Abstract
Objective
There is increasing interest in using electronic health records (EHRs) to identify subjects for genomic association studies, due in part to the availability of large amounts of clinical data and the expected cost efficiencies of subject identification. We describe the construction and validation of an EHR-based algorithm to identify subjects with age-related cataracts.
Materials and methods
We used a multi-modal strategy consisting of structured database querying, natural language processing on free-text documents, and optical character recognition on scanned clinical images to identify cataract subjects and related cataract attributes. Extensive validation on 3657 subjects compared the multi-modal results to manual chart review. The algorithm was also implemented at participating electronic MEdical Records and GEnomics (eMERGE) institutions.
Results
An EHR-based cataract phenotyping algorithm was successfully developed and validated, resulting in positive predictive values (PPVs) >95%. The multi-modal approach increased the identification of cataract subject attributes by a factor of three compared to single-mode approaches while maintaining high PPV. Components of the cataract algorithm were successfully deployed at three other institutions with similar accuracy.
Discussion
A multi-modal strategy incorporating optical character recognition and natural language processing may increase the number of cases identified while maintaining similar PPVs. Such algorithms, however, require that the needed information be embedded within clinical documents.
Conclusion
We have demonstrated that algorithms to identify and characterize cataracts can be developed utilizing data collected via the EHR. These algorithms provide a high level of accuracy even when implemented across multiple EHRs and institutional boundaries.
Keywords: Cataract, electronic health record, intelligent character recognition, natural language processing, phenotyping, bioinformatics, NLP, information systems, software engineering, clinical research informatics, natural-language processing, linking the genotype and phenotype, improving the education and skills training of health professionals, translational research, application of biological knowledge to clinical care, genomics, pharmacogenomics, genome wide association studies, clinical phenotyping, ritu and pupu and 12, medical informatics, infection control
Introduction
There is a growing interest in utilizing the electronic health record (EHR) to identify clinical populations for genome-wide association studies (GWAS)1–3 and pharmacogenomics research.4–6 This interest results, in part, from the availability of extensive clinical data found within the EHR and the expected cost efficiencies that can result when using computing technology. As in all research that attempts to identify and quantify relationships between exposures and outcomes, rigorous characterization of study subjects is essential and often challenging.7 8
Marshfield Clinic is one of five institutions participating in the electronic MEdical Records and GEnomics (eMERGE) Network.1 4 9 One of the goals of eMERGE is to demonstrate the viability of using EHR systems as a resource for selecting subjects for GWAS. Marshfield's GWAS focused on identifying genetic markers that predispose subjects to the development of age-related cataracts. Cataract subtypes and severity are also important attributes to consider, and possibly bear different genetic signatures.10 Often, clinically relevant information on conditions such as cataracts is buried within clinical notes or in scanned, hand-written documents created during office visits, making this information difficult to extract.
Cataracts are the leading cause of blindness in the world,11 the leading cause of vision loss in the USA,12 and account for approximately 60% of Medicare costs related to vision.13 Prevalence estimates indicate that 17.2% of Americans residing in the USA aged 40 years and older have a cataract in at least one eye, and 5.1% have a pseudophakia/aphakia (previous cataract surgery).14 Age is the primary risk factor for cataracts. With increasing life expectancy, the number of cataract cases and cataract surgeries is expected to increase dramatically unless primary prevention strategies can be developed and successfully implemented.
In this paper, we describe the construction and validation of a novel algorithm that combines multiple techniques and heuristics to identify subjects with age-related cataracts and the associated cataract attributes using only information available in the EHR. We also describe our current multi-modal phenotyping strategy that combines conventional data mining (CDM) with natural language processing (NLP) and optical character recognition (OCR) to increase the detection of subjects with cataract subtypes and optimize the phenotyping algorithm accuracy for case detection. The use of NLP and OCR methods was influenced by previous work that has shown great success in pulling concepts and information from textual and image documents.15–17 We were able to quantify the accuracy and recall of the multi-modal phenotyping components. Finally, this algorithm was implemented at four other eMERGE institutions, thereby validating the transportability of the algorithm.
Background and significance
EHR-based phenotyping is a process that uses computerized analysis to identify subjects with particular traits as captured in an EHR. This process provides the efficiency of utilizing existing clinical data but also introduces obstacles, since those data were collected primarily for patient care rather than research purposes. Previously described EHR data issues include a lack of standardized data entered by clinicians, inadequate capture of absence of disease, and wide variability among patients with respect to data availability (this availability itself may be related to the patient's health status).3 18 Careful phenotyping is critical to the validity of subsequent genomic analyses,7 and a source of great challenge due to the variety of phenotyping options and approaches that can be employed with the same data.8
Previous investigators have demonstrated successful use of billing codes and NLP for biomedical research.19–23 Most often, the focus in the informatics domain is on the application and evaluation of one specific technique in the context of a disease or domain, with a goal of establishing that technique's utility and performance. For example, Savova et al23 24 evaluated the performance of the clinical Text Analysis and Knowledge Extraction System (cTAKES)24 for the discovery of peripheral arterial disease cases from radiology notes. Peissig et al24a evaluated the results of FreePharma (Language and Computing, Inc., http://www.landcglobal.com/) for the construction of atorvastatin dose–response.
Existing research has also demonstrated the ability to use multiple techniques as part of the implementation of a phenotyping algorithm,22 but few have attempted to quantify the benefits of a multi-modal approach (CDM, NLP, and OCR). Those that have were able to demonstrate the benefits of two approaches (commonly coded data in conjunction with NLP) over a single approach that was limited to a single domain.22 25 Although the use of multiple modes for phenotyping is practical, no known work has explored beyond a bimodal approach. The research presented here demonstrates the ability to implement a tri-modal phenotyping algorithm including quantification of the performance of the algorithm as additional modes are implemented.
