's diabetes treated | Binary | 408 (57.7%) |
| 41 | Donor's diabetes duration | Real | 414 (58.6%) |
| 42 | Donor's hypertension | Binary | 193 (27.3%) |
| 43 | Donor's hypertension treated | Binary | 304 (43.1%) |
| 44 | Donor's hypertension duration | Real | 414 (58.6%) |
| 45 | Donor's days (intensive care unit) | Real | 202 (28.6%) |
| 46 | Donor's last 24 h diuresis | Real | 230 (32.5%) |
| 47 | Donor's urine erythrocytes | Real | 242 (34.2%) |
| 48 | Donor's urine glucose | Real | 283 (40%) |
| 49 | Donor's urine protein | Real | 283 (40%) |
| 50 | Donor's kidney ultrasound | Discrete | 295 (41.7%) |
| 51 | Donor's Hb (g/dl) | Real | 221 (31.3%) |
| 52 | Donor's leucocytes (nl) | Real | 219 (31%) |
| 53 | Donor's C-reactive protein (mg/dl) | Real | 267 (37.8%) |
| 54 | Donor's pH | Real | 281 (39.7%) |
| 55 | Donor's O2 pressure (mm Hg) | Real | 279 (39.5%) |
| 56 | Donor's O2 saturation | Real | 286 (40.5%) |
The 56 imput variables, together with the outcome variable (estimated glomerular filtration rate). Details are provided in appendix section 1.
HLA, human leucocyte antigen.
Out of this matrix, two datasets were created. The full dataset, with all 56 features, has dimensions of 707×75. The robust dataset, with the 36 features that originally contained <10% missing values, has dimensions of 707×48. The difference between the number of features and the number of columns stems from the discrete features being binarized.
Regressors
To learn a model that predicts the eGFR, four different regressors were considered. The first two, LR and NNs, are the most commonly used in this kind of application, though NNs were reported to perform better.
LR is a linear model that assumes the targets follow a Gaussian distribution. A prediction on a transplantation x is made using y(x)=wTx, where w is the weight vector being learned. LR was implemented using the R function lm.
NNs20 can be represented mentally by a layered loop-free network of linear models. This alleviates the linearity of LR, thereby allowing any function to be learned. NNs were implemented using the R package nnet.
The other two, SVMs and RFs, are relatively new in the field of transplantation.
SVMs21 are also related to linear models. However, instead of building networks, they suppress linearity by changing the space of action.22 Indeed, if we apply a transformation φ to our data before training the regressor, then the function is linear in φ (x) but no longer in x. The space can be changed explicitly by defining φ or implicitly via a kernel. We chose a Gaussian kernel. The similarity between two points is given by , where σ is the width of the kernel. G-SVMs were implemented using the toolbox shogun.23
RFs24 are also non-linear regressors. A random forest is a collection of binary trees, where each node is associated with a test on a feature, and each leaf contains a different eGFR prediction. In each tree, a datapoint falls into a particular leaf depending on its features, and is assigned a prediction. The datapoint's different predictions are then averaged. RFs have a built-in feature-selection system, and allow for joint features (it is not only an additive model but also a multiplicative one). RFs were implemented using the R package randomForest.
Since data were scarce, we used 10-fold cross-validation to obtain a better estimate of the performance. A resulting regressor is therefore made of 10 subregressors, each tested on a different 10% of the data (and trained on the remaining 90%). Moreover, each sub-NN and sub-G-SVM had parameters that needed tuning, so we used nested cross-validation, which is thought to be a solid estimator of the true error when parameters have to be selected.31 Details on (nested) cross-validation may be found in appendix section 2. It is important to understand that every subregressor is tested on unseen data, and that this unseen data is different for each subregressor. The regressor's performance is the average of these 10 test performances.
In addition to RFs, we investigated two regressors that allow for joint features: SVMs with a polynomial kernel (P-SVMs) and multivariate adaptive regression splines (MARSs). MARSs also have the advantage of performing piecewise regression, thereby accounting for potential subgroups in the data. The results for these two regressors are only reported in appendix section 8.
Feature selection
Several subsets of features were used, some preselected by hand, some preselected automatically, and some selected automatically during training. The subsets preselected by hand are:
The donor's age, because it is the most informative variable.
The number of HLA mismatches and the CIT, because they are the main variables used by ETKAS. This set of variables performed very poorly.
