Abstract
MicroRNAs, small non-coding RNAs, may act as tumor suppressors or oncogenes, and each regulate their own transcription and that of hundreds of genes, often in a tissue-dependent manner. This creates a tightly interwoven network regulating and underlying oncogenesis and cancer biology. Although protein-coding gene signatures and single protein pathway markers have proliferated over the past decade, routine adoption of the former has been hampered by interpretability, reproducibility, and dimensionality, whereas the single molecule–phenotype reductionism of the latter is often overly simplistic to account for complex phenotypes. MicroRNA-derived biomarkers offer a powerful alternative; they have both the flexibility of gene expression signature classifiers and the desirable mechanistic transparency of single protein biomarkers. Furthermore, several advances have recently demonstrated the robust detection of microRNAs from various biofluids, thus providing an additional opportunity for obtaining bioinformatically derived biomarkers to accelerate the identification of individual patients for personalized therapy.
Keywords: MicroRNA signatures, gene expression, biomarkers, bioinformatics, knowledge representations, uncertain reasoning and decision theory, languages and computational methods, prostate cancer, protein networks, pathway analysis, network modeling, machine learning, predictive modeling, statistical learning, privacy technology
Overview of microRNAs, classification, biology, and role in cancer
Since the awarding of the 2006 Nobel Prize in Medicine to the discoverers of microRNAs (miRNAs),1 there has been an enhanced interest in these molecules and an explosion of data. miRNAs are 19–24 nucleotides in length, and to date 1424 human mature and immature forms have been cataloged (http://www.mirbase.org).2 They are omnipresent, stable, and increasingly recognized as critical modulators of biological, in particular oncogenic, pathways. They are often tissue specific3 and developmental stage specific.4 miRNAs participate in RNA interference (RNAi) pathways, which affect the level of expression of mRNA transcripts. miRNAs, which have multiple targets in contrast with other RNAi molecules such as small interfering RNAs (siRNAs), induce degradation of specific mRNAs by bearing fully homologous target sites.5 miRNAs are located in fragile chromosome areas prone to deletions, amplifications, and translocations.6 They interact with their target mRNAs in multiple ways, and, although they only represent 1% of the human genome, it has been estimated that at least a third of the genome is probably regulated by miRNAs.7 Additionally, 60% of their 3′ untranslated region (UTR) target sites are evolutionary conserved, suggesting an equally conserved role for miRNAs.8 Most miRNAs are derived from precursor RNA and function by base pairing with their target mRNAs to modulate protein translation by directly preventing either mRNA repression or degradation typically through 3′ UTR binding. However, multiple other mechanisms have been biologically described and insufficiently bioinformatically modeled in an excellent review by Garzon et al9: they include binding to the 5′ UTR or the open reading frame or in fact activating, instead of repressing, translation. One miRNA can target up to 500 mRNAs, and multiple mRNAs can be targeted by multiple miRNAs.
Although miRNAs only modestly inhibit gene expression, they preferentially target hubs and bottleneck proteins, which amplifies their regulatory impact on dynamic protein networks.10 In oncology, some tumor suppressor miRNAs target multiple oncogenes,10 whereas others have been reported to be deleted in leukemia and subsequently found to be altered in multiple cancers.6 In fact, miRNA expression patterns have been demonstrated to have superior accuracy for classifying tumors of unknown origin compared with mRNA-based patterns.3 They have been shown to initiate carcinogenesis (so-called “oncomirs”) or drive progression of disease. Indeed, simply querying a database predicting miRNA targets using sequence alignment of miRNA and mRNA with the keyword “cancer” will return ∼20% of available human miRNAs.2 To put this in perspective, only ∼1% of the protein-coding genome has been directly implicated in cancer11; therefore, miRNAs are far more enriched for oncogenic pathways than their mRNA counterparts.