Methods
Marshfield's study population
The Personalized Medicine Research Project (PMRP),26 27 sponsored by Marshfield Clinic, is one of the largest population-based biobanks in the USA. The PMRP cohort consists of approximately 20 000 consented individuals who provided DNA, plasma, and serum samples along with access to health information from the EHR and questionnaire data relating to health habits, diet, activity, environment, and family history of disease. Participants in this cohort generally receive most, if not all, of their primary, secondary, and tertiary care from the Marshfield Clinic system, which provides health services throughout Central and Northern Wisconsin. This study was approved by the Marshfield Clinic's Institutional Review Board.
Electronic medical record
Founded in 1916, Marshfield Clinic is one of the largest comprehensive medical systems in the nation. CattailsMD, an internally developed EHR at Marshfield Clinic, is the primary source of EHR data for this investigation. The EHR is deployed on wireless tablets and personal computers to over 13 000 users, including over 800 primary and specialty care physicians in both inpatient and outpatient healthcare settings. Medical events including diagnoses, procedures, medications, clinical notes, radiology, laboratory, and clinical observations, are captured for patients within this system. EHR-coded data are transferred daily to Marshfield Clinic's data warehouse (DW) and integrated with longitudinal patient data, currently providing a median of 23 years of diagnosis history for PMRP participants. In addition to the coded data, Marshfield has over 66 million electronic clinical narrative documents, notes, and images that are available back to 1988, with supporting paper clinical charts available back to 1916. Manual review of the electronic records (and clinical charts as needed) was used as the gold standard when validating the EHR-based algorithms.
Cataract code-based phenotyping
Cataract ‘cases’ were identified using an electronic algorithm that interrogated the EHR-coded data found within the DW (figure 1). A goal of the electronic algorithm development was to increase the number of subjects identified for the study (sensitivity), while maintaining a positive predictive value (PPV) of 95% or greater. Cases had to have at least one cataract Current Procedural Terminology (CPT) surgery code or multiple International Classification of Diseases (ICD-9-CM) cataract diagnostic codes. In cases where only one cataract diagnostic code existed for a subject, NLP and/or OCR were used to corroborate the diagnosis. Cataract ‘controls’ had to have an optical exam in the previous 5 years with no evidence of cataract surgery or a cataract diagnostic code or indication of a cataract when using either NLP and/or OCR. Since the focus of the eMERGE study was limited to age-related cataracts, subjects were excluded if they had any diagnostic code for congenital, traumatic, or juvenile cataract. We further restricted cases to be at least 50 years old at the time of either cataract surgery or first cataract diagnosis, and controls had to be at least 50 years old at their most recent optical exam.
Figure 1.
eMERGE cataract phenotyping algorithm. Overview of the cataract algorithm logic used when selecting the cataract cases and controls for the electronic MEdical Records and GEnomics (eMERGE) genome-wide association study. Cataract cases were selected if the subject had either a cataract surgery, or two or more cataract diagnoses, or one cataract diagnosis with either an indication found using natural language processing (NLP) or optical character recognition (OCR). Controls had to have an optical exam within 5 years with no evidence of a cataract. Both cataract cases and controls had to be age 50 or older with controls requiring the absence of the exclusion criteria. The details of this algorithm are published on the eMERGE website.1 Dx, diagnosis; PMRP, Personalized Medicine Research Project.
Cataract subtype multi-modal phenotyping
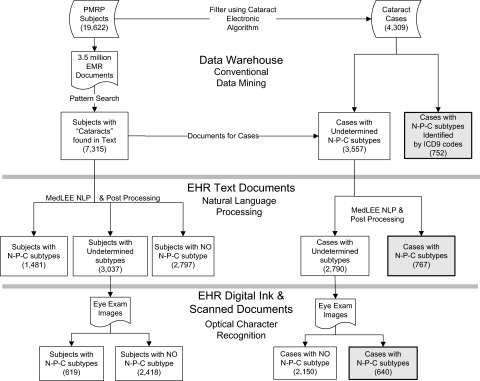
A multi-modal phenotyping strategy was applied to the EHR data and documents to identify information pertaining to nuclear sclerotic, posterior sub-capsular, and cortical (N-P-C) cataract subtypes, severity (numeric grading scale), and eye. Over 3.5 million documents for the PMRP cohort were pre-processed using a pattern search mechanism for the term ‘cataract.’ The strategy (figure 2) consisted of three methods to identify additional cataract attributes: CDM using coded data found in the DW, NLP used on electronic text documents, and OCR used on scanned image documents. CDM was used to identify all subjects having a documented N-P-C subtype (ICD-9-CM codes 366.14–366.16).
Figure 2.
Multi-modal cataract subtype processing. Overview of the information extraction strategy used in multi-modal phenotyping to identify nuclear sclerotic, posterior sub-capsular, and/or cortical (N-P-C) cataract subtypes. This figure depicts the N-P-C subtype yield using two populations: (1) the left-most path of the figure denotes unique subject counts for the entire Personalized Medicine Research Project (PMRP) cohort; and (2) the right-most path denotes unique subject counts for the identified cataract cases. A hierarchical extraction approach was used to identify the N-P-C subtypes. If a subject had a cataract subtype identified by an ICD-9-CM code, natural language processing (NLP) or optical character recognition (OCR) was not utilized. Cataract subtypes identified using NLP had no subsequent OCR processing. EHR, electronic health record; EMR, electronic medical record; MedLEE, Medical Language Extraction and Encoding.