To preselect features automatically, principal-component analysis (PCA) and regressional Relief-F were run:
PCA is a statistical method used to reduce dimensionality. It aims at finding the subspace of dimension M<D that preserves the most variance in the data (thereby also reducing noise). The data are then projected onto this subspace, which gives a new representation using M variables instead of D. PCA was run on each dataset, and M was chosen so as to retain at least 65% of the total variance (34 for the full dataset, 23 for the robust one).
Relief-F is a filter method and selects features that take different values in different classes and similar values within the same class, in other words features that are discriminant. Regressional Relief-F25 is the version of Relief-F for regression problems. Note that Relief-F uses the information of the targets so, unlike unsupervised methods such as PCA, it cannot be run on the whole dataset. Instead it is run on the training set, separately for each fold.
To select features automatically during training, forward feature selection (FFS), recursive feature elimination (RFE) and L1 regularization were implemented. Because cross-validation was performed, feature selection was repeated for each fold on the training set only, using another level of cross-validation. Each fold therefore induced a different subset of variables, which came in handy when assessing the reliability of these selected variables.
FFS is a wrapper method and was performed on LR. Indeed, being the simplest, LR was the most likely to bear insufficient data and greedy algorithms, and thus to select reliable features. Once chosen, the features were used to train any type of regressor (referred to as as LR-selected features). The principle of FFS and pseudo-code are given in appendix section 3.
RFE26 is an SVM-specific wrapper method and was performed on G-SVM. A G-SVM a is learned so as to minimize the margin-related cost function aTKa, where K is the Gaussian kernel. An irrelevant feature is expected not to change the cost function much (ie, not to degrade the margin much). For each feature f in F, a kernel K′ is built on F\{f}, and the feature minimizing aT Ka−aT K′a is then discarded. The principle of RFE and pseudo-code are given in appendix section 4.
L1 regularization penalizes the L1-norm of a vector, essentially forcing its entries to be 0 except where absolutely necessary. This technique combined with LR is called Lasso and penalizes the regression weights. For G-SVMs, L1 regularization was used via multikernel learning,27 whereby one subkernel per feature is created. The resulting kernel is then a weighted sum of all subkernels, where the weights are subject to the L1-norm.
Performance evaluation
All models were trained to minimize the mean square error between predictions and real targets (given eGFR). However, for clarity, the Pearson correlation coefficient between predictions and real targets (COR) is reported. Reporting the performance of our cross-validated regressors means averaging the performance of each subregressor on its test set over the 10-folds, and plotting it with error bars representing the standard deviation of this performance. All results are therefore reported on test sets, that is, on data that were not used for training.
Results
The goal of this study is to predict the eGFR of a patient one year after transplantation, and to identify which model can do it best. The workflow is summarized in figure 1. A different model was trained for each combination of dataset/regressor, once with all features, once with LR-selected features, once with RFE when available (G-SVMs), once with L1 regularization when available (LR and G-SVMs), once with PCA, Relief-F, once with HLA+CIT and once with the most predictive feature (donor's age). The combination HLA+CIT is studied because it has the main variables used by ETKAS. Although ETKAS does not aim at predicting the eGFR, it allocates a graft to a patient, so its features constitute a very reasonable base to compare our dataset with. However, HLA+CIT performed so poorly (maximum correlation of 0.11) that it was discarded from the plots.
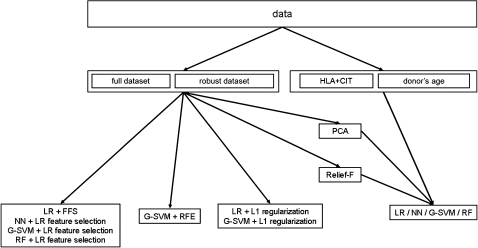
Figure 1.
Workflow. Out of the complete data, two datasets are built and referred to as full and robust, and two subsets of features are extracted: human leucocyte antigen (HLA)+cold ischemia time (CIT) and donor's age (middle row). The four regressors are run on each of these datasets (bottom row, rightmost box). Additionally, the four regressors are applied on the full and robust datasets with principal component analysis (PCA) and Relief-F, L1 regularization (bottom row, third box), recursive feature elimination (RFE) (bottom row, second box), and linear regression (LR) feature selection (bottom row, first box). Note that PCA and Relief-F are pretreatments, as opposed to L1 regularization, RFE, and LR feature selection that are part of the training. G-SVM, support vector machine with a Gaussian kernel; NN, neural network; RF, random forest.