Despite our increased knowledge of the oncological functions of miRNAs and our ability to assay them, miRNA knowledge translation from the laboratory bench to the patient bedside has been limited at best. A simple PubMed search (performed May 1, 2011) with search terms “microRNA or miRNA” and “signature” returned only 310 hits, whereas a similar query using “mRNA or gene and signature” returned nearly 7000 entries. In this perspective, we contend that bioinformatics can and should play an essential role in bridging this translational gap. In particular, we explore how: (1) a similar length signature provides better genomic coverage than mRNAs; (2) the probability of a statistically and clinically significant finding is greater than from conventional mRNA-based results because of reduced dimensionality; (3) miRNA stability and ability to be extracted from multiple biofluids and pre-genomics era tumor banks are a boon to cancer researchers for validation and training samples.
Detection of miRNAs
A practical consideration in favor of miRNAs is their stability in many types of tissue. Old tumor banks consist primarily of formalin-fixed, paraffin-embedded (FFPE) tissue, as this was the method of choice for preserving tissue morphology. However, this procedure limits their usage in genomic studies. Formalin fixation causes cross-linking of tissue proteins with one another and other DNA and RNA molecules. Although the protein architecture is preserved, nucleic acid extraction is difficult. Furthermore, an analysis of RNA from FFPE specimens demonstrated modification of the RNA structure, rendering reverse transcription difficult.12 Consequently, for mRNA expression analysis, flash-frozen specimens have been the tissue of choice.
Fortunately, these limitations are not necessarily relevant for miRNA. miRNA extracted from FFPE specimens even after 10 years showed similar expression patterns to flash-frozen tissue.13 Even laser dissection of tumor from FFPE blocks after immunohistostaining has generated robust miRNA expression patterns.13 14 Further, miRNA quality does not correlate with RNA quality as assayed by capillary electrophoresis. Thus, tissue banks of FFPE specimens previously thought to be of insufficient quality for genomic profiling may be a treasure trove for high-throughput miRNA transcriptome profiling and subsequent bioinformatics analysis.
Equally exciting is the discovery that, unlike mRNAs which are rapidly degraded, miRNAs are not only found in tissue, but are ubiquitously found in all body fluids, including blood, urine, and saliva.15 In the future, miRNA biofluid profiling may become the de facto non-invasive standard for evaluating tumor status. Indeed, the first paper on plasma-based miRNAs demonstrated its ability to differentiate patients with prostate cancer from those who did not.16 Furthermore, it was recently demonstrated that blood-based miRNAs from historic tumor/blood banks could predict lung cancer recurrence 1–2 years before radiographic detection17 with similar prognostic abilities demonstrated in colon cancer.18 However, the concordance between plasma and tissue miRNAs may be poor.17 These plasma/serum biomarkers may be indicative of cellular processes that are not cancer-related, and thus identifying biofluid-based miRNAs that are tumor specific will be critical to proper interpretation.
Biomarker challenges in cancer
Cancer is an extremely heterogeneous disease. Not only does the derivative tissue (eg, breast, prostate) matter, but the origin of the transformed cells also has tremendous importance in tumor behavior and prognosis. There are multiple publications demonstrating that miRNA biomarkers are associated with these transformed cell subtypes. In breast cancer, miRNAs can classify estrogen receptor, progesterone receptor, and HER2/neu status, which are critical for treatment-related decisions.19 For a given cell of origin, the genetic alterations conferring its malignant phenotype are equally diverse. Epigenetic silencing, gene mutations, amplifications, and deletions are but a few of the modalities that alter the molecular pathways. Fortunately, miRNAs, too, are effective in predicting mutational status. Work by the Croce laboratory demonstrated that miRNA expression patterns could classify translocation-specific subtypes of multiple myeloma.20
As a general rule, tumors adapt to treatment (eg, chemotherapy) pressure and will select clonal variations of the tumor that have different activated pathways. For example, in biochemically recurrent prostate cancer, hormone therapy is effective in 80–90% of men; however, most men will have transcriptome changes and become resistant to this therapy in 1–2 years.21 Even assuming that one has somehow managed to select the proper medication for the patient, germ-line differences may affect drug metabolism and thus drug efficacy. By definition, a biomarker is a measured quality that is an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses.22 Achieving the goal of personalized medicine will require a multitude of biomarkers constantly capturing the changing state of the patient and the tumor.