Prior to using NLP to identify N-P-C subtypes, a domain expert was consulted regarding documentation practices surrounding cataracts, who determined that the term ‘cataract’ should always appear within a document for it to be considered relevant to the N-P-C subtypes. The reasoning behind this was to avoid any potential ambiguity when terms related to cataract subtype (ie, ‘NS’ as an abbreviation for ‘nuclear sclerotic’) appeared in a document with no further support related to a cataract. As all clinical documents were of interest, not just ones from ophthalmology, this rule enabled the application of a filter to the documents to be processed by NLP. The Medical Language Extraction and Encoding (MedLEE) NLP engine,28 developed by Friedman and colleagues29 at Columbia University, was tuned to the ophthalmology domain for this specific task to process documents from PMRP patients. MedLEE was chosen for its demonstrated performance in other studies29 and also given the experience of one of the authors (JS) with MedLEE in previous studies.30–33 While MedLEE was chosen for Marshfield's implementation, the multi-modal approach was created to be NLP engine-neutral, and other sites utilized the cTAKES24 system.
The NLP engine tuning involved iterative changes to the underlying lexicon and rules based on a training corpus of 100 documents. MedLEE parses narrative text documents and outputs eXtensible Markup Language (XML) documents which associate clinical concepts with Unified Medical Language System (UMLS) Concept Unique Identifiers (CUIs)34 with relevant status indicators, such as negation status. To identify general cataract concepts and specific cataract subtypes, we queried MedLEE's output for specific CUIs. Additional CUIs were used to determine in which eye the cataract was found. A regular expression pattern search was performed on MedLEE attributes to identify the severity of the cataract and the certainty of the information provided. Refer to figure 3 for an overview of the NLP process.
Figure 3.
Natural language processing of clinical narratives. Textual documents containing at least one reference of a cataract term were fed into the MedLEE NLP engine and then tagged with appropriate Unified Medical Language System (UMLS) Concept Unique Identifiers (CUIs) before being written to an eXtensible Markup Language (XML) formatted file. Post-processing consisted of identifying the relevant UMLS cataract CUIs and then writing them along with other patient and event identifying data to a file that was used in the phenotyping process. MedLEE, Medical Language Extraction and Encoding; NLP, natural language processing.
For subjects with no cataract subtype coded or detected through NLP processing, ophthalmology image documents were processed using an OCR pipeline developed by the study team. The software loaded images and encapsulated the Tesseract35 and LEADTOOLS36 OCR engines into a single pipeline that processed either digital or scanned images for detecting cataract attributes (figure 4). From each image an identification number and region of interest were extracted and stored in a Tagged Image File Format (TIFF) image. The TIFF image was then passed through both OCR engines with the results being recorded independently. Neither the Tesseract nor LEADTOOLS engine was 100% accurate, and often both engines misclassified characters in the same way (ie, ‘t’ instead of ‘+’). Regular expression processing was used to correct these misclassifications. The output from this process yielded a new string that was matched against a list of acceptable results. If after two iterations the string could not be matched, it was discarded. Otherwise, the result was loaded into a database, similar to the NLP results. Details of this process are the subject of another paper.25
Figure 4.
Optical character recognition utilizing electronic eye exam images. Image documents were processed using the LEADTOOLS and Tesseract OCR engines. A tagged image file format (TIFF) image was pasted through the engines with results being recorded independently. Common misclassifications were corrected using regular expressions, and final determinations were made regarding subtype and severity. OCR, optical character recognition.
Validation
All electronically identified potential surgical cataract cases were manually reviewed by a trained research coordinator who verified the presence or absence of a cataract by using the medical chart and supporting documentation. A board-certified ophthalmologist resolved questions surrounding classifications. In addition, stratified random samples were selected to validate potential cases with diagnoses but no surgery, potential controls, and each cataract attribute for the multi-modal approach.
Analysis
Validation results for the algorithms are summarized with standard statistical information retrieval metrics, sensitivity (or recall), specificity, PPV (or precision), and negative predictive value (NPV).37 38 Sampling for the validation was stratified with different sampling fractions in different strata (eg, 100% of surgeries were reviewed). Accuracy and related statistics have been weighted based on the sampling fractions to reflect the test properties as estimated for the full PMRP cohort. Final cataract algorithm and overall cataract subtype 95% confidence interval statistics in tables 1 and 2 are based on 1000 bootstrap samples.39 40
Table 1.
Cataract phenotype validation results
| Electronic approaches | Cohort size | Manually reviewed | Electronic case yield | Manual case yield | True positives | PPV | NPV | Sensitivity | Specificity |
| Surgery | 19 622 | 3657 | 1800 | 3270 | 1747 | 100% | 82.8% | 36.9% | 100% |
| 1 or more Dx (no surgery) | 17 822 | 1910 | 3035 | 1523 | 1507 | 86.6% | 97.0% | 85.6% | 97.2% |
| 2 or more Dx (no surgery) | 17 822 | 1910 | 2047 | 1523 | 1046 | 92.1% | 92.5% | 61.4% | 98.9% |
| 3 or more Dx (no surgery) | 17 822 | 1910 | 1592 | 1523 | 842 | 94.4% | 90.3% | 44.8% | 99.4% |
| NLP and/or OCR plus 1 Dx (no surgery) | 1470 | 350 | 462 | 286 | 272 | 93.4% | 76.8% | 64.8% | 96.2% |
| Combined definition* (95% CI)† | 19 622 | 3657 | 4309 | 3270 | 3065 | 95.6% (94.7% to 96.4%) | 95.1% (92.6% to 96.9%) | 84.6% (78.3% to 89.7%) | 98.7% (98.5% to 99.0%) |
Validation results for the cataract phenotyping effort are based on unique subject counts. Results are presented in a step-wise fashion starting with Surgery. The surgery cases were removed prior to presenting the Dx results. The combined electronic phenotype definition consists of subjects having a cataract surgery or two or more Dx or one Dx with a confirmed recognition from either NLP or OCR.