The results, shown in appendix sections 5 and 6 show that:
It is unclear which dataset should be preferred, indicating that variables with many missing values do not bring much.
Unlike previously reported results, LR and G-SVMs consistently perform best.
A few features are sufficient
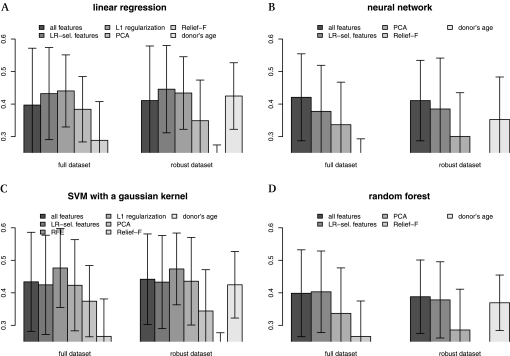
Figure 2 shows the impact of feature selection on performance. HLA+CIT did so poorly (maximum correlation of 0.11) that they are not reported.
Figure 2.
Effects of feature selection. The y-axis shows the Pearson correlation coefficient between predicted and given estimated glomerular filtration rate (eGFR). HLA+cold ischemia time (the 'Eurotransplant's kidney allocation system features') performed so poorly (COR=0.11) that they were discarded from the plots. In the case of linear regression (LR) (A) and support vector machines with a Gaussian kernel (G-SVMs) (C), feature selection with a wrapper is clearly the better choice: forward feature selection (FFS) for LR and recursive feature elimination (RFE) for G-SVM. In the case of neural networks (NNs) (B) and random forests (RFs) (D), using all features is the best option, but they do more poorly. LR and G-SVM benefit from feature selection, suggesting that only a few factors actually influence the outcome as investigated in section “Relevant features”. In particular, the donor's age, which is the best feature, carries most of the predictive power. However, it never performs best, indicating that it is not the only factor. Using principal-component analysis (PCA) or Relief-F as a filter for the features does not seem a good idea, since it consistently performs worse.
In the case of LR and G-SVM, feature selection with a wrapper is clearly the better choice: FFS for LR, and RFE for G-SVM. In the case of NN and RF, using all features is the best option, but they do more poorly.
LR and G-SVM benefit from feature selection, suggesting that only a few factors actually influence the outcome as investigated in section Relevant features. In particular, donor's age, which is the best feature, carries most of the predictive power. However, it never performs best, indicating that it is not the only factor.
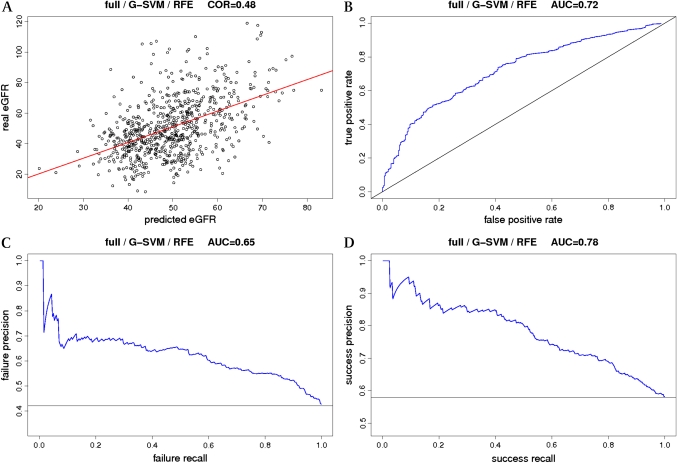
G-SVM with RFE is very effective. In fact, on the full dataset, it is the best regressor we could produce (COR=0.48). In the rest of the article, we will refer to it as full (F)-G-SVM-RFE. Figure 3 and appendix section 7 summarize all the information about F-G-SVM-RFE.
Figure 3.