As miRNAs are part of the transcriptome, they provide chronological snapshots of the transcriptome. In contrast, static germ-like markers, such as germ-line single-nucleotide polymorphisms, are not reflective of the current disease state, and thus their association or correlation with therapeutically important disease characteristics is likely to be more indirect. miRNAs are often biologically interpretable and may offer causal relationships to aspects of the somatic transcriptome that can be targeted by therapy.
Bioinformatics considerations in reducing the dimensionality of heterogeneous expression profiles
By definition, a transcriptome signature is a classifier that provides identification of a class of tumor (eg, good vs poor risk) based on the expression levels of the component mRNA or miRNA. In most cases, these signatures are derived by examining the differential expression of the transcriptome from discrete cancer states. In our experience, there may be upwards of 500–1000 significantly differentially expressed genes in each phenotypic comparison, constituting ∼4% (1000 of 25 000 transcripts) of the genome surveyed. For either technical or cost reasons, most researchers will eventually winnow down this list to a smaller set of 10–100 features (genes) by pruning among co-expressed groups of genes. This approach may well prioritize bystander or passenger genes rather than the co-expressed genes driving the oncogenesis. This said, these smaller signatures are really now only evaluating 0.2–0.4% of the mRNA transcriptome. As this reduction in dimensionality is generally performed with supervised learning algorithms on oftentimes limited datasets, these smaller feature sets can become unstable (their membership may change with repeated analysis), become less informative, or, more likely, become overfitted to the training data.23 Multiple researchers have demonstrated that short gene signatures are inherently unstable, and repeated permutations will choose different genes and are prone to making random associations.24 25 Further, gene expression signature classifiers have been shown to have poor overlap when developed independently in distinct datasets.26
With miRNA expression signatures, the typical length is between 15 and 100 features without the need for extensive post-processing. Although this signature length may seem comparable to the length of mRNA signatures mentioned above, there is a fundamental difference. The miRNA space is considerably smaller, and, at these gene signature lengths, there is coverage of up to 10% of the genomic space (100 out of ∼1000 mature miRNAs). Simply put, for a similarly sized signature, miRNA signatures provide better coverage and the increased probability of statistically more informative features.
As of the writing of this perspective, the cost to process an mRNA microarray for the entire human genome is equivalent to higher-quality real time-PCR (RT-PCR) based technology for profiling all known miRNAs. RT-PCR technology remains the gold standard for expression profiling because of its superior detection sensitivity, superior assay specificity, and wider linear dynamic range.27 28 Indeed, microarray expression data varied by as much as 60% when compared with RT-PCR-derived data.28 Therefore, for a similar cost, RT-PCR-derived differentially expressed miRNAs are more likely to be accurate with lower variance than their microarray mRNA counterparts. In other words, miRNA measures are both highly accurate and highly precise (reproducibility), while expression arrays yield mRNA measures with less precision (high variance on repeated measures). Therefore, even before statistical analysis is performed, miRNA profiling may provide a more reliable signal than mRNA via microarray.
What is often overlooked in the clinical literature regarding genomic signatures is that the statistical power for detecting differentially expressed features (miRNA or mRNA) is dependent on the genomic space that is being evaluated. Given a set of m features from an independently developed classifier, assume m0 are the differentially expressed features between paired samples in a validation dataset. Let m1 be the non-differentially expressed features. The number of classifier features that are declared differentially expressed (true positive (TP)) divided by the sum of TP and the number of differentially expressed features represents the sensitivity (TP/(TP+m1)). Similarly, TN/(TN+m0) represents the specificity where the true negative (TN) is the number of true null hypotheses correctly identified by the classifier. Thus, on the basis of this simple math, we can see that, in large transcriptome profiling data, the greater the number of features profiled, the lower the sensitivity and specificity and the greater the number of samples required.