Combined electronic phenotype definition: surgery, and/or two or more inclusion criteria, and/or (one inclusion Dx plus NLP/OCR).
95% CI based on 1000 bootstrap samples.
DX, diagnosis; NLP, natural language processing; NPV, negative predictive value; OCR, optical character recognition; PPV, positive predictive value.
Table 2.
Detailed results for the cataract subtype multi-modal validation
| Relevant cohort size* | Manually reviewed* | Electronic subtype yield† | Manual subtype yield† | True positives | Weighted statistics based on sampling strata | Statistics based on relevant data | |||||||
| PPV‡ | NPV‡ | Sensitivity‡ | Specificity‡ | PPV | NPV | Sensitivity | Specificity | ||||||
| Nuclear sclerotic§ | |||||||||||||
| CDM on coded diagnosis | 752 | 629 | 493 | 612 | 410 | 99.5% | 1.9% | 13.8% | 96.2% | 99.5% | 3.8% | 67.0% | 80.0% |
| Overall (CDM/NLP/OCR)¶ (95% CI)** | 4236 | 3118 | 1849 | 2946 | 1654 | 99.9% (99.7% to 100%) | 3.7% (2.7% to 4.7%) | 55.8% (53.9% to 57.6%) | 96.2% (9.03% to 100%) | 99.9% (99.7% to 100%) | 3.7% (2.6% to 4.7%) | 56.1% (54.2% to 58.0%) | 96.1% (90.0% to 100%) |
| Posterior sub-capsular§ | |||||||||||||
| CDM on coded diagnosis | 752 | 629 | 287 | 287 | 224 | 97.0% | 66.5% | 21.5% | 99.6% | 97.0% | 81.5% | 78.0% | 97.5% |
| Overall (CDM/NLP/OCR)¶ (95% CI)** | 4236 | 3118 | 529 | 1036 | 425 | 95.1% (92.9% to 97.1%) | 72.3% (70.6% to 74.2%) | 40.9% (37.7% to 43.8%) | 98.6% (98.0% to 99.2%) | 95.1% (93.0% to 96.8%) | 72.3% (70.4% to 74.0%) | 41.0% (38.0% to 43.9%) | 98.6% (98.0% to 99.1%) |
| Cortical§ | |||||||||||||
| CDM on coded diagnosis | 752 | 630 | 3 | 384 | 1 | 100.0% | 25.1% | 0.0% | 100.0% | 100.0% | 32.3% | 0.3% | 100.0% |
| Overall (coded/NLP/OCR)¶ (95% CI)** | 4236 | 3119 | 765 | 2102 | 674 | 95.7% (94.1% to 97.2%) | 32.0% (29.9% to 34.1%) | 31.9% (30.0% to 33.8%) | 95.8% (94.3% to 97.2%) | 95.7% (94.3% to 97.3%) | 32.1% (30.0% to 34.01%) | 32.1% (29.9% to 34.1%) | 95.7% (94.2% to 97.2%) |
Shown are the multi-modal cataract subtype validation statistics. Statistics were calculated using two different approaches: (1) weighted based on the sampling strata, to reflect the properties as would be expected for the full PMRP cohort; and (2) based on the availability of relevant data. A multi-modal cataract subtyping strategy, consisting of CDM, NLP on electronic clinical narratives, and OCR utilizing eye exam images was used to electronically classify cataract subtypes. The electronic subtype classifications were then compared to manually abstracted subtype classifications to determine the accuracy of the multi-modal components.
Unique case counts.
Yield represents total number of cataract subtypes identified. There may be up to two subtypes noted for each subject, but the subject will only be counted once.
Statistics are weighted based on sampling fractions to reflect the properties as would be expected for the full PMRP cohort.
On 4309 cases meeting cataract phenotype definition.
Weighted average using CDM, NLP, and OCR strategies to reflect the properties as would be expected for the full PMRP cohort.
95% CI based on 1000 bootstrap samples.
CDM, conventional data mining; NLP, natural language processing; NPV, negative predictive value; OCR, optical character recognition; PMRP, Personalized Medicine Research Project; PPV, positive predictive value.
Cross-site algorithm implementation
Group Health Research Institute/University of Washington (GH), Northwestern University (NU), and Vanderbilt University (VU) belong to the eMERGE consortium and are performing GWAS using EHR-based phenotyping algorithms to identify patients for research. The GWAS activities conducted by each institution had approval from their respective internal review boards. GH conducted GWAS on dementia; NU utilized type 2 diabetics for their GWAS; and VU conducted GWAS on determinants of normal cardiac conduction.19 EHRs used by these institutions included an in-house developed EHR (VU) and the vendors Epic and Cerner (GH and NU). The cataract algorithm (figure 1) was implemented at three additional sites. Due to lack of use of digital forms at the other institutions, only CDM using billing codes and NLP portions of the algorithm were implemented, with adjustments to some of the diagnostic and procedural codes representing inclusion or exclusion criteria.
Results
The PMRP cohort consisted of 19 622 adult subjects as described previously.41 The number of years of available EHR data for subjects within the PMRP cohort ranged from <1 year to 46 years, with a median of 23 years. Interrogating the coded DW identified 4835 subjects who had either a cataract surgery and/or a cataract diagnosis code. Of the 1800 subjects who had a cataract surgery code, 1790 subjects (99.4%), had a corresponding cataract diagnosis code recorded in the EHR.
We manually validated the cataract status and clinical attributes of 3657 subjects during the course of this study. The cataract phenotype validation results are shown in table 1. The final algorithm developed to identify cataract cases and controls is shown in figure 1, with the full algorithm publically available at http://gwas.org.1 We identified cases for eMERGE using the ‘combined definition’ from table 1. Controls had to meet the definition previously provided. This algorithm yielded 4309 cases with a PPV of 95.6% and 1326 controls with an NPV of 95.1%.