Best model. This figure summarizes all the performance information on F-G-SVM-RFE, the model using a support vector machine with a Gaussian kernel (G-SVM) and recursive feature elimination (RFE) on the full dataset (F). To provide a more straightforward measure of performance of this regressor, the test data were divided according to the estimated glomerular filtration rate (eGFR) into two balanced classes: one with eGFR<45 ml/min and the other with GFR>45 ml/min, and the targets replaced by labels representing the classes. The task is now classification, and all common measures can be applied. A) scatter plot of real eGFR against predicted eGFR, together with the regression line. The correlation achieved is 0.486±0.12. B) receiver-operating characteristic (ROC) curve, together with the random performance (diagonal line). The achieved area under the ROC curve is 0.726±0.08. C) precision-recall (PR) curve for failure detection (eGFR<45 ml/min), together with the random performance (horizontal line). The area obtained under the PR curve is 0.656±0.12. D) PR curve for success detection (eGFR>45 ml/min), together with the random performance (horizontal line). The area obtained under the PR curve is 0.786±0.07. Additional performance measures: the accuracy is 0.686±0.07. For transplantation failure detection (eGFR<45 ml/min), the sensitivity is 0.516±0.08, the specificity 0.86±0.08, and the precision 0.656±0.13. For transplantation-success detection (eGFR>45 ml/min), the sensitivity is 0.86±0.08, the specificity 0.516±0.08, and the precision 0.696±0.06. Details on these measures and the PR curve may be found in appendix section 7.
Note that using PCA or Relief-F as a filter for the features does not seem a good idea at all, since it consistently performs worse. PCA does not aim at making predictions easier though; it simply tries to retain as much variance as possible in a lower-dimensional space, so it is not necessarily beneficial.
Relevant features
Figure 2 shows that the best model is F-G-SVM-RFE (COR=0.48), which strongly suggest that only a few variables are really necessary for the bulk of prediction. It is worth looking at these features in detail.
FFS was performed using LR and nested cross-validation on both datasets. The webpage shows, for the various training/test splits, the performance of the growing feature subset against the number of features. A different subset of features was obtained for each fold. The features appearing in at least 5 of these subsets are listed, which gives one list for the full dataset and one list for the robust dataset. The intersection of both lists contains the following robust features:
− for the donor: age (10/10), weight (10/10), glucose (10/10), hepatitis C status (10/10), creatinine (9/10) and hepatitis B status (9/10);
− for the recipient: weight (10/10), sex (10/10), number of previous heart transplants (10/10) and cytomegalovirus (7/10);
− CIT (10/10) and HLA mismatches (DR-broad and total-split) (9/10).
None of the non-robust features were found more than 5 times in the full dataset's list.
RFE was performed using G-SVM and nested cross-validation on both datasets. Following the procedure above, the following robust features were retrieved:
− for the donor: age (10/10) and death code (5/10);
− for the recipient: weight (8/10), height (5/10) and sex (5/10).
Additionally, among the non-robust features, the donor's ultrasound was found 5 times.
A ranking of the variables by importance is provided by the RFs. Every variable is taken in turn and permuted. This induces a new error for each variable, supposedly higher than the normal error. The feature leading to the highest difference with the normal error disrupts the regressor the most and therefore should be the most important. Features that came out 5 out of 10 times within the top 25/15 for the full/robust datasets were extracted:
− for the donor: age (10/10), death code (10/10), creatinine (10/10), height (9/10), sodium (9/10), urea (6/10) and weight (6/10);
− for the recipient: age (10/10), weight (10/10), height (9/10) and sex (5/10);
− HLA mismatches (B-broad (9/10) and total-split (6/10)).
Additionally, among the non-robust features, the donor's hypertension status (10/10), O2 pressure (10/10), O2 saturation (10/10), leukocytes (9/10), pH (8/10), C-reactive protein (7/10), diabetes status (6/10), and Hb (5/10) were found more than 5 times. However, RFs perform more poorly than the other regressors, therefore care must be taken when interpreting those results.
In all cases, the most important variable is the age of the donor. It is known to influence outcomes,7 28 but in our dataset it surprisingly outweighs the influence of the other established factors by far. Other variables that are present in all three selection methods, or that appear many times (>8 times) in at least two selection methods, are the donor's creatinine as well as the recipient's weight and sex.
Discussion
The analysis presented here simulates the process of decision-making that takes place when an organ is allocated, and is based exclusively on data provided by Eurotransplant at the time of organ allocation. As a result, it is very similar to what happens in reality and could be implemented in a clinical setting without much effort. A few differences with a physician's decision can be found. For example, some factors most probably influencing allograft function (such as recipient presensitization to alloantigen) were not taken into account. Moreover, while commonly used in kidney studies, the eGFR is biased by the influence of muscle mass on creatinine production. Even though the MDRD formula for eGFR reduces this bias, it cannot be said whether the recipient's variables (age, sex, height and weight) have a stronger impact on allograft function or on muscle mass. Clinicians have to consider a large number of parameters that may carry conflicting information, and an automatic prediction based on the most relevant factors may be of help when taking the decision. We hope to have made this help more valuable by keeping the experimental setting as close as possible to the real situations experienced at hospital. Our study identified the donor's age as the most important factor on allograft function. This may directly influence medical decision-making, if allocation programs were to increase the impact of this feature.