This problem of dimensionality carries over to corrections for multiplicity. Inherently, with the greater number of features, there is a greater risk of a random false positive. Several methods have been developed to address this. For example, the popular Benjamini and Hochberg29 FDR approach controls this probability by finding a comparison-wise error level such that the error is proportional to the number of significant features and the desired overall p value from the observed dataset. More concretely, let us take, for example, differentially expressed genes from a paired microarray experiment. Assume that we would like to have an overall t test p value of 0.05 adjusted for multiple comparisons to derive a biomarker from a given feature. We would like to calculate the threshold for significance based on a straightforward Bonferroni-type correction.30 This consists of multiplying the unadjusted p value by the total number of features evaluated to obtain an adjusted p value. Comparing 20 000 mRNA features with 1000 miRNA features, there is a 20-fold difference in the adjusted p value: an unadjusted p value of 0.00001 becomes adjusted-pmRNA=1 (not significant), while adjusted-pmiRNA=0.01 (significant). Therefore, with limited patient samples, a previously underpowered study in the mRNA space may now be statistically feasible in the miRNA space.
Bioinformatics-derived miRNA classifiers anchored in biological mechanisms: opportunities and challenges
miRNA-based biomarkers are currently limited by the scale of the number of interactions and thus offer a unique modeling challenge that lends them to computational analysis. The power of miRNAs lies in their stunning array of functions and potential targets and deep infiltration of critical pathways. A subtle change in miRNA expression has potentially far-reaching effects on cellular signaling. However, if identifying key protein coding mRNAs is the ultimate research goal, miRNA targeting multiplicity would be disadvantageous. If one is interested in the miRNAs themselves (sometimes described as non-coding genes or non-coding RNA), the advantage is certain. Nevertheless, in comparison with mRNA-based expression signature classifiers, changes in miRNA expression have a greater chance of being biologically relevant. Unlike traditional protein-based regulatory pathways where each mRNA-coding protein is constrained to usually no more than one or two regulatory pathways, the miRNA regulatory network exists in multidimensional space. While straightforward statistics can control for the set of potential interactions, these unsupervised approaches are generally insufficient to reach statistical significance and are not designed to unveil the underpinning molecular mechanisms. Indeed, each miRNA exponentially increases the network complexity, as they exist in a many-to-many relationship, with multiple miRNAs regulating target genes in a potentially overlapping manner. Second, each mRNA–miRNA interaction is specific to the cell-type context and does not necessarily result in a uniform, consistently predictable outcome.3
However, miRNAs are non-protein-coding genes and thus cannot be appraised by pathologists using the common immunohistochemistry approach. A plethora of biological functions can be assigned to each miRNA via its hundreds of gene targets. Part of the challenge that we and other researchers have attempted to address in the protein-coding mRNA space is to develop a computational means of identifying the subset of primary “driver” alterations (alterations that confer a clonal growth advantage on cancers) or passengers, a byproduct of a magnified downstream effect in genome-wide mRNA changes.31 32 This is a non-trivial task as, for example, even after examination of over 746 cancer cell lines, fewer than 0.3% were discovered to be driver mutations.33 To focus further on these driver mechanisms, we have used knowledge from multiple biological domains to further prioritize within the miRNA–mRNA network inferred from expression patterns10: sequence alignment between miRNA and its putative mRNA targets, genetic oncogenes from the Online Mendelian Inheritance in Man,34 protein–protein interaction networks, canonical molecular pathways, and biological mechanisms from Gene Ontology annotations. While miRNA–mRNA sequence alignment databases generally report precisions of ∼30% in identifying any miRNA targets, we confirmed through biological experiments ∼85% precision in identifying miRNA targets as well as its predicted tumor suppressor functions in head and neck cancer.10 This study not only uncovered connections between miRNA regulation, protein network topology, and expression dynamics, but also demonstrated a workflow for translating an in silico miRNA-regulated network into biomarker development in clinical trials.