The multi-modal phenotyping strategy was used on all PMRP subject data to identify cataract subtype attributes (figure 1). Overall, 7315 of the 19 622 subjects (37%) had a cataract reference that triggered further NLP or OCR processing to identify the N-P-C subtypes. Of the 4309 subjects identified as cataract cases (figure 2), 2159 subjects (50.1%), had N-P-C subtypes identified using the multi-modal methods. Of these, CDM identified 752 unique subjects (34.8%), NLP recognized an additional 767 distinct subjects (35.5%), and OCR further added 640 unique subjects (29.6%) with N-P-C subtypes. Table 2 reports summary validation results for each of the N-P-C cataract subtypes with respect to the 4309 cases that were identified using the combined cataract algorithm (table 1). We present statistics that are weighted to the full PMRP cohort based on the sampling fractions and statistics that are based on the available relevant data. For each group, all approaches (CDM, NLP, OCR) had high predictive values meeting our threshold guidelines. Combining all multi-modal strategies produced promising results and met our threshold for quality. Online supplementary appendix 1 presents detailed validation statistics for all cataract subtypes by all multi-modal methods for the weighted and relevant data sampling fractions.
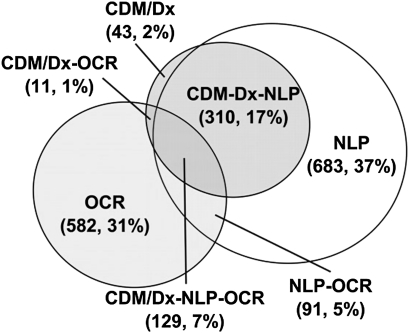
Figure 5 illustrates the overlap of data sources for classifying cases with the nuclear sclerotic (NS) cataract subtype, which is by far the most common. The NLP approach identified the majority, 1213 (65.6%) subjects with the subtype, OCR methods identified 813 (44%) subjects, and CDM identified 493 (26.7%) subjects.
Figure 5.
Nuclear sclerotic (NS) cataract subtype multi-modal approaches. Illustrates the overlap between multi-modal phenotyping approaches when phenotyping for NS cataract subtypes. The largest subtype yield comes from natural language processing (NLP) with NS being identified for 1213 unique subjects. This is followed by the optical character recognition (OCR) approach, which identified 813 unique subjects. Conventional data mining using diagnostic codes (CDM/Dx) identified NS subtypes in only 493 unique subjects.
Figure 6 shows the contribution of multi-modal phenotyping in increasing the number of subjects with NS subtypes from 493 (11.6%) to 1849 (43.6%). In actual practice, the CDM approach would be used first to identify subtypes because it requires the least effort. The remaining documents would then be processed using NLP to identify more subtypes, and if there were subjects remaining, the OCR approach would be used to identify subtypes. In this study, OCR required the most effort to set up and train the OCR engine to gain subtype information. All of the methods had a high PPV, which was required.
Figure 6.
Multi-modal yield and accuracy for the nuclear sclerotic (NS) cataract subtype. Illustrates a step-wise approach that was used to identify subjects with NS subtypes. We started with conventional data mining using ICD-9-CM diagnostic codes (CDM/Dx) because it required the least effort. Natural language processing (NLP) was applied to the remaining subjects if an NS subtype was not identified and optical character recognition (OCR) was used if the previous approaches did not yield a subtype. Out of a possible 4309 subjects having cataracts, 1849 had indication of an NS subtype. Both yield (represented by the number of unique subjects having an NS subtype) and accuracy (represented by positive predictive value (PPV), specificity, sensitivity, and negative predictive value (NPV)) are presented for each approach. This study used PPV as the accuracy indicator. The number of unique subjects with NS subtypes increased when the multi-modal approach was used while maintaining a high PPV.
Validation results for cataract severity and location are shown in table 3. There were no coded values for either cataract severity or location found in the EHR, so the only way to obtain that information was to rely solely on NLP or OCR. NLP identified cataract location (left or right eye) with high reliability in 53% of the subjects. The OCR yield presented in table 3 is low because only the remaining subjects who did not have NLP-identified locations were processed.
Table 3.
Severity and location validation results
| Relevant cohort size | Manually reviewed | Electronic attribute yield | Manual attribute yield | True positives | PPV | NPV | Sensitivity | Specificity | |
| Severity of grade 2+ or higher detected* † | |||||||||
| NLP | 3862 | 3144 | 198 | 2510 | 189 | 98.6% | 27.1% | 6.9% | 99.7% |
| OCR (no NLP) | 3236 | 2952 | 180 | 2321 | 150 | 91.4% | 29.3% | 6.9% | 98.4% |
| Location detected* | |||||||||
| NLP | 3862 | 2502 | 2265 | 2422 | 1529 | 99.7% | 7.7% | 63.1% | 93.8% |
| OCR (no NLP) | 3236 | 968 | 21 | 893 | 10 | 100% | 7.8% | 1.1% | 100.0% |
Shown are the cataract severity and location (left or right eye) validation results for the 4309 cataract subjects who were classified as cases using the electronic cataract phenotyping definition. NLP was utilized first on electronic clinical narratives to identify cataract severity and/or location. Eye exam document images were processed using OCR if the severity and location could not be determined using NLP. There were no coded severities or location data found within the electronic health record.
On 4309 cases meeting cataract phenotype definition.
If cataracts found via abstraction and severity unknown, then severity (per abstraction) set to ‘No.’
NLP, natural language processing; NPV, negative predictive value; OCR, optical character recognition; PPV, positive predictive value.