We performed a thorough analysis of the data at hand using four established regressors and different subsets of features, and were able to build a regressor that achieved a 0.48 correlation on our dataset. First, we want to emphasize the great care that was taken to ensure the machine learning was sound. Every experiment was carried out using 10-fold cross-validation, that is, each reported performance is an average and comes with error bars, so the results are much more reliable in this study than in most of the literature. Moreover, data that were used for selecting features or for tuning parameters were never used for testing. Indeed, features and parameters were chosen using cross-validation on the training set only, so each reported performance range should reflect the real performance.
We would also like to point out the benefits of running such an extensive analysis. When data are scarce, models might not behave as expected and it is not obvious which regressor should perform best, so it is more informative to show all the results rather than to report the best performance only. Our particular case shows that, even if we could identify a superior model, there is actually not much difference between the various set-ups, indicating that non-linearity is not critical for this dataset. For example, F-G-SVM-RFE is not significantly better than LR on the robust dataset with FFS (COR=0.48 vs COR=0.45; one-tailed Steiger test29: p=0.08) or than G-SVM on the robust dataset with RFE (COR=0.48 vs COR=0.45; one-tailed Steiger test: p=0.394). The models with a similar performance to F-G-SVM-recursive feature elimination are highlighted in appendix section 9.
Choosing the best outcome parameter is critical for the design of such a study. In renal transplantation, a variety of outcome parameters are regularly used, such as acute rejection, DGF, graft survival and renal functions (estimated GFR or, more rarely, measured GFR). Here the GFR estimated with the MDRD formula was chosen, because this formula is the base for the classification of chronic kidney diseases, and is associated with morbidity and mortality in ESRD. Additionally, the eGFR is a valid surrogate parameter for long-term graft survival.
The correlation obtained between predicted and real eGFR is 0.48 (R2=0.23), and the scatter plot in figure 3 looks very encouraging. The analysis only includes information available at the time of surgery, which means a small subset of all the possible parameters. Indeed, after transplantation, the patient is subject to a large number of influencing factors, such as occurrence of acute rejection, patient's adherence level to therapy, adverse effects of immunosuppression, infections, and so on. A perfect performance therefore can not be achieved. Additionally, the analysis only includes the grafts that were accepted for transplantation. Since Charité rejects roughly 10–20% of kidneys (due to organ quality, safety reasons, etc), our dataset is a biased selection.
It is tricky to compare our results to others'. The methods used in the literature (linear models and NNs) are also investigated here, but the critical differences between the various contributions lie in the data and in the outcome parameter which is usually binary as opposed to real-valued. Moreover, most published results are unclear, it is often hard to find critical details (such as which samples were included, whether the results are reported on training or test data, how many samples were in each set, etc), and every author uses a different measure of performance. However, when recast into a classifier (see figure 3 or appendix section 7 for details), our model had an area under the ROC curve of 0.72±0.08, which is fairly close to that of Lin et al16 (0.73), but with much fewer samples. It also achieved an accuracy of 0.68±0.07, which is better than that of Shadabi et al15 (0.62). For transplantation failure detection (eGFR<45 ml/min), which is the usual point of view in this kind of application, it achieved a sensitivity of 0.51±0.08 and a specificity of 0.8±0.08, which is better than that of Brier et al14 (0.3/0.7).
Part of the interest of this work was to extract important features. The variables that came out as important are known to influence the eGFR after transplantation. While donor's age and creatinine are directly related to the donor's renal function before transplantation, the influence of the recipient's weight is less evident. Some data suggest an adverse impact of donor/recipient weight mismatch.30 Our analysis reveals that the age of the donor is by far the strongest factor (included in the data) for allograft function. This is consistent with earlier published data.28 The clinical relevance of this variable suggests paying special attention to it when adapting allocation strategies. The minor impact of the HLA mismatches, which used to be regarded as one of the most important factors, probably reflects the higher efficiency of modern immunosuppressive agents in preventing graft rejection.