Other bioinformatics approaches have focused on imputing the miRNA–mRNA, miRNA–miRNA, and miRNA–pathway networks.2 10 35–37 Although useful, existing algorithms for scoring or imputing miRNA–mRNA interactions only calculate probability of downregulation of the target mRNA transcript protein. These network models of miRNA interactions lack clinical translation and resolution to clinically meaningful biomarkers. Additionally, the computational modeling of miRNA-related mechanisms merits attention. For example, to our knowledge, there are no provisions for considering the recently discovered upregulation of mRNA expression with increased miRNA expression9 and interactions between epigenetic and miRNA regulation of mRNA expression. Indeed, miRNA transcription can be regulated by both methylation and direct methylation of target sites.38 39 New opportunities arise from new molecular measurements such as (1) modeling post-transcriptional regulation using multiplexing measures of protein-level expression such as microWesterns40 and improved MS,41 or (2) discovering individualized regulatory patterns using high-throughput sequencing of DNA or RNA. While high-throughput sequencing databases of miRNA regulatory regions of mRNA transcripts are emerging and may reveal clinically informative patterns,42 combined mRNA/miRNA transcription profiles are only interpretable in the context of full integration with clinical phenotyping data, an exercise that exceeds the storage and computing requirements of standard computing approaches and may require computation in GRID or CLOUD space.43 As pharmacogenetic knowledge of common variants was informative for imputing the pathophysiology of rare variants,44 so too will the knowledge of miRNA–mRNA regulatory networks.
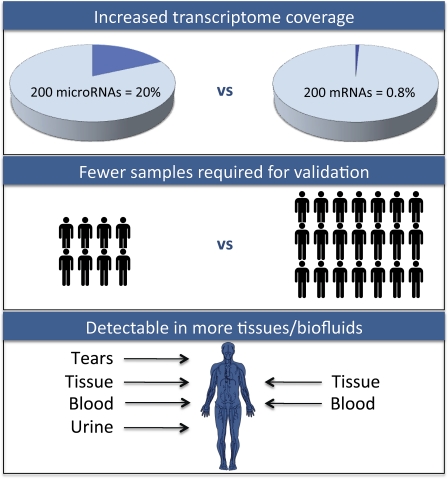
Perhaps an even greater bioinformatics opportunity is integrating phenotypic or domain knowledge into these miRNA networks. Incorporating this additional information will be essential to improve the accuracy of predictions.45 Indeed, taken together, miRNA modeling is clearly at its nascence. Indeed, there are no biofluid-based miRNA networks. More robust, domain-constrained, multi-scale informatics models will be essential to properly identify miRNAs and their targets in an efficient, unbiased manner for the personalization of medicine. Figure 1 provides a summary of the advantages of miRNA signatures compared with mRNA signatures.
Figure 1.
Advantages of microRNA signatures compared with similar-length mRNA signatures.
Conclusion
Taken together, bioinformatics has an outstanding opportunity to accelerate the development of miRNA-based biomarkers in cancer for the personalization of medicine. Although miRNA-based biomarkers have the advantages of manageable dimensionality, ease of testing, and clinical import, they are ultimately hampered by the sheer complexity of their interactions. Without doubt, computational models, currently underdeveloped, are desirable and high impact. Indeed, we firmly hold that network models will ultimately function as the critical bridge between miRNA tumor biology and the individualized care of cancer patients.
Footnotes
Funding: This work was supported in part by the following NIH grants: T32 CA09566, LM008308, and 5UL1RR024999.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: This is a perspective paper with no data sharing.