External validation
The other eMERGE institutions were able to contribute 1386 cases and 1360 controls to Marshfield's cataract GWAS study. GH did not explicitly validate CDM-based phenotype case and control definitions by manual chart review. Instead, a separate review using NLP-assisted chart abstraction methods was conducted for 118 subjects selected from the GH cohort of 1042 (931 cases and 111 controls) who had at least one chart note associated with a comprehensive eye exam in the EHR. All 118 (100%) were in the CDM-based eMERGE phenotype case group, and all had cataract diagnoses documented in their charts. Additionally, cataract type was recorded for 108 (92%) of the subjects. The remaining 10 (8%) did not specify cataract type at all or with insufficient detail to satisfy study criteria. Although the 118 charts reviewed for the cataract type analysis does not constitute a random sample for validation sample, the fact that 100% were in the cataract case group is reassuring. VU sampled and validated 50 cases and 42 controls that were electronically identified against classifications determined by a physician when reviewing the clinical notes. Results from this validation indicated a PPV of 96%.
Discussion
The EHR represents a valuable resource for obtaining a longitudinal record of a patient's health and treatments through on-going interactions with a healthcare system. The health information obtained from those interactions can be stored in multiple formats within the EHR, which may vary between institutions and may present significant challenges when trying to retrieve and utilize the information for research.3 This work demonstrates that electronic phenotyping using data derived from the EHR can be successfully deployed to characterize cataract cases and controls with a high degree of accuracy. It also demonstrates that a multi-modal phenotyping approach, which includes the use of CDM, NLP, and OCR, increases the number of subjects identified with N-P-C subtypes from 752 (17.5%) to 2159 (50.1%) of the available cataract cases. Multi-modal phenotyping increases the number of subjects with NS subtypes from 493 to 1849 (figure 6) while maintaining a high PPV.
These multi-modal methods can be applied to a variety of studies beyond the current one. In many clinical trials, the actual patients who are sought cannot be accurately identified by the presence of an ICD code. As a simple example, squamous and basal cell skin cancer can be billed using the same ICD code, yet have different pathophysiology, treatment, and prognosis. Efficiently identifying patients with one or the other diagnosis requires using text documents or scanned documents. Similarly, many studies include historical inclusion or exclusion criteria. Such historical data are poorly incorporated in standard codes, but are frequently noted in textual or scanned documents. The addition of multi-modal methods can improve the efficiency of both interventional and observational studies. These techniques can also be applied to quality and safety issues to identify adverse events or process failures. When used in these domains, the tools will need to be biased toward sensitivity, rather than toward specificity, as they were in this study.
A commonly repeated saying in the EHR phenotyping domain is that ‘The absence of evidence is not evidence of absence’ (Carl Sagan). In the case of this study, that could be due to cataract surgery at an outside institution or the presence of an early cataract that was not detected on routine examination. Therefore, in this and other phenotyping studies, it is critical to actively document the absence of the target phenotype when defining the control group. In the case of this study, intraocular lenses that were placed during cataract surgery were observed and documented on clinical eye exams. Because we required everyone to have had an eye exam, we believe it is highly unlikely that our algorithm would miss any prior cataract surgeries, regardless of where they took place, when identifying controls.
An extensive validation comparison between the multi-modal results and manually abstracted results (considered the gold standard) has been presented. Although the validation statistics for the final electronic algorithms met our accuracy threshold (PPV ≥95%), the potential exists to improve the accuracy by modifying the algorithm to exclude additional misclassified subjects. For example, when classifying cataract cases, we chose the ‘combined definition’ (table 1) to maximize the yield of cases at the expense of lowering our accuracy threshold to a PPV of 95.6%, when compared to using only subjects having a cataract surgery (PPV of 100% and case yield of 1800; table 1). We could achieve 100% accuracy, albeit with small numbers (low sensitivity). However, excluding subjects to improve accuracy is something that would have to be discussed when conducting individual studies. Further, improvement in the statistics would have to be balanced against the possibility that the resulting cases would be less representative of the population of interest.
The portability of the cataract algorithm to other EHRs has also been demonstrated. Although EHRs differed between eMERGE institutions, basic information captured as part of patient care is similar across all of the institutions.9 An important outcome of this study is the demonstrated performance of the cataract algorithm at three institutions with separate EHR systems using only structured billing codes. The cataract algorithm was validated at two other institutions with similarly high PPVs. We found that the ability to successfully identify N-P-C subtype information is improved with the availability of electronic textual and image documents. This work adds to other recent work from the eMERGE Network demonstrating the portability of an algorithm to identify primary hypothyroidism cases and controls using solely EHR-derived data.
One weakness of this study was the use of a single subject matter expert (SME) for the development of NLP and OCR tools. The recording of cataract information tends to be more uniform in the medical record than many other clinical conditions; therefore, the role of the SME was primarily to assist in identifying any coding or documentation nuances particular to ophthalmologists. However, multiple informatics/phenotyping experts, including at least one at each eMERGE institution, helped implement and refine the algorithm. The subsequent high accuracy of the NLP tool when applied to other institutions affirmed the consistency of documentation. Clinical domains with higher variability may require multiple SMEs.
A limitation of this study is that the multi-modal strategy was applied to a single clinical domain area. Although this study demonstrates the utility of this approach for cataracts, there may be other clinical domains where the multi-modal strategy may not be cost-effective or even possible. For example, if much of the domain information is in coded formats, the resource to implement a multi-modal algorithm would not justify the use of this strategy. There have been other studies that utilized a bimodal approach to phenotyping (CDM and NLP) which have shown an increase in PPV values.19 21 22 We realize that the application of a multi-modal strategy should be evaluated on a study-by-study basis with consideration given to the time and effort of developing tools, the number of subjects required to conduct a meaningful study, and the sources of available data. Utilizing a multi-modal strategy for identifying information that is not often reliably coded is an excellent use of resources when compared to the alternative manual abstraction method. Benefits of using automation over manual record abstraction are reuse and the portability of the electronic algorithms to other EHRs.