The dataset is the source of two limitations, which leaves room for improvement and hope for the future. First, the information content seems quite low. Indeed, the age of the donor contains much of the predictive power, which is odd in itself. Consequently, the flexibility of the model becomes optional rather than necessary, and G-SVMs are only neck and neck with LR. Furthermore, some error bars are quite wide, showing that performance is sensitive to a particular subset of the data. High variance is usually a sign of insufficient amounts of data. This can be fixed, as time will provide more samples.
Another limitation of this analysis lies in the imputation method. For now, missing values are predicted with linear models using complete variables. In our case, imputation should not be too harmful, though, as the robust dataset contains at least 90% real values. However, a much better and more elegant way would be to design a generative probabilistic model that would suppress the need for imputation. There are too many features and too few samples to design such a model reliably just yet, but as datasets grow larger, and dependencies between variables are better understood, generative graphical models should lead the way.
Conclusion
We obtained data from Eurotransplant and Charité to create a learning database in order to predict the outcome of a transplant for a given donor-patient pair. We had 707 transplantations, for which targets were the estimated glomerular filtration rate of the recipient one year after the transplantation. Each transplant was described using classic clinical data (weight, size, age, etc) as well as data specific to this kind of application (number of previous transplantations, creatinine levels, etc).
We built a regressor (COR=0.48) that performs much better than random (COR=0, one-tailed t test: p=2.87×10−7), than the features used by ETKAS (COR=0.11, one-tailed Steiger test: p=1.34×10−20) and than linear regression on all features (COR=0.4, one-tailed Steiger test: p=0.001), and we were able to extract a subset of features that were consistently picked up by several models, indicating that they may be the main factors influencing the outcome.
Renal function is subject to many factors after surgery, therefore perfect performance is unlikely to be achieved, even if all the presurgery factors were available. The accuracies reported in the literature are not high enough to be convincing to humans, but it is very hard to estimate how much better or worse clinicians perform, so the computational community should not be discouraged in its efforts. Additionally, more and more transplantations will be performed, and datasets will become larger and larger. This is hopeful for machine-learning techniques that generally benefit from a large amount of data, it should increase their performance, perhaps even to a level acceptable to humans.
Supplementary Material
Acknowledgments
JL would like to gratefully acknowledge funding from Max Planck Society—Fraunhofer Society. CH would like to gratefully acknowledge funding from Berlin School of Regenerative Therapies.
Footnotes
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Data sharing is subject to the approval of Charité Hospital, and is by no means guaranteed.
References
- 1.Oosterlee A, Rahmel A. Eurotransplant International Foundation Annual report The Netherlands: Eurotransplant International Foundaton, 2008. http://www.eurotransplant.org/cms/mediaobject.php?file=ar_2008.pdf (accessed 21 Jun 2011). 2008. [Google Scholar]
- 2.Mayer G, Persijn GG. Eurotransplant kidney allocation system (ETKAS): rationale and implementation. Nephrol Dial Transplant 2006;21:2–3 [DOI] [PubMed] [Google Scholar]
- 3.Smits JM, Persijn GG, van Houwelingen HC, et al. Eurotransplant Senior Program Centers Evaluation of the Eurotransplant Senior Program. The results of the first year. Am J Transplant 2002;2:664–70 [DOI] [PubMed] [Google Scholar]
- 4.Bartels MC, Otten H, van Gelderen BE, et al. Influence of HLA-A, HLA-B and HLA-DR matching on rejection of random corneal grafts using corneal tissue for retrospective DNA HLA typing. Br J Ophthalmol 2001;85:1341–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall CL, Sansom JR, Obeid M, et al. Agonal phase, ischaemic times, and renal vascular abnormalities and outcome of cadaver kidney transplants. BMJ 1975;3:667–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed K, Ahmad N, Khan MS, et al. Influence of number of retransplants on renal graft outcome. Transplant Proc 2008;40:1349–52 [DOI] [PubMed] [Google Scholar]