References
- 1.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806–11 [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011;39:D152–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 2008;26:462–9 [DOI] [PubMed] [Google Scholar]
- 4.Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol 2008;28:6609–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002;297:2056–60 [DOI] [PubMed] [Google Scholar]
- 6.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20 [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 2010;9:775–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Yang X, Huang Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol 2010;6:e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer 2004;4:177–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda N, Ohnishi T, Kawamoto S, et al. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 1999;27:4436–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 2007;13:1668–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster C, Budczies J, Faber C, et al. MicroRNA expression profiling of specific cells in complex archival tissue stained by immunohistochemistry. Lab Invest 2011;91:157–65 [DOI] [PubMed] [Google Scholar]
- 15.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 2011;108:3713–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118–26 [DOI] [PubMed] [Google Scholar]
- 19.Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 2007;8:R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez NC, Sarasquete ME, Misiewicz-Krzeminska I, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia 2010;24:629–37 [DOI] [PubMed] [Google Scholar]
- 21.Denis L, Murphy GP. Overview of phase III trials on combined androgen treatment in patients with metastatic prostate cancer. Cancer 1993;72:3888–95 [DOI] [PubMed] [Google Scholar]
- 22.Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001;69:89–95 [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Li J, Stadler WM, et al. Protein-network modeling of prostate cancer gene signatures reveals essential pathways in disease recurrence. J Am Med Inform Assoc 2011;18:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis CA, Gerick F, Hintermair V, et al. Reliable gene signatures for microarray classification: assessment of stability and performance. Bioinformatics 2006;22:2356–63 [DOI] [PubMed] [Google Scholar]
- 25.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet 2005;365:488–92 [DOI] [PubMed] [Google Scholar]
- 26.Massague J. Sorting out breast-cancer gene signatures. N Engl J Med 2007;356:294–7 [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Gelfond JA, McManus LM, et al. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 2009;10:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canales RD, Luo Y, Willey JC, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol 2006;24:1115–22 [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat 2000;25:60–83 [Google Scholar]
- 30.Kirk R. Experimental Design: Procedure for Behavioral Science. 2nd edn Belmont, CA: Brooks/Cole, 1982 [Google Scholar]
- 31.Chen J, Sam L, Huang Y, et al. Protein interaction network underpins concordant prognosis among heterogeneous breast cancer signatures. J Biomed Inform 2010;43:385–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bozic I, Antal T, Ohtsuki H, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A 2010;107:18545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bignell GR, Greenman CD, Davies H, et al. Signatures of mutation and selection in the cancer genome. Nature 2010;463:893–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKusick VA. Mendelian inheritance in man and its online version, OMIM. Am J Hum Genet 2007;80:588–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Backes C, Meese E, Lenhof HP, et al. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res 2010;38:4476–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010;11:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betel D, Wilson M, Gabow A, et al. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008;36:D149–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008;105:13556–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L, Zhou H, Zhang Q, et al. DNA methylation mediated by a microRNA pathway. Mol Cell 2010;38:465–75 [DOI] [PubMed] [Google Scholar]
- 40.Ciaccio MF, Wagner JP, Chuu CP, et al. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat Methods 2010;7:148–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brusniak MY, Kwok ST, Christiansen M, et al. ATAQS: A computational software tool for high throughput transition optimization and validation for selected reaction monitoring mass spectrometry. BMC Bioinformatics 2011;12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang JH, Li JH, Shao P, et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 2011;39:D202–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudley JT, Pouliot Y, Chen R, et al. Translational bioinformatics in the cloud: an affordable alternative. Genome Med 2010;2:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet 2010;375:1525–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitra R, Bandyopadhyay S, Maulik U, et al. SFSSClass: an integrated approach for miRNA based tumor classification. BMC Bioinformatics 2010;11(Suppl 1):S22. [DOI] [PMC free article] [PubMed] [Google Scholar]