A second limitation is the lack of validation of a third mode (specifically, image documents containing handwritten notes) at other eMERGE institutions that implemented this algorithm. Results describing the benefits of applying a third mode are then localized to Marshfield Clinic. This does, however, demonstrate that algorithms developed with multiple phenotyping modes may do so in a way that modularizes each mode such that it can be removed from the algorithm if a replicating institution does not have the same information in their EHR.
A criticism of this study may be the use of a domain expert to identify features for model development in the CDM, NLP, and OCR approaches. Other studies have used logistic regression to identify attributes for CDM and NLP. Other machine learning techniques may also prove beneficial in classification tasks; however, the involvement of domain experts is the current standard in studies involving identification of research subjects or samples. The ophthalmology domain expert not only provided guidance on where to find the information, but also provided features that were representative and commonly used in Marshfield Clinic's ophthalmology practice. We believe that domain expert knowledge was critical to the success of this study, although introducing automatic classification in conjunction with a domain expert would be of interest in future research. Implementing the cataract algorithm at other eMERGE institutions required EHR domain knowledge rather than ophthalmology domain knowledge.
This study underscores a previously stated caution regarding the use of coded clinical data for research: just because a code exists in the code set does not guarantee that clinicians use it. Cataract subtype codes exist in ICD-9-CM coding, but as seen in figure 5, fewer than half of the NS cases identified were coded as such. From a clinical standpoint, this is not surprising, because cataract subtype has little impact on either billing or clinical management. As a result of this study, the Marshfield ophthalmology specialty is considering workflows that will emphasize coded data capture of subtypes. Current documentation practices support ink-over-forms (figure 3). A hybrid approach utilizing ink-over-forms combined with coded data capture to document cataract subtypes could be developed. Although NLP and OCR are viable strategies for obtaining subtype information, utilizing CDM would be most cost-effective for obtaining cataract subtypes.
Conclusion
The utilization of ICD-9-CM codes and CPT codes for the identification of cataract cases and controls is generally accurate. This study demonstrated that phenotyping yield improves by using multiple strategies with a slight trade-off in accuracy, when compared to the results of a single strategy. Most EHRs capture similar types of data for patient care. Because of this, the portability of electronic phenotyping algorithms to other EHRs can be accomplished with minor changes to the algorithms. The decision to use a multi-modal strategy should be evaluated on an individualized study basis. Not all clinical conditions will require the multiple approaches that were applied to our study.
Acknowledgments
The authors thank the Marshfield Clinic Research Foundation's Office of Scientific Writing and Publication for editorial assistance with this manuscript.
Footnotes
Funding: The eMERGE Network was initiated and funded by NHGRI, with additional funding from NIGMS through the following grants: U01-HG-004610 (Group Health Cooperative), U01-HG-004608 (Marshfield Clinic), U01-HG-04599 (Mayo Clinic), U01-HG-004609 (Northwestern University), U01-HG-04603 (Vanderbilt University, also serving as the Coordinating Center); and the State of Washington Life Sciences Discovery Fund award to the Northwest Institute of Genetic Medicine.
Competing interests: None.
Ethics approval: Ethics approval was provided by Marshfield Clinic Institutional Review Board.
Contributors: PP prepared the initial draft of the paper. RB carried out the statistical analyses. JL developed the electronic algorithm to identify cases/controls and created the databases and data sets. CW completed the data abstraction. LR configured and executed the NLP and OCR programs. PP and JS oversaw the informatics components of the study. LC, RW, and CMcC were the content experts and provided training for data abstraction. CMcC was Principal Investigator and is responsible for the conception, design, and analysis plan. JD, JP, DC, and AK oversaw informatics activities at other eMERGE institutions. All authors read and approved the final manuscript.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Electronic Medical Records and Genomics (eMERGE) Network. http://www.gwas.org/ (accessed 21 Jul 2010).