- 7.Resende L, Guerra J, Santana A, et al. Impact of donor age on renal allograft function and survival. Transplant Proc 2009;41:794–6 [DOI] [PubMed] [Google Scholar]
- 8.Cho YW, Cecka JM, Gjertson DW, et al. Prolonged hypertension (>10 years) is a significant risk factor in older cadaver donor renal transplants. Transplant Proc 1999;31:1283. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad M, Cole EH, Cardella CJ, et al. Impact of deceased donor diabetes mellitus on kidney transplant outcomes: a propensity score-matched study. Transplantation 2009;88:251–60 [DOI] [PubMed] [Google Scholar]
- 10.Armstrong KA, Campbell SB, Hawley CM, et al. Impact of obesity on renal transplant outcomes. Nephrology (Carlton) 2005;10:405–13 [DOI] [PubMed] [Google Scholar]
- 11.Matis S, Doyle H, Marino I, et al. Use of neural networks for prediction of graft failure following liver transplantation. Proceedings of the 8th Annual Symposium on Computer-Based Medical Systems. Lubbock, TX, 1995:133–40 [Google Scholar]
- 12.Petrovsky N, Tam SK, Brusic V, et al. Use of artificial neural networks in improving renal transplantation outcomes. Graft 2002;5:6–13 [Google Scholar]
- 13.Shoskes DA, Ty R, Barba L, et al. Prediction of early graft function in renal transplantation using a computer neural network. Transplant Proc 1998;30:1316–17 [DOI] [PubMed] [Google Scholar]
- 14.Brier ME, Ray PC, Klein JB. Prediction of delayed renal allograft function using an artificial neural network. Nephrol Dial Transplant 2003;18:2655–9 [DOI] [PubMed] [Google Scholar]
- 15.Shadabi F, Cox R, Sharma D, et al. Use of artificial neural networks in the prediction of kidney transplant outcomes. Knowledge-Based Intelligent Information and Engineering Systems 2004;3215:566–72 [Google Scholar]
- 16.Lin RS, Horn SD, Hurdle JF, et al. Single and multiple time-point prediction models in kidney transplant outcomes. J Biomed Inform 2008;41:944–52 [DOI] [PubMed] [Google Scholar]
- 17.Krikov S, Khan A, Baird BC, et al. Predicting kidney transplant survival using tree-based modeling. ASAIO J 2007;53:592–600 [DOI] [PubMed] [Google Scholar]
- 18.Akl A, Ismail AM, Ghoneim M. Prediction of graft survival of living-donor kidney transplantation: nomograms or artificial neural networks? Transplantation 2008;86:1401–6 [DOI] [PubMed] [Google Scholar]
- 19.Levey AS, Bosch JP, Lewis JB, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med 1999;130:461–70 [DOI] [PubMed] [Google Scholar]
- 20.Widrow B, Hoff ME. Adaptive switching circuits. Proceedings of the IRE Western Electronic Show and Convention. Vol. 4. Los Angeles, 1960:96–104 [Google Scholar]
- 21.Vapnik VN, Lerner A. Pattern recognition using generalized portrait method. Automation and Remote Control 1963;24:774–80 [Google Scholar]
- 22.Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. Proceedings of the 5th ACM Annual Workshop on Computational Learning Theory. Pittsburgh, PA, 1992:144–52 [Google Scholar]
- 23.Sonnenburg S, Raetsch G, Schaefer C, et al. Large scale multiple kernel learning. J Mach Learn Res 2006;7:1531–65 [Google Scholar]
- 24.Breiman L. Random forests. Mach Learn 2001;45:5–32 [Google Scholar]
- 25.Robnik-Sikonja M, Kononenko I. An adaptation of RELIEF for attribute estimation in regression. Proceedings of the International Conference on Machine Learning 1997:296–304 [Google Scholar]
- 26.Mao Y, Zhou X, Pi D, et al. Parameters selection in gene selection using Gaussian kernel support vector machines by genetic algorithm. J Zhejiang Univ Sci B 2005;10:961–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanckriet G, Cristianini N, Bartlett P, et al. Learning the kernel matrix with semi-definite programming. J Mach Learn Res 2004;5:27–72 [Google Scholar]
- 28.Moers C, Kornmann NS, Leuvenink HG, et al. The influence of deceased donor age and old-for-old allocation on kidney transplantation outcome. Transplantation 2009;88:542–52 [DOI] [PubMed] [Google Scholar]
- 29.Steiger JH. Tests for comparing elements of a correlation matrix. Psychol Bull 1980;87:245–51 [Google Scholar]
- 30.Goldberg R, Smits G, Wiseman AC. Long-term impact of donor–recipient size mismatching in deceased donor kidney transplantation and in expanded criteria donor recipients. Transplantation 2010;90:867–74 [DOI] [PubMed] [Google Scholar]
- 31.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics 2006;7:91–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.