- 2.Manolio TA. Collaborative genome-wide association studies of diverse diseases: programs of the NHGRI's office of population genomics. Pharmacogenomics 2009;10:235–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wojczynski MK, Tiwari HK. Definition of phenotype. Adv Genet 2008;60:75–105 [DOI] [PubMed] [Google Scholar]
- 4.McCarty CA, Chisholm RL, Chute CG, et al. ; eMERGE Team The eMERGE network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther 2011;89:378–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarty CA, Wilke RA. Biobanking and pharmacogenetics. Pharmacogenomics 2010;11:637–41 [DOI] [PubMed] [Google Scholar]
- 7.Bickeböller H, Barrett JH, Jacobs KB, et al. Modeling and dissection of longitudinal blood pressure and hypertension phenotypes in genetic epidemiological studies. Genet Epidemiol 2003;25(Suppl 1):S72–7 [DOI] [PubMed] [Google Scholar]
- 8.Schulze TG, McMahon FJ. Defining the phenotype in human genetic studies: forward genetics and reverse phenotyping. Hum Hered 2004;58:131–8 [DOI] [PubMed] [Google Scholar]
- 9.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011;3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty CA, Mukesh BN, Dimitrov PN, et al. The incidence and progression of cataract in the Melbourne Visual Impairment Project. Am J Ophthalmol 2003;136:10–17 [DOI] [PubMed] [Google Scholar]
- 11.Thylefors B, Négrel AD, Pararajasegaram R, et al. Available data on blindness (update 1994). Ophthalmic Epidemiol 1995;2:5–39 [DOI] [PubMed] [Google Scholar]
- 12.Congdon N, O'Colmain B, Klaver CC, et al. ; Eye Diseases Prevalence Research Group Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol 2004;122:477–85 [DOI] [PubMed] [Google Scholar]
- 13.Ellwein LB, Urato CJ. Use of eye care and associated charges among the Medicare population: 1991–1998. Arch Ophthalmol 2002;120:804–11 [DOI] [PubMed] [Google Scholar]
- 14.Congdon N, Vingerling JR, Klein BE, et al. ; Eye Diseases Prevalence Research Group Prevalence of cataract and pseudophakia/aphakia among adults in the United States. Arch Ophthalmol 2004;122:487–94 [DOI] [PubMed] [Google Scholar]
- 15.Govindaraju V. Emergency medicine, disease surveillance, and informatics. Proceedings of the 2005 National Conference on Digital Government Research. Digital Government Society of North America, 2005:167–8 [Google Scholar]
- 16.Milewski R, Govindaraju V. Handwriting analysis of pre-hospital care reports. Proceedings of CBMS. Los Alamitos, CA: IEEE Computer Society, 2004:428–33 [Google Scholar]
- 17.Piasecki M, Broda B. Correction of medical handwriting OCR based on semantic similarity. In: Yao X, Tino P, Corchado E, et al., eds. Intelligent Data Engineering and Automated Learning - IDEAL 2007. Vol. 4881. Berlin/Heidelberg: Springer-Verlag, 2007:437–46 [Google Scholar]
- 18.Gurwitz D, Pirmohamed M. Pharmacogenomics: the importance of accurate phenotypes. Pharmacogenomics 2010;11:469–70 [DOI] [PubMed] [Google Scholar]
- 19.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation 2010;122:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peissig P, Sirohi E, Berg RL, et al. Construction of atorvastatin dose-response relationships using data from a large population-based DNA biobank. Basic Clin Pharmacol Toxicol 2006;100:286–8 [DOI] [PubMed] [Google Scholar]
- 21.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010;86:560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc 2010;17:568–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savova GK, Fan J, Ye Z, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annu Symp Proc 2010;2010:722–6 [PMC free article] [PubMed] [Google Scholar]
- 24.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Sirohi E, Peissig P. Study of effect of drug lexicons on medication extraction from electronic medical records. In: Altman RB, Dunker AK, Hunter L, et al., eds. Biocomputing 2005: Proceedings of the Pacific Symposium. World Scientific Publishing Company, 2005;308–17 [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen LV, Peissig PL, McCarty CA, et al. Development of an optical character recognition pipeline for handwritten form fields from an electronic health record. J Am Med Inform Assoc. Published Online First: 2 September 2011. doi:10.1136/amiajnl-2011-000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarty C, Wilke RA, Giampietro PF, et al. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Pers Med 2005;2:49–79 [DOI] [PubMed] [Google Scholar]
- 27.McCarty CA, Peissig P, Caldwell MA, et al. The Marshfield Clinic Personalized Medicine Research Project: 2008 scientific update and lessons learned in the first 6 years. Pers Med 2008;5:529–42 [DOI] [PubMed] [Google Scholar]
- 28.MedLEETM Natural Language Processing. http://www.nlpapplications.com/index.html (accessed 23 Jan 2011).
- 29.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman C, Johnson SB, Forman B, et al. Architectural requirements for a multipurpose natural language processor in the clinical environment. In: Gardner RM, ed. Proceedings of Ninteenth SCAMC. Philadelphia, PA: Hanley & Belfus, 1995:238–42 [PMC free article] [PubMed] [Google Scholar]
- 31.Starren J, Johnson SB. Notations for high efficiency data presentation in mammography. In: Cimino JJ, ed. Proceedings of the American Medical Informatics Association Annual Fall Symposium (formerly SCAMC). Philadelphia, PA: Hanley & Belfus, 1996:557–61 [PMC free article] [PubMed] [Google Scholar]
- 32.Starren J, Friedman C, Johnson SB. Design and Development of the Columbia Integrated Speech Interpretation System (CISIS). (abstract). Proceedings of the 1995 AMIA Spring Congress. 1995 [Google Scholar]
- 33.Starren J, Friedman C, Johnson SB. The Columbia Integrated Speech Interpretation System (CISIS). (abstract and poster). In: Gardner RM, ed. Proceedings of Ninteenth SCAMC. Philadelphia, PA: Hanley & Belfus, 1995:985 [Google Scholar]
- 34.National Library of Medicine Fact Sheet UMLS® Metathesaurus®. http://www.rsna.org/radlex/committee/UMLSMetaThesaurusFactSheet.doc (accessed 13 Jan 2003).
- 35.Tesseract-OCR Engine Homepage. Google Code. http://code.google.com/p/tesseract-ocr/ (accessed 30 Sep 2010).
- 36.LEADTOOLS® Developer's Toolkit: ICR Engine. http://www.leadtools.com/sdk/ocr/icr.htm (accessed 30 Sep 2010).
- 37.Altman DG, Bland JM. Diagnostic tests 1: sensitivity and specificity. BMJ 1994;308:1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altman DG, Bland JM. Diagnostic tests 2: predictive values. BMJ 1994;309:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Efron B, Tibshrani R. An Introduction to the Bootstrap. Monographs on Statistics and Applied Probability. New York, NY: Chapman and Hall, 1993 [Google Scholar]
- 40.Zhou XH, Obuchowski NA, McClish DK. Statistical Methods in Diagnostic Medicine. New York, NY: John Wiley and Sons, Inc., 2002 [Google Scholar]
- 41.Waudby CJ, Berg RL, Linneman JG, et al. Cataract research using electronic health records. BMC Ophthalmol 2011;